Transcriptional Activation of Ecdysone-Responsive Genes Requires H3K27 Acetylation at Enhancers
Abstract
:1. Introduction
2. Results
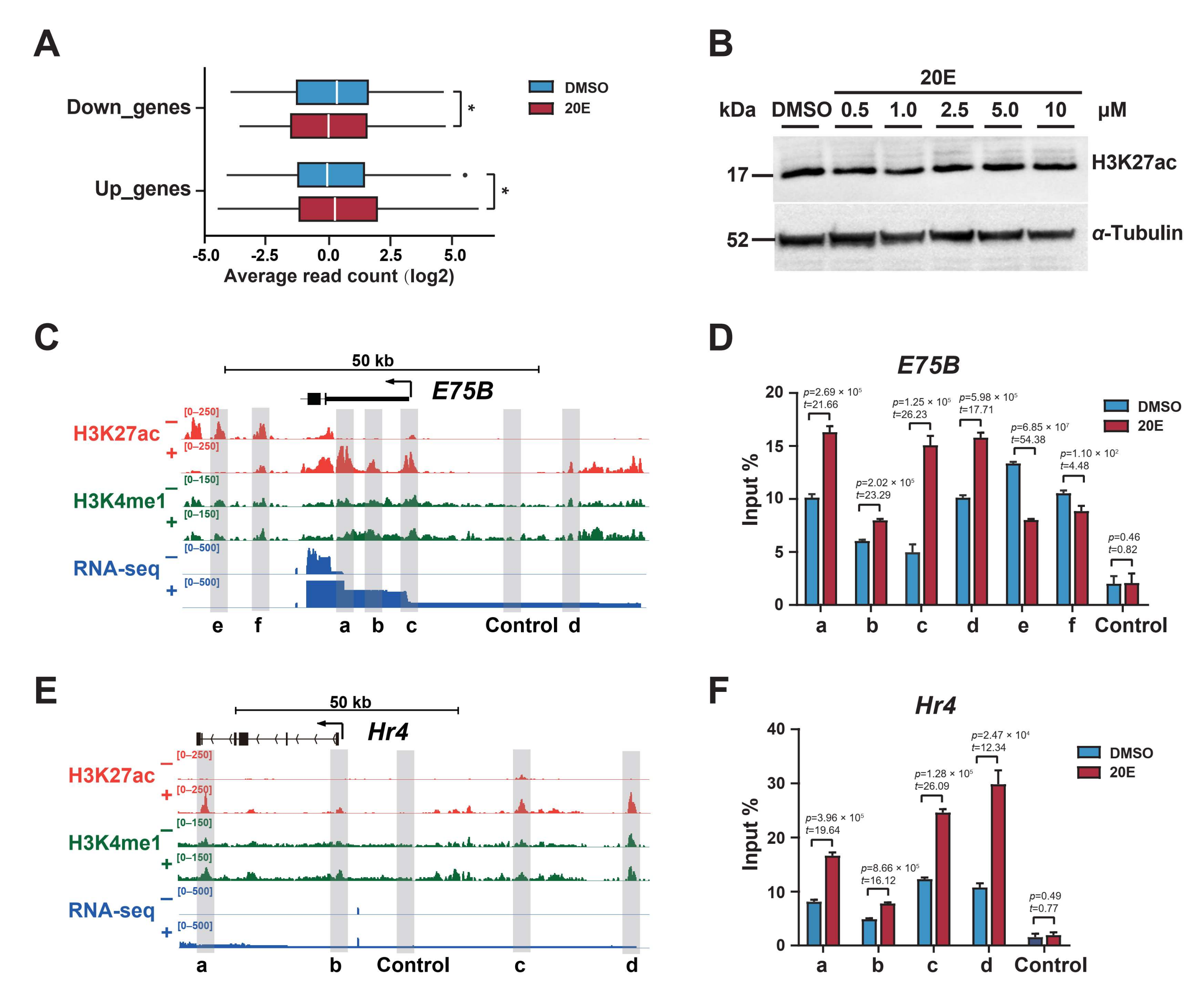
2.1. Ecdysone Induces Local Dynamic Changes in H3K27 Acetylation
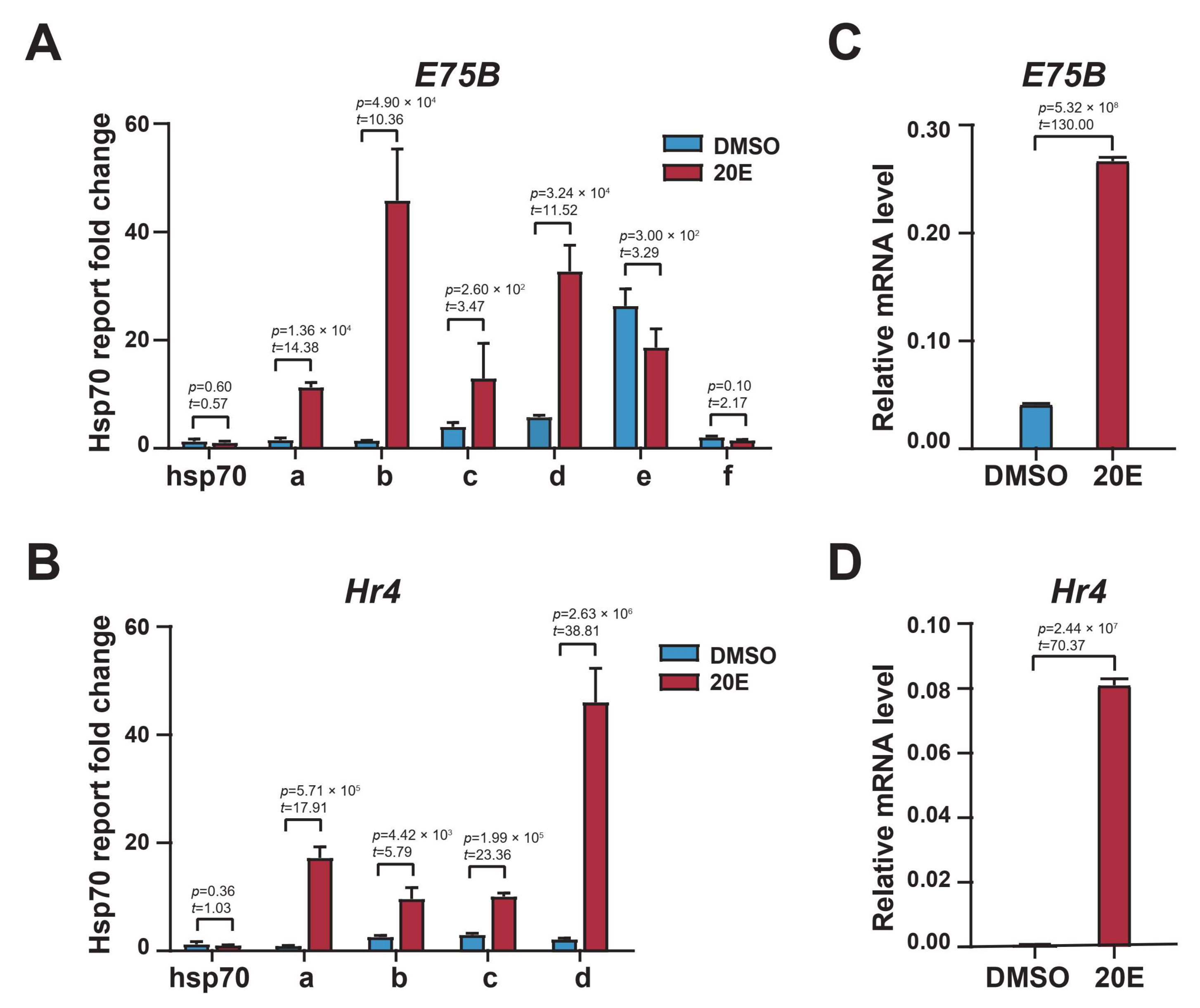
2.2. Enhancer H3K27 Acetylation Enables Ecdysone-Responsive Gene Transcription Activation
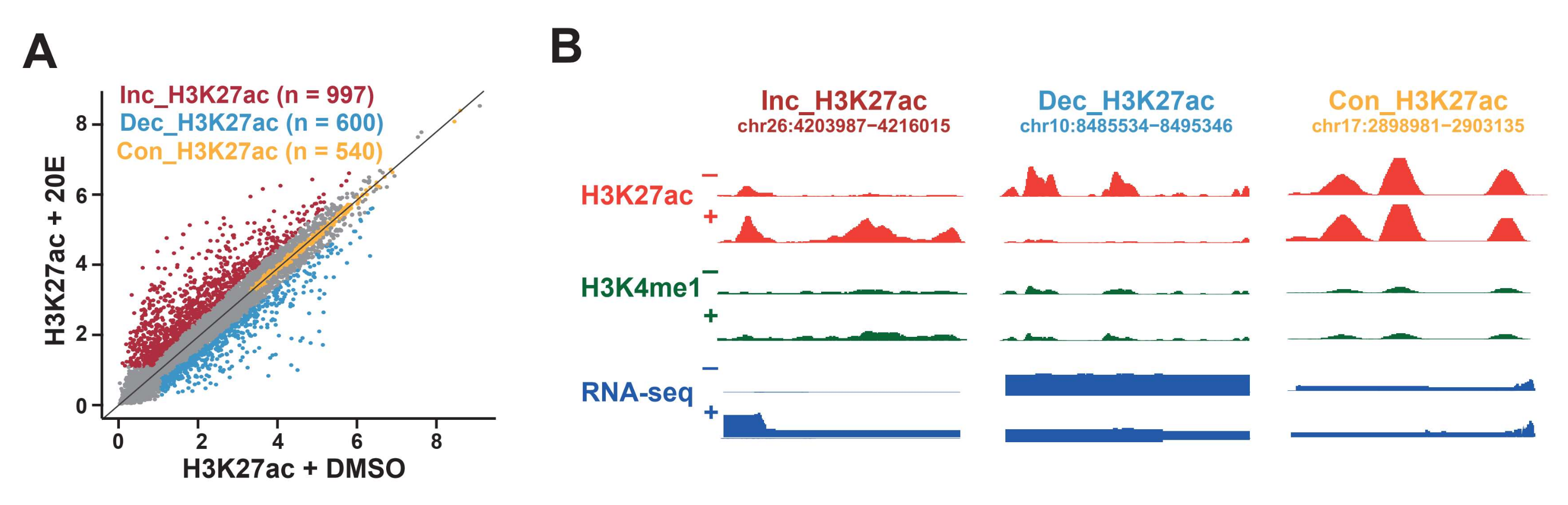
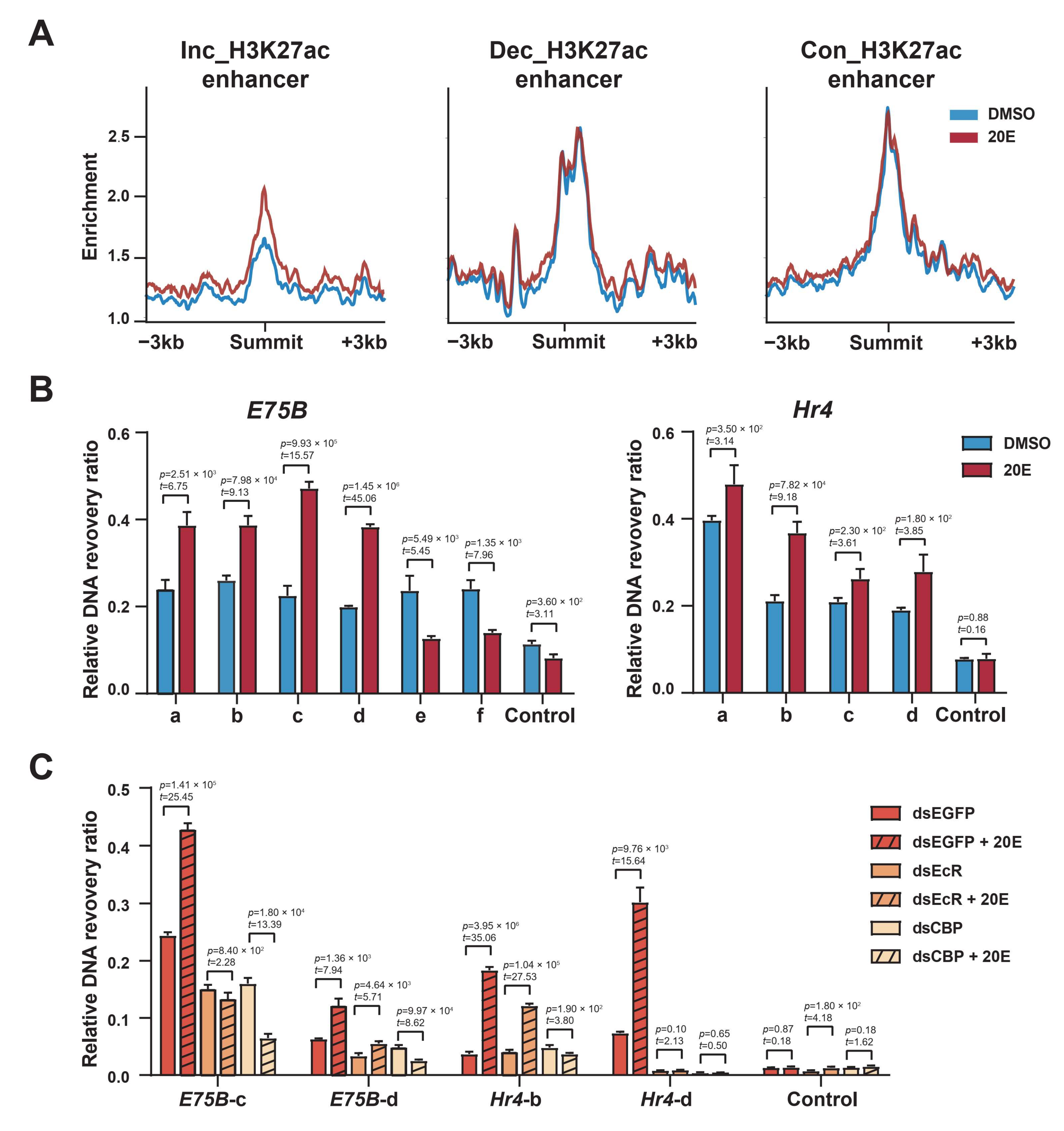
2.3. Enhancer Classification Based on H3K27 Acetylation Dynamics in Response to 20E
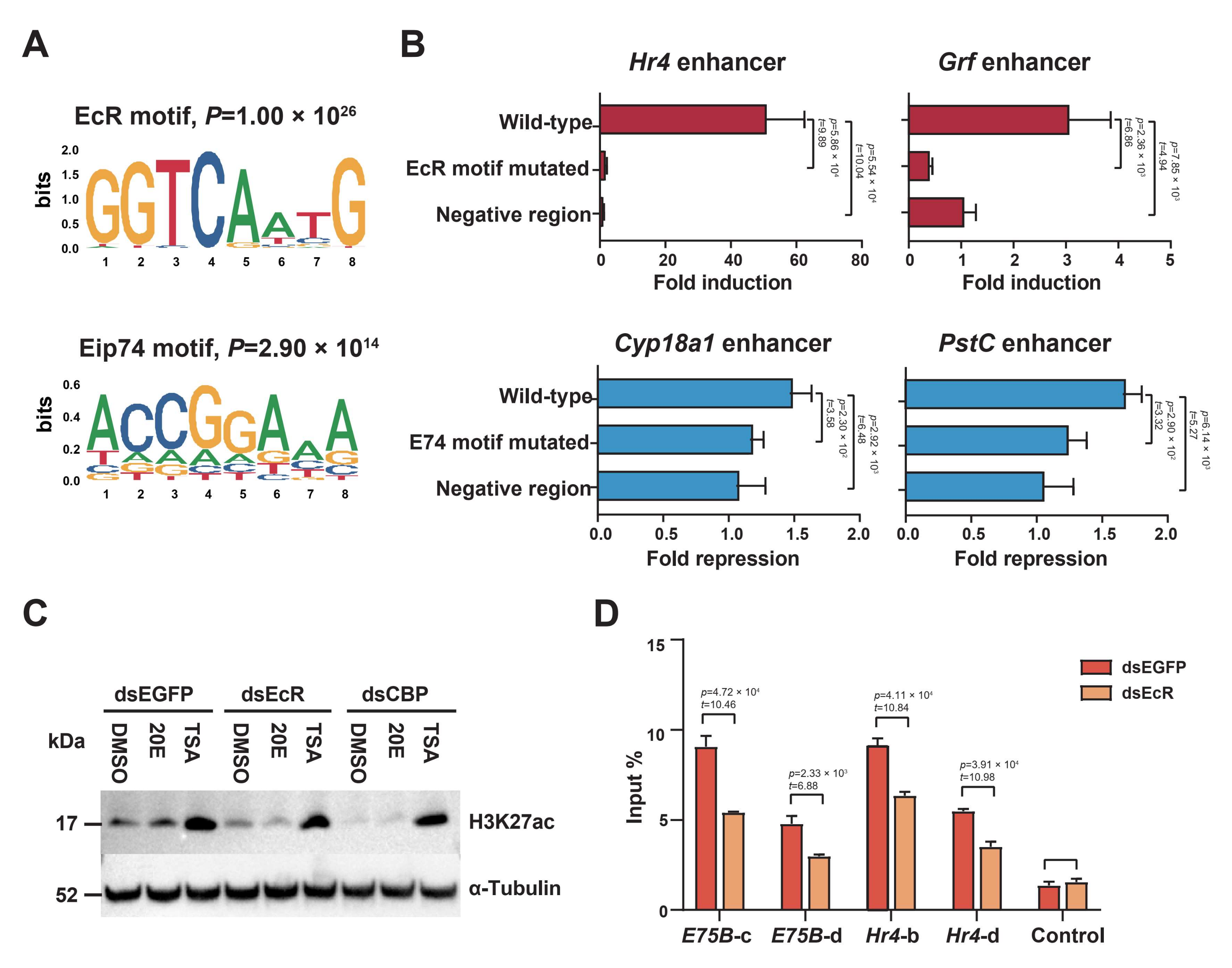
2.4. EcR Binding Is Highly Enriched in H3K27 Acetylation-Enriched Enhancers
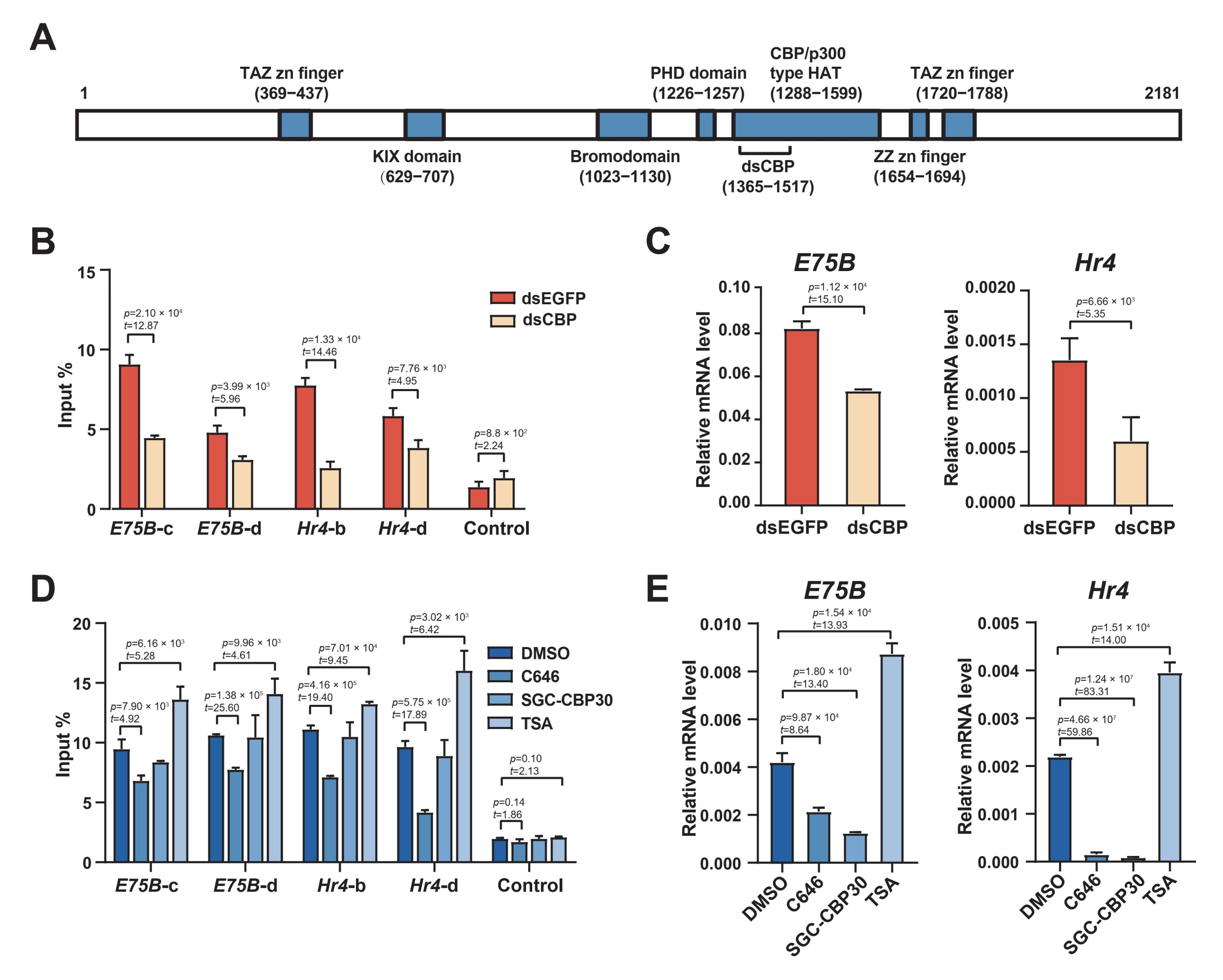
2.5. The HAT Domain of CBP Is Required to Acetylate H3K27
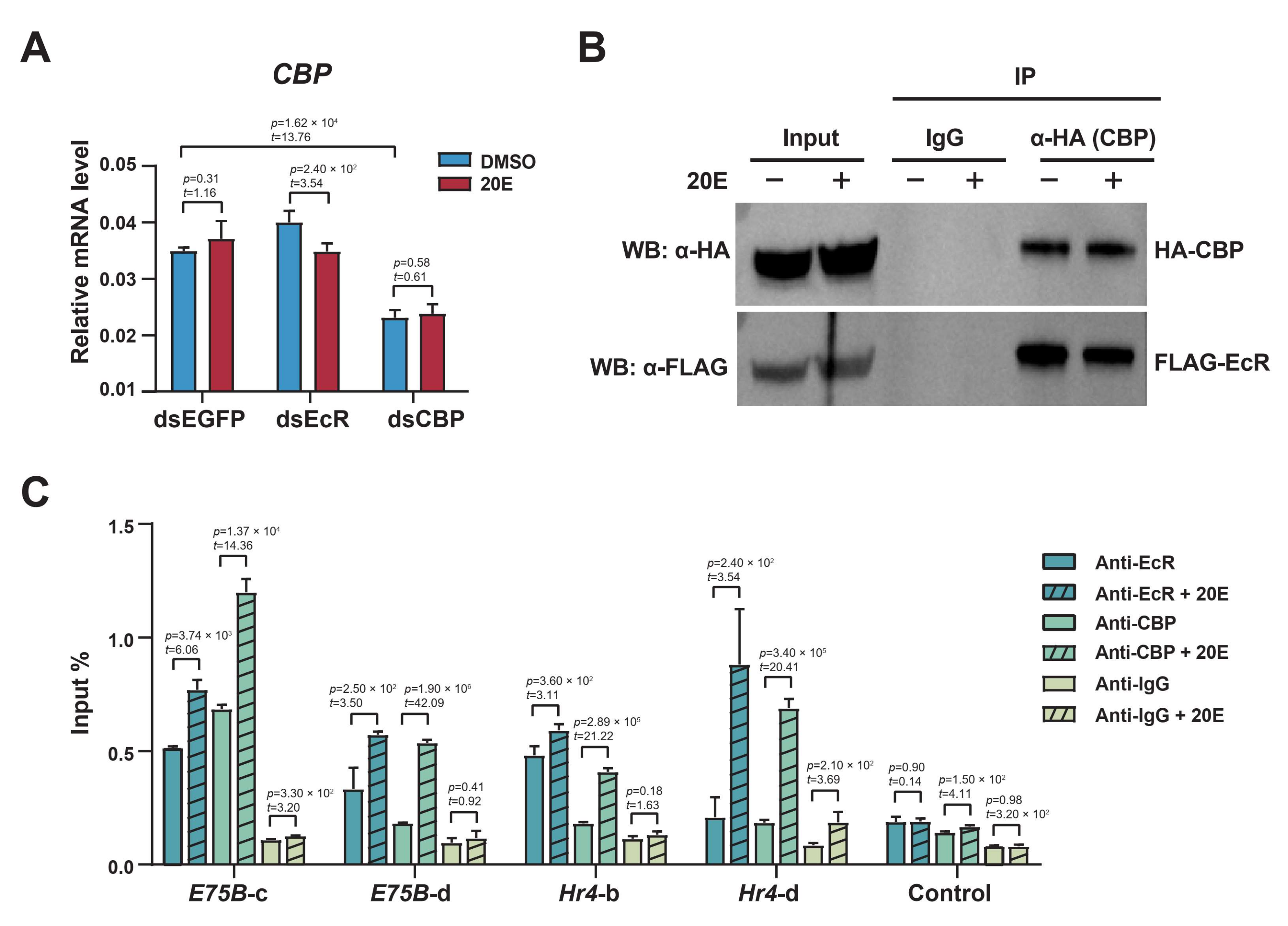
2.6. The Ecdysone Receptor Recruits CBP as a Coactivator to Acetylate H3K27 in Response to 20E
2.7. Ecdysone-Induced Chromatin Opening
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Ecdysone Incubation, and Inhibitor Treatment
4.2. qRT-PCR
4.3. Transfection and Luciferase Reporter Assay
4.4. RNAi
4.5. Western Blotting
4.6. Coimmunoprecipitation (Co-IP)
4.7. ChIP-qPCR
4.8. FAIRE-Seq and FAIRE-qPCR
4.9. Computational Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamanaka, N.; Rewitz, K.F.; O’Connor, M.B. Ecdysone control of developmental transitions: Lessons from Drosophila research. Annu. Rev. Entomol. 2013, 58, 497–516. [Google Scholar] [CrossRef]
- Schwedes, C.C.; Carney, G.E. Ecdysone signaling in adult Drosophila melanogaster. J. Insect Physiol. 2012, 58, 293–302. [Google Scholar] [CrossRef]
