A Wild Rice Rhizobacterium Burkholderia cepacia BRDJ Enhances Nitrogen Use Efficiency in Rice
Abstract
:1. Introduction
2. Results
2.1. Isolation of Rhizobacteria from DXWR
2.2. Identification of the Strain BRDJ
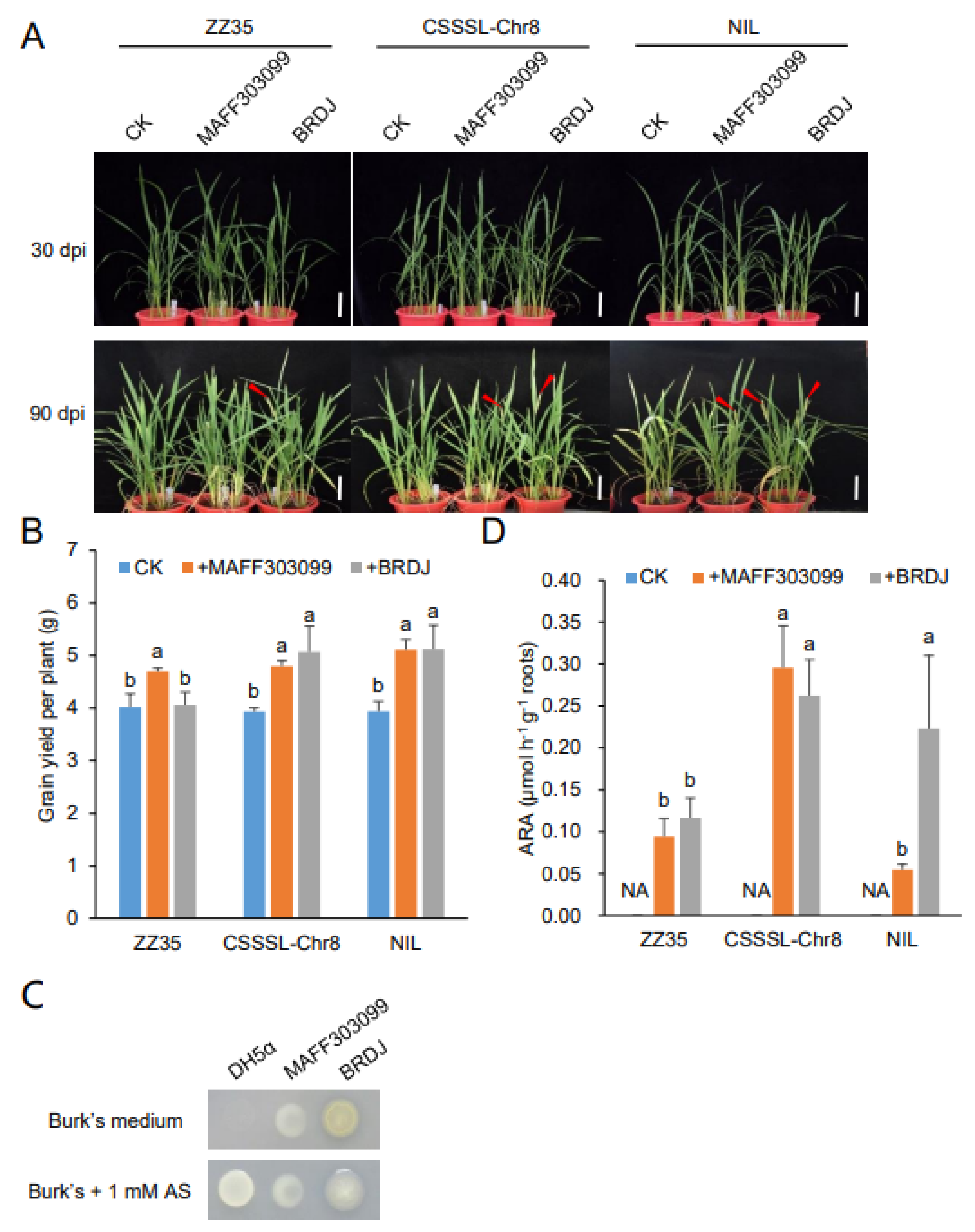
2.3. Plant Growth–Promoting Activity of BRDJ
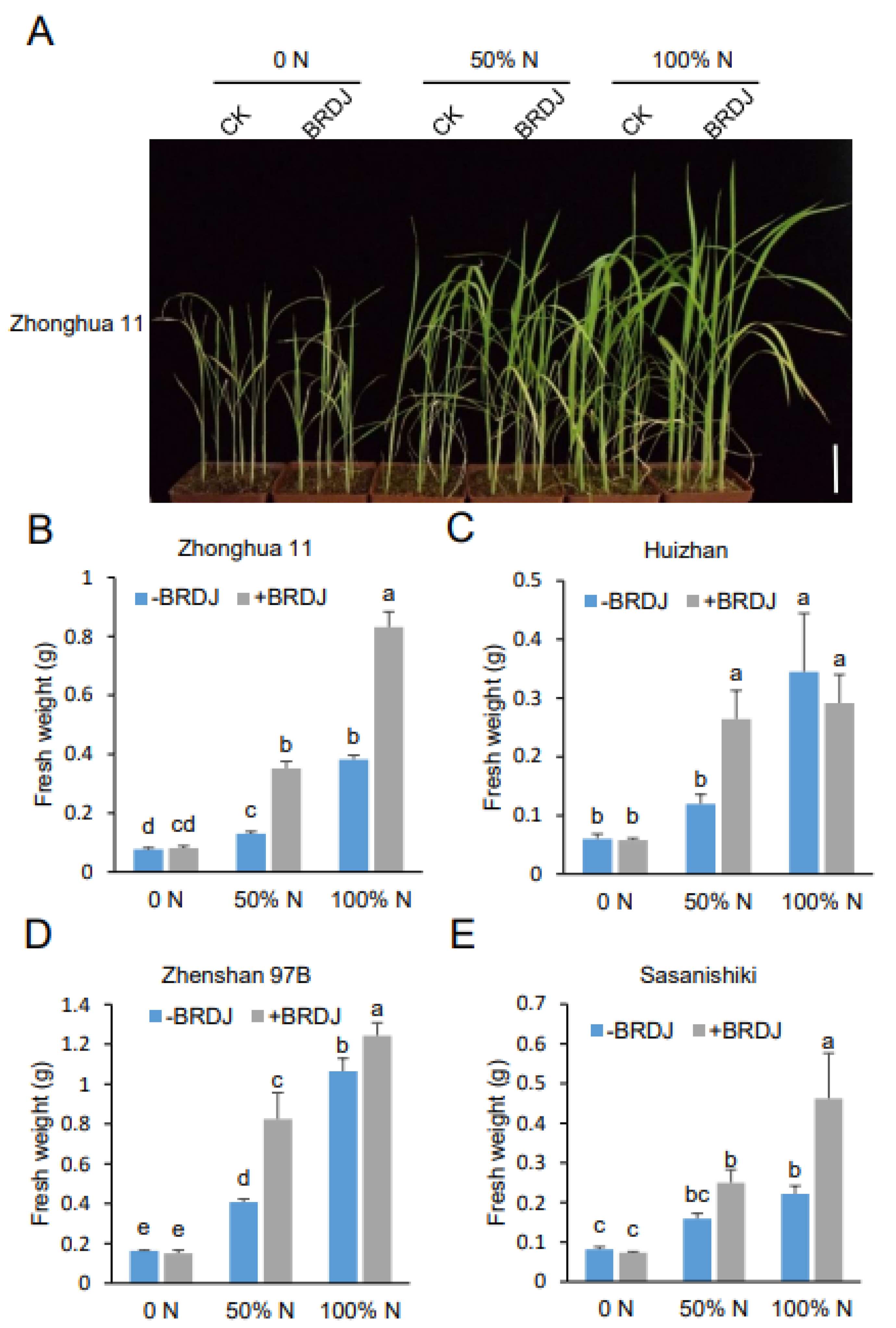
2.4. Plant Growth–Promoting Activity of the BRDJ Strain on Different Rice Cultivars
2.5. Identification of Bacterial Plant Growth–Promoting Genes
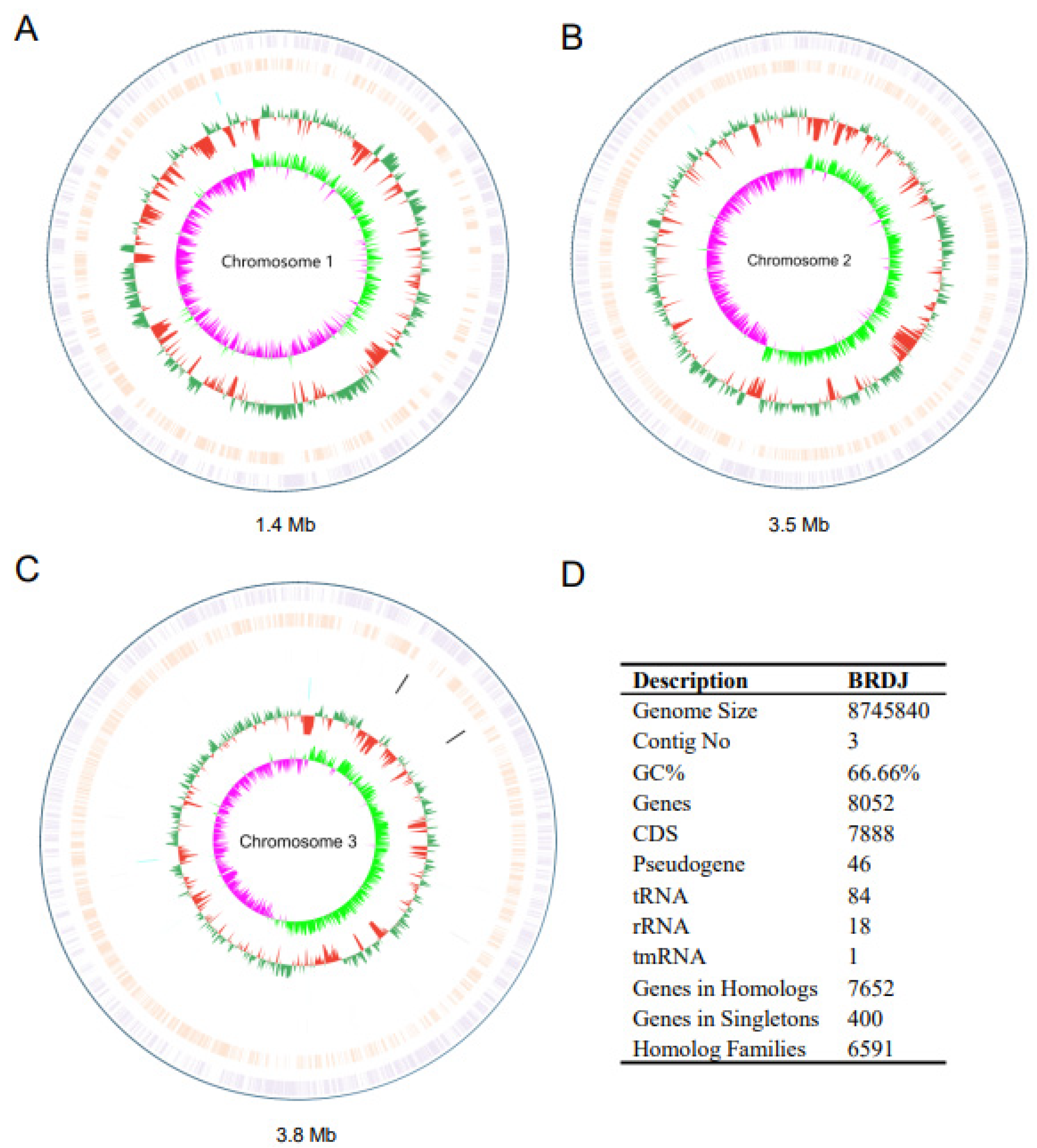
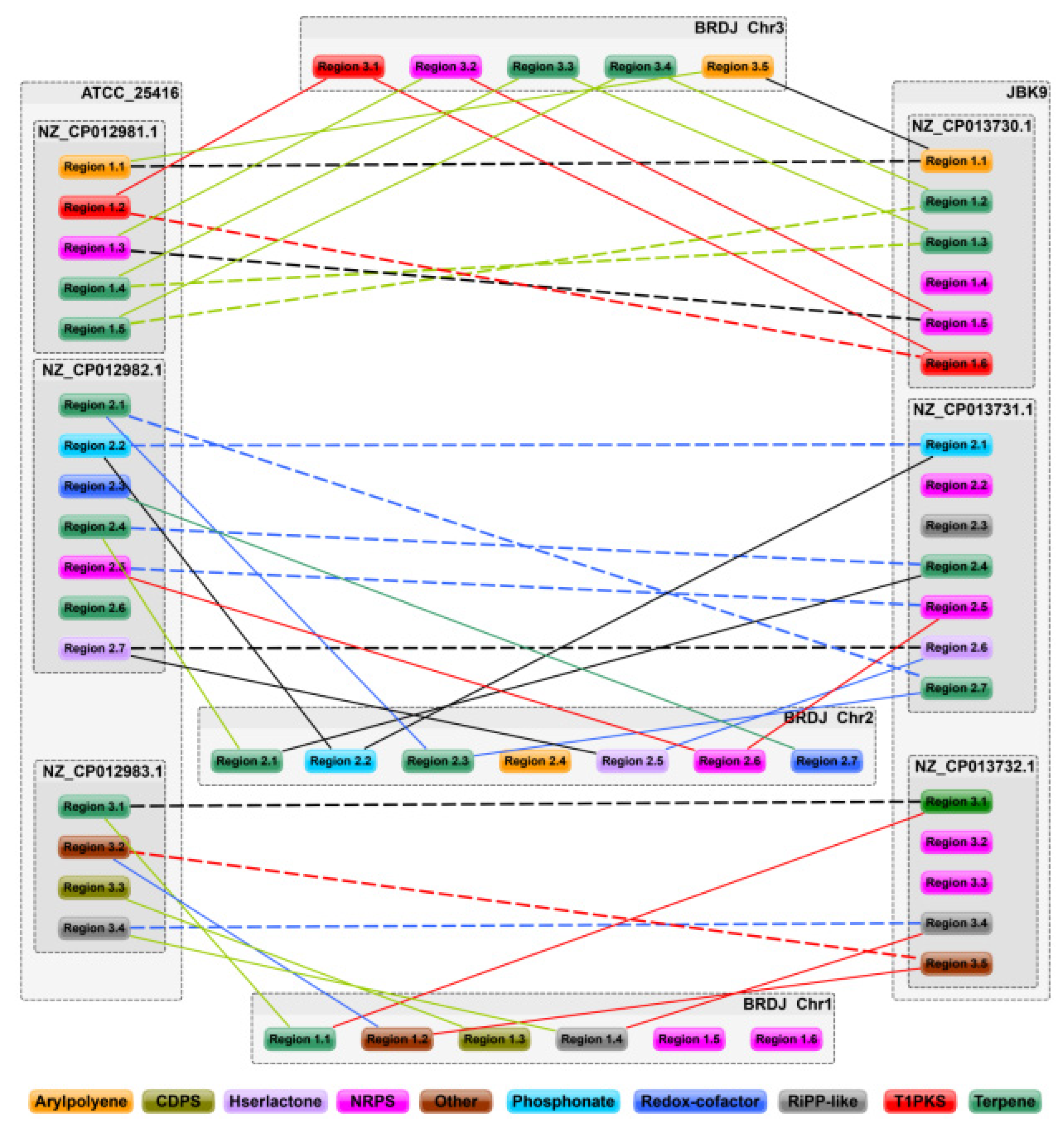
2.6. Comparative Genomics Analysis
2.7. Pathogenicity Test in Onions and Nematodes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Isolation of Rhizobacterium Strains
4.3. Inoculation Experiments in Rice
4.4. Nitrogenase Activity Assay
4.5. Whole-Genome Sequencing
4.6. Genome Assembly
4.7. Genome Component Prediction
4.8. Gene Function Prediction
4.9. Comparative Genomics Analysis
4.10. Pathogenicity Test on Onion and Nematode
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | 1-aminocyclopropane-1-carboxylic acid |
| AS | Ammonium sulfate |
| AMF | Arbuscular mycorrhizal fungi |
| Bcc | Burkholderia cepacia complex |
| CDPS | tRNA-dependent cyclodipeptide synthases |
| COGs | Clusters of Orthologous Groups |
| DXWR | Dongxiang wild rice |
| IAA | Indole-3-acetic acid |
| NRPS | Non-ribosomal peptide synthetase |
| RiPP | Ribosomally synthesized and post-translationally modified peptide |
| T1PKS | Type I polyketide synthase |
| YEB | Yeast Extract Beef |
References
- Luo, W.; Yang, Y.; Fang, F.; Li, W.; Hu, F.; Zhang, J.; Chen, D.; Yu, L. Chronology of ancient Dongxiang wild rice (Oryza rufipogon Griff.), and the morphologies of grains, double-peaked phytoliths, and starch, in the middle Yangtze river region, China. Rev. Palaeobot. Palynol. 2017, 244, 140–147. [Google Scholar] [CrossRef]