- Hill, R.J.; Billas, I.M.; Bonneton, F.; Graham, L.D.; Lawrence, M.C. Ecdysone receptors: From the Ashburner model to structural biology. Annu. Rev. Entomol. 2013, 58, 251–271. [Google Scholar] [CrossRef] [PubMed]
- Aranda, A.; Pascual, A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001, 81, 1269–1304. [Google Scholar] [CrossRef]
- Nishita, Y. Ecdysone response elements in the distal promoter of the Bombyx Broad-Complex gene, BmBR-C. Insect Mol. Biol. 2014, 23, 341–356. [Google Scholar]
- Cherbas, L.; Lee, K.; Cherbas, P. Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev. 1991, 5, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Uyehara, C.M.; McKay, D.J. Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. Proc. Natl. Acad. Sci. USA 2019, 116, 9893–9902. [Google Scholar] [CrossRef] [PubMed]
- Antoniewski, C.; Laval, M.; Dahan, A.; Lepesant, J.A. The ecdysone response enhancer of the Fbp1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol. Cell. Biol. 1994, 14, 4465–4474. [Google Scholar]
- Kato, S.; Yokoyama, A.; Fujiki, R. Nuclear receptor coregulators merge transcriptional coregulation with epigenetic regulation. Trends Biochem. Sci. 2011, 36, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Glastad, K.M.; Hunt, B.G.; Goodisman, M.A.D. Epigenetics in Insects: Genome Regulation and the Generation of Phenotypic Diversity. Annu. Rev. Entomol. 2019, 64, 185–203. [Google Scholar] [CrossRef]