- Mao, D.; Yu, L.; Chen, D.; Li, L.; Zhu, Y.; Xiao, Y.; Zhang, D.; Chen, C. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor. Appl. Genet. 2015, 128, 1359–1371. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Chen, W.; Jiang, W.; Ge, S.; Hong, D.; Wang, X. Genetic erosion in northern marginal population of the common wild rice Oryza rufipogon Griff. and its conservation, revealed by the change of population genetic cstructure. Hereditas 2000, 133, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lei, J.; Kong, H.; Guo, Y.; Li, M.; Yu, L.; Yu, C.; Guo, A. Flowering characteristics of Dongxiang common wild rice in Nanchang ex-situ environment. Chin. J. Trop. Crops 2010, 31, 1063–1067. [Google Scholar]
- Zhang, F.T.; Xu, T.; Mao, L.Y.; Yan, S.Y.; Chen, X.W.; Wu, Z.F.; Chen, R.; Luo, X.D.; Xie, J.K.; Gao, S. Genome-wide analysis of Dongxiang wild rice (Oryza rufipogon Griff.) to investigate lost/acquired genes during rice domestication. BMC Plant Biol. 2016, 16, 103. [Google Scholar] [CrossRef]
- Qi, W.D.; Chen, H.P.; Yang, Z.Z.; Hu, B.L.; Luo, X.D.; Ai, B.; Luo, Y.; Huang, Y.; Xie, J.K.; Zhang, F.T. Systematic characterization of long non-coding RNAs and their responses to drought stress in Dongxiang wild rice. Rice Sci. 2020, 27, 21–31. [Google Scholar]
- Huang, R.; Li, Z.; Mao, C.; Zhang, H.; Sun, Z.; Li, H.; Huang, C.; Feng, Y.; Shen, X.; Bucher, M.; et al. Natural variation at OsCERK1 regulates arbuscular mycorrhizal symbiosis in rice. New Phytol. 2020, 225, 1762–1776. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Hanson, P.J.; Edwards, N.T.; Garten, C.T.; Andrews, J.A. Separating root and soil microbial contributions to soil respiration: A review of methods and observations. Biogeochemistry 2000, 48, 115–146. [Google Scholar] [CrossRef]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 2001, 237, 173–195. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Boyce, A.N. Role of plant growth promoting rhizobacteria in agricultural sustainability-a review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [Green Version]
- Bharti, N.; Pandey, S.S.; Barnawal, D.; Patel, V.K.; Kalra, A. Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 2016, 6, 34768. [Google Scholar] [CrossRef] [PubMed]
- Kuan, K.B.; Othman, R.; Rahim, K.A.; Shamsuddin, Z.H. Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS ONE 2016, 11, e0152478. [Google Scholar] [CrossRef]
- Zhu, R.M.; Cao, Y.T.; Li, G.Z.; Guo, Y.; Ma, L.J.; Bu, N.; Hao, L. Paraburkholderia sp. GD17 improves rice seedling tolerance to salinity. Plant Soil 2021, 467, 373–389. [Google Scholar] [CrossRef]
- Haque, M.M.; Mosharaf, M.K.; Khatun, M.; Haque, M.A.; Biswas, M.S.; Islam, M.S.; Islam, M.M.; Shozib, H.B.; Miah, M.M.U.; Molla, A.; et al. Biofilm producing rhizobacteria with multiple plant growth-promoting traits promote growth of tomato under water-deficit stress. Front. Microbiol. 2020, 11, 542053. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Mañero, F.J.; Ramos-Solano, B.; Probanza, A.; Mehouachi, J.; Tadeo, F.R.; Talon, M. The plant-growth-promoting rhizobacteria Bacillus pumilus and Bacillus licheniformis produce high amounts of physiologically active gibberellins. Physiol. Plant. 2001, 111, 206–211. [Google Scholar] [CrossRef]
- Remans, R.; Beebe, S.; Blair, M.; Manrique, G.; Tovar, E.; Rao, I.; Croonenborghs, A.; Torres-Gutierrez, R.; El-Howeity, M.; Michiels, J.; et al. Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean (Phaseolus vulgaris L.). Plant Soil 2008, 302, 149–161. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Burkholder, W.H. Sour skin, a bacterial rot of onion bulbs. Phytopathology 1950, 40, 115–117. [Google Scholar]
- Yabuuchi, E.; Kosako, Y.; Oyaizu, H.; Yano, I.; Hotta, H.; Hashimoto, Y.; Ezaki, T.; Arakawa, M. Proposal of Burkholderia gen. nov. and Transfer of Seven Species of the Genus Pseudomonas Homology Group II to the New Genus, with the Type Species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 1992, 36, 1251–1275. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandamme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Moreno, Z.R.; Caballero-Mellado, J.; Coutinho, B.G.; Mendonça-Previato, L.; James, E.K.; Venturi, V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012, 63, 249–266. [Google Scholar] [CrossRef]