- Mazina, M.Y.; Kovalenko, E.V.; Derevyanko, P.K.; Nikolenko, J.V.; Krasnov, A.N.; Vorobyeva, N.E. One signal stimulates different transcriptional activation mechanisms. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Kohzaki, H.; Asano, M.; Murakami, Y.; Mazo, A. Epigenetic regulation affects gene amplification in Drosophila development. Front. Biosci. 2020, 25, 632–645. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Calo, E.; Wysocka, J. Modification of Enhancer Chromatin: What, How, and Why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef]
- Niederhuber, M.J.; McKay, D.J. Mechanisms underlying the control of dynamic regulatory element activity and chromatin accessibility during metamorphosis. Curr. Opin. Insect Sci. 2021, 43, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Fischer, R.; Vilcinskas, A. Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front. Zool. 2012, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Palli, S.R. Epigenetic modifications acetylation and deacetylation play important roles in juvenile hormone action. BMC Genom. 2018, 19, 934. [Google Scholar] [CrossRef] [PubMed]
- Borsos, B.N.; Pankotai, T.; Kovacs, D.; Popescu, C.; Pahi, Z.; Boros, I.M. Acetylations of Ftz-F1 and histone H4K5 are required for the fine-tuning of ecdysone biosynthesis during Drosophila metamorphosis. Dev. Biol. 2015, 404, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Bodai, L.; Zsindely, N.; Gaspar, R.; Kristo, I.; Komonyi, O.; Boros, I.M. Ecdysone induced gene expression is associated with acetylation of histone H3 lysine 23 in Drosophila melanogaster. PLoS ONE 2012, 7, e40565. [Google Scholar]
- Hassig, C.A.; Schreiber, S.L. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1997, 1, 300–308. [Google Scholar] [CrossRef]