- Ganesh, P.S.; Vishnupriya, S.; Vadivelu, J.; Mariappan, V.; Vellasamy, K.M.; Shankar, E.M. Intracellular survival and innate immune evasion of Burkholderia cepacia: Improved understanding of quorum sensing-controlled virulence factors, biofilm, and inhibitors. Microbiol. Immunol. 2020, 64, 87–98. [Google Scholar] [CrossRef]
- Noureddini, H.; Gao, X.; Philkana, R.S. Immobilized Pseudomonas cepacia lipase for biodiesel fuel production from soybean oil. Bioresour. Technol. 2005, 96, 769–777. [Google Scholar] [CrossRef]
- Tabacchioni, S.; Bevivino, A.; Dalmastri, C.; Chiarini, L. Burkholderia cepacia complex in the rhizosphere: A minireview. Ann. Microbiol. 2002, 52, 103–117. [Google Scholar]
- Matthaiou, D.K.; Chasou, E.; Atmatzidis, S.; Tsolkas, P. A case of bacteremia due to Burkholderia cepacia in a patient without cystic fibrosis. Respir. Med. CME 2011, 4, 144–145. [Google Scholar] [CrossRef]
- Zhao, K.; Penttinen, P.; Zhang, X.; Ao, X.; Liu, M.; Yu, X.; Chen, Q. Maize rhizosphere in Sichuan, China, hosts plant growth promoting Burkholderia cepacia with phosphate solubilizing and antifungal abilities. Microbiol. Res. 2014, 169, 76–82. [Google Scholar] [CrossRef]
- Jung, B.K.; Hong, S.-J.; Park, G.-S.; Kim, M.-C.; Shin, J.-H. Isolation of Burkholderia cepacia JBK9 with plant growth-promoting activity while producing pyrrolnitrin antagonistic to plant fungal diseases. Appl. Biol. Chem. 2018, 61, 173–180. [Google Scholar] [CrossRef]
- Matthews, S.; Suhaimi, M. Selection of suitable growth medium for free-living diazotrophs isolated from compost. J. Trop. Agric. Food Sci. 2010, 38, 211–219. [Google Scholar]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef]