- Peserico, A.; Simone, C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011, 2011, 371832. [Google Scholar] [CrossRef]
- Kalkhoven, E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004, 68, 1145–1155. [Google Scholar] [CrossRef]
- Gaddelapati, S.C.; Dhandapani, R.K.; Palli, S.R. CREB-binding protein regulates metamorphosis and compound eye development in the yellow fever mosquito, Aedes aegypti. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194576. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, T.; Gartner, S.M.K.; Rathke, C.; Renkawitz-Pohl, R. Nejire/dCBP-mediated histone H3 acetylation during spermatogenesis is essential for male fertility in Drosophila melanogaster. PLoS ONE 2018, 13, e0203622. [Google Scholar]
- Roy, A.; George, S.; Palli, S.R. Multiple functions of CREB-binding protein during postembryonic development: Identification of target genes. BMC Genom. 2017, 18, 996. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Roy, A.; Palli, S.R. CREB-binding protein plays key roles in juvenile hormone action in the red flour beetle, Tribolium Castaneum. Sci. Rep. 2018, 8, 1426. [Google Scholar] [CrossRef]
- George, S.; Palli, S.R. Histone Deacetylase 11 Knockdown Blocks Larval Development and Metamorphosis in the Red Flour Beetle, Tribolium castaneum. Front. Genet. 2020, 11, 683. [Google Scholar] [CrossRef]
- Kellner, W.A.; Ramos, E.; Van Bortle, K.; Takenaka, N.; Corces, V.G. Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters. Genome Res. 2012, 22, 1081–1088. [Google Scholar] [CrossRef]
- Kirilly, D.; Wong, J.J.; Lim, E.K.; Wang, Y.; Zhang, H.; Wang, C.; Liao, Q.; Wang, H.; Liou, Y.C.; Wang, H.; et al. Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila. Neuron 2011, 72, 86–100. [Google Scholar] [CrossRef]
- George, S.; Gaddelapati, S.C.; Palli, S.R. Histone deacetylase 1 suppresses Kruppel homolog 1 gene expression and influences juvenile hormone action in Tribolium castaneum. Proc. Natl. Acad. Sci. USA 2019, 116, 17759–17764. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, L.; Sun, G.; Raikhel, A.S. The competence factor beta Ftz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol. Cell. Biol. 2006, 26, 9402–9412. [Google Scholar] [CrossRef]
- Cheng, D.; Cheng, T.; Yang, X.; Zhang, Q.; Fu, J.; Feng, T.; Gong, J.; Xia, Q. The genome-wide transcriptional regulatory landscape of ecdysone in the silkworm. Epigenetics Chromatin 2018, 11, 48. [Google Scholar] [CrossRef]
- Ou, Q.; Magico, A.; King-Jones, K. Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS Biol. 2011, 9, e1001160. [Google Scholar] [CrossRef]
- Segraves, W.A.; Hogness, D.S. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990, 4, 204–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fletcher, J.C.; D’Avino, P.P.; Thummel, C.S. A steroid-triggered switch in E74 transcription factor isoforms regulates the timing of secondary-response gene expression. Proc. Natl. Acad. Sci. USA 1997, 94, 4582–4586. [Google Scholar] [CrossRef] [PubMed]
- Kirfel, P.; Vilcinskas, A.; Skaljac, M. Lysine Acetyltransferase p300/CBP Plays an Important Role in Reproduction, Embryogenesis and Longevity of the Pea Aphid Acyrthosiphon pisum. Insects 2020, 11, 265. [Google Scholar] [CrossRef]