- Payne, G.W.; Vandamme, P.; Morgan, S.H.; Lipuma, J.J.; Coenye, T.; Weightman, A.J.; Jones, T.H.; Mahenthiralingam, E. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 2005, 71, 3917–3927. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.W.; Holsten, R.D.; Jackson, E.K.; Burns, R.C. The acetylene-ethylene assay for N2 fixation: Laboratory and field evaluation. Plant Physiol. 1968, 43, 1185–1207. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Agrama, H.A.; Kong, D.; Zhuang, J.; Hu, B.; Wan, Y.; Yan, W. Genetic diversity associated with conservation of endangered Dongxiang wild rice (Oryza rufipogon). Genet. Resour. Crop Evol. 2009, 57, 597–609. [Google Scholar] [CrossRef]
- Keswani, C.; Singh, S.P.; Cueto, L.; García-Estrada, C.; Mezaache-Aichour, S.; Glare, T.R.; Borriss, R.; Singh, S.P.; Blázquez, M.A.; Sansinenea, E. Auxins of microbial origin and their use in agriculture. Appl. Microbiol. Biotechnol. 2020, 104, 8549–8565. [Google Scholar] [CrossRef]
- Babalola, O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010, 32, 1559–1570. [Google Scholar] [CrossRef]
- Aguilar, C.; Bertani, I.; Venturi, V. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 2003, 69, 1739–1747. [Google Scholar] [CrossRef]
- Gondry, M.; Sauguet, L.; Belin, P.; Thai, R.; Amouroux, R.; Tellier, C.; Tuphile, K.; Jacquet, M.; Braud, S.; Courçon, M.; et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat. Chem. Biol. 2009, 5, 414–420. [Google Scholar] [CrossRef]
- Mahajan-Miklos, S.; Tan, M.-W.; Rahme, L.G.; Ausubel, F.M. Molecular Mechanisms of Bacterial Virulence Elucidated Using a Pseudomonas aeruginosa–Caenorhabditis elegans Pathogenesis Model. Cell 1999, 96, 47–56. [Google Scholar] [CrossRef]
- Conery, A.L.; Larkins-Ford, J.; Ausubel, F.M.; Kirienko, N.V. High-throughput screening for novel anti-infectives using a C. elegans pathogenesis model. Curr. Protoc. Chem. Biol. 2014, 6, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Gyaneshwar, P.; Hirsch, A.M.; Moulin, L.; Chen, W.M.; Elliott, G.N.; Bontemps, C.; Estrada-de los Santos, P.; Gross, E.; dos Reis, F.B.; Sprent, J.I.; et al. Legume-nodulating betaproteobacteria: Diversity, host range, and future prospects. Mol. Plant Microbe Interact. 