- Fernandez-Nicolas, A.; Belles, X. CREB-binding protein contributes to the regulation of endocrine and developmental pathways in insect hemimetabolan pre-metamorphosis. Biochim. Biophys. Acta 2016, 1860, 508–515. [Google Scholar] [CrossRef]
- Ferrari, K.J.; Scelfo, A.; Jammula, S.; Cuomo, A.; Barozzi, I.; Stutzer, A.; Fischle, W.; Bonaldi, T.; Pasini, D. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 2014, 53, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Heinz, S.; Romanoski, C.E.; Benner, C.; Glass, C.K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015, 16, 144–154. [Google Scholar] [CrossRef]
- Shlyueva, D.; Stelzer, C.; Gerlach, D.; Yanez-Cuna, J.O.; Rath, M.; Boryn, L.M.; Arnold, C.D.; Stark, A. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol. Cell 2014, 54, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Ratman, D.; Vanden Berghe, W.; Dejager, L.; Libert, C.; Tavernier, J.; Beck, I.M.; De Bosscher, K. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol. Cell. Endocrinol. 2013, 380, 41–54. [Google Scholar] [CrossRef]
- Grontved, L.; Waterfall, J.J.; Kim, D.W.; Baek, S.; Sung, M.H.; Zhao, L.; Park, J.W.; Nielsen, R.; Walker, R.L.; Zhu, Y.J.; et al. Transcriptional activation by the thyroid hormone receptor through ligand-dependent receptor recruitment and chromatin remodelling. Nat. Commun. 2015, 6, 7048. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef] [Green Version]
- Libbrecht, R.; Nadrau, D.; Foitzik, S. A Role of Histone Acetylation in the Regulation of Circadian Rhythm in Ants. iScience 2020, 23, 100846. [Google Scholar] [CrossRef] [PubMed]
- Lyu, H.; Xu, G.; Chen, P.; Song, Q.; Feng, Q.; Yi, Y.; Zheng, S. 20-Hydroxyecdysone receptor-activated Bombyx mori CCAAT/enhancer-binding protein gamma regulates the expression of BmCBP and subsequent histone H3 lysine 27 acetylation in Bo. mori. Insect Mol. Biol. 2020, 29, 256–270. [Google Scholar] [CrossRef]
- Chakravarti, D.; LaMorte, V.J.; Nelson, M.C.; Nakajima, T.; Schulman, I.G.; Juguilon, H.; Montminy, M.; Evans, R.M. Role of CBP/P300 in nuclear receptor signalling. Nature 1996, 383, 99–103. [Google Scholar] [CrossRef]
- Khan, O.Y.; Nawaz, Z. Nuclear hormone receptor co-regulators. Curr. Opin. Drug Discov. Dev. 2003, 6, 692–701. [Google Scholar]
- Badenhorst, P.; Xiao, H.; Cherbas, L.; Kwon, S.Y.; Voas, M.; Rebay, I.; Cherbas, P.; Wu, C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005, 19, 2540–2545. [Google Scholar] [CrossRef]
- Kreher, J.; Kovac, K.; Bouazoune, K.; Macinkovic, I.; Ernst, A.L.; Engelen, E.; Pahl, R.; Finkernagel, F.; Murawska, M.; Ullah, I.; et al. EcR recruits dMi-2 and increases efficiency of dMi-2-mediated remodelling to constrain transcription of hormone-regulated genes. Nat. Commun. 2017, 8, 14806. [Google Scholar] [CrossRef]
- Tsai, C.C.; Kao, H.Y.; Yao, T.P.; McKeown, M.; Evans, R.M. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell 1999, 4, 175–186. [Google Scholar] [CrossRef]
- Carbonell, A.; Mazo, A.; Serras, F.; Corominas, M. Ash2 acts as an ecdysone receptor coactivator by stabilizing the histone methyltransferase Trr. Mol. Biol. Cell 2013, 24, 361–372. [Google Scholar] [CrossRef]