2011, 24, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Bontemps, C.; Elliott, G.N.; Simon, M.F.; Dos Reis, F.B.D.; Gross, E.; Lawton, R.C.; Neto, N.E.; Loureiro, M.D.; De Faria, S.M.; Sprent, J.I.; et al. Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 2010, 19, 44–52. [Google Scholar] [CrossRef]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-solubilizing microorganisms: Mechanism and their role in phosphate solubilization and uptake. J. Soil Sci. Plant Nutr. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Silva, H.; Cunha, A. Siderophore-producing rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: A review. Pedosphere 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Naing, A.H.; Maung, T.T.; Kim, C.K. The ACC deaminase-producing plant growth-promoting bacteria: Influences of bacterial strains and ACC deaminase activities in plant tolerance to abiotic stress. Physiol. Plant. 2021, 173, 1992–2012. [Google Scholar] [CrossRef]
- Zhang, J.; Yao, Z.; Chen, Y.; Zhang, J.; Luo, S.; Tian, C.; Tian, L. Study of rhizosphere microbial community structures of asian wild and cultivated rice showed that cultivated rice had decreased and enriched some functional microorganisms in the process of domestication. Diversity 2022, 14, 67. [Google Scholar] [CrossRef]
- Ferreira, M.R.; Gomes, S.C.; Moreira, L.M. Mucoid switch in Burkholderia cepacia complex bacteria: Triggers, molecular mechanisms and implications in pathogenesis. Adv. Appl. Microbiol. 2019, 107, 113–140. [Google Scholar]
- Gimenez-Ibanez, S.; Ntoukakis, V.; Rathjen, J.P. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 2009, 4, 539–541. [Google Scholar] [CrossRef] [PubMed]
- Willmann, R.; Lajunen Heini, M.; Erbs, G.; Newman, M.-A.; Kolb, D.; Tsuda, K.; Katagiri, F.; Fliegmann, J.; Bono, J.-J.; Cullimore, J.V.; et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, S.I.; Yun, S.H.; Shin, M.; Lee, Y.C.; Lee, J.C. Proteins in outer membrane vesicles produced by Burkholderia cepacia are responsible for pro-inflammatory responses in epithelial cells. J. Bacteriol. Virol. 2020, 50, 227–234. [Google Scholar] [CrossRef]
- Siefert, J.L. Defining the Mobilome. In Horizontal Gene Transfer: Genomes in Flux; Gogarten, M.B., Gogarten, J.P., Olendzenski, L., Eds.; Humana Press: New York, NY, USA, 2009; Volume 532, pp. 13–27. [Google Scholar]
- Lasek, R.; Szuplewska, M.; Mitura, M.; Decewicz, P.; Chmielowska, C.; Pawlot, A.; Sentkowska, D.; Czarnecki, J.; Bartosik, D. Genome structure of the opportunistic pathogen Paracoccus yeei (Alphaproteobacteria) and identification of putative virulence factors. Front. Microbiol. 2018, 9, 2553. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Trudel, M.V.; Freschi, L.; Nagar, V.; Gagné-Thivierge, C.; Levesque, R.C.; Charette, S.J. Increasing genomic diversity and evidence of constrained lifestyle evolution due to insertion sequences in Aeromonas salmonicida. BMC Genom. 2016, 17, 44. [Google Scholar] [CrossRef] [Green Version]
- Mebrhatu, M.T.; Cenens, W.; Aertsen, A. An overview of the domestication and impact of the Salmonella mobilome. Crit. Rev. Microbiol. 2014, 40, 63–75. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Zamora-Lagos, M.A.; Blettinger, M.; Yeroslaviz, A.; Dahl, A.; Gruber, S.; Habermann, B.H. The complete and fully assembled genome sequence of Aeromonas salmonicida subsp. pectinolytica and its comparative analysis with other Aeromonas species: Investigation of the mobilome in environmental and pathogenic strains. BMC Genom. 2018, 19, 20. [Google Scholar] [CrossRef]
- Lim, H.J.; Lee, E.H.; Yoon, Y.; Chua, B.; Son, A. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 2016, 120, 379–387. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009, 37, D136–D140. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Brinkman, F.S.L. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 2018, 34, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]

| PGP Activities | Gene Name | Gene Annotation | Chromosomal Location |
|---|---|---|---|
| Nodulation | |||
| nodW | two component transcriptional regulator, Nodulation protein W | Chr1:564266-564919− | |