- Gorisch, S.M.; Wachsmuth, M.; Toth, K.F.; Lichter, P.; Rippe, K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005, 118 Pt 24, 5825–5834. [Google Scholar] [CrossRef] [PubMed]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef] [PubMed]
- Paakinaho, V.; Swinstead, E.E.; Presman, D.M.; Grontved, L.; Hager, G.L. Meta-analysis of Chromatin Programming by Steroid Receptors. Cell Rep. 2019, 28, 3523–3534.e2. [Google Scholar] [CrossRef] [Green Version]
- Simon, J.M.; Giresi, P.G.; Davis, I.J.; Lieb, J.D. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012, 7, 256–267. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Feng, J.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Thomas-Chollier, M.; Herrmann, C.; Defrance, M.; Sand, O.; Thieffry, D.; van Helden, J. RSAT peak-motifs: Motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 2012, 40, e31. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, D.; Dong, Z.; Lin, P.; Shen, G.; Xia, Q. Transcriptional Activation of Ecdysone-Responsive Genes Requires H3K27 Acetylation at Enhancers. Int. J. Mol. Sci. 2022, 23, 10791. https://doi.org/10.3390/ijms231810791
Cheng D, Dong Z, Lin P, Shen G, Xia Q. Transcriptional Activation of Ecdysone-Responsive Genes Requires H3K27 Acetylation at Enhancers. International Journal of Molecular Sciences. 2022; 23(18):10791. https://doi.org/10.3390/ijms231810791
Chicago/Turabian StyleCheng, Dong, Zhaoming Dong, Ping Lin, Guanwang Shen, and Qingyou Xia. 2022. "Transcriptional Activation of Ecdysone-Responsive Genes Requires H3K27 Acetylation at Enhancers" International Journal of Molecular Sciences 23, no. 18: 10791. https://doi.org/10.3390/ijms231810791