| nodT | NodT family protein | Chr1:957928-959382+ | |
| nodS | Nodulation protein S | Chr3:1174329-1175027− | |
| nodJ | Nodulation protein J | Chr3:1903018-1903851− | |
| nodI | Nod factor export ATP-binding protein I | Chr3:1903858-1904772− | |
| nfeD | Nodulation efficiency protein NfeD | Chr3:2432325-2432759− | |
| Nitrogen Fixation | |||
| fixX | Ferredoxin-like protein FixX | Chr1:906003-907661− | |
| nifQ | nitrogen fixation protein NifQ | Chr1:1372743-1373342− | |
| fixB, etfA | electron transfer flavoprotein alpha | Chr2:2094197-2095141− | |
| fixA, etfB | subunit electron transfer flavoprotein beta subunit | Chr2:2095158-2095913− | |
| rnfH | Protein RnfH | Chr3:2429820-2430143− | |
| iscU, nifU | nitrogen fixation protein NifU and related proteins | Chr3:2612558-2612965− | |
| iscS, NFS1 | cysteine desulfurase, Nitrogen Fixation 1 | Chr3:2613034-2614224− | |
| fixJ | Transcriptional regulatory protein FixJ | Chr3:2631016-2631654+ | |
| rnfB | electron transport complex protein RnfB | Chr3:2897345-2898232+ | |
| nifH | 4Fe-4S iron sulfur cluster binding proteins, NifH/frxC family | Chr3:2990786-2991844− | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Henawy, A.R.; Halema, A.A.; Fan, Q.; Duanmu, D.; Huang, R. A Wild Rice Rhizobacterium Burkholderia cepacia BRDJ Enhances Nitrogen Use Efficiency in Rice. Int. J. Mol. Sci. 2022, 23, 10769. https://doi.org/10.3390/ijms231810769
Li Z, Henawy AR, Halema AA, Fan Q, Duanmu D, Huang R. A Wild Rice Rhizobacterium Burkholderia cepacia BRDJ Enhances Nitrogen Use Efficiency in Rice. International Journal of Molecular Sciences. 2022; 23(18):10769. https://doi.org/10.3390/ijms231810769
Chicago/Turabian StyleLi, Zheng, Ahmed R. Henawy, Asmaa A. Halema, Qiuling Fan, Deqiang Duanmu, and Renliang Huang. 2022. "A Wild Rice Rhizobacterium Burkholderia cepacia BRDJ Enhances Nitrogen Use Efficiency in Rice" International Journal of Molecular Sciences 23, no. 18: 10769. https://doi.org/10.3390/ijms231810769





