A Comprehensive Evolutionary Study of Chloroplast RNA Editing in Gymnosperms: A Novel Type of G-to-A RNA Editing Is Common in Gymnosperms
Abstract
:1. Introduction
2. Results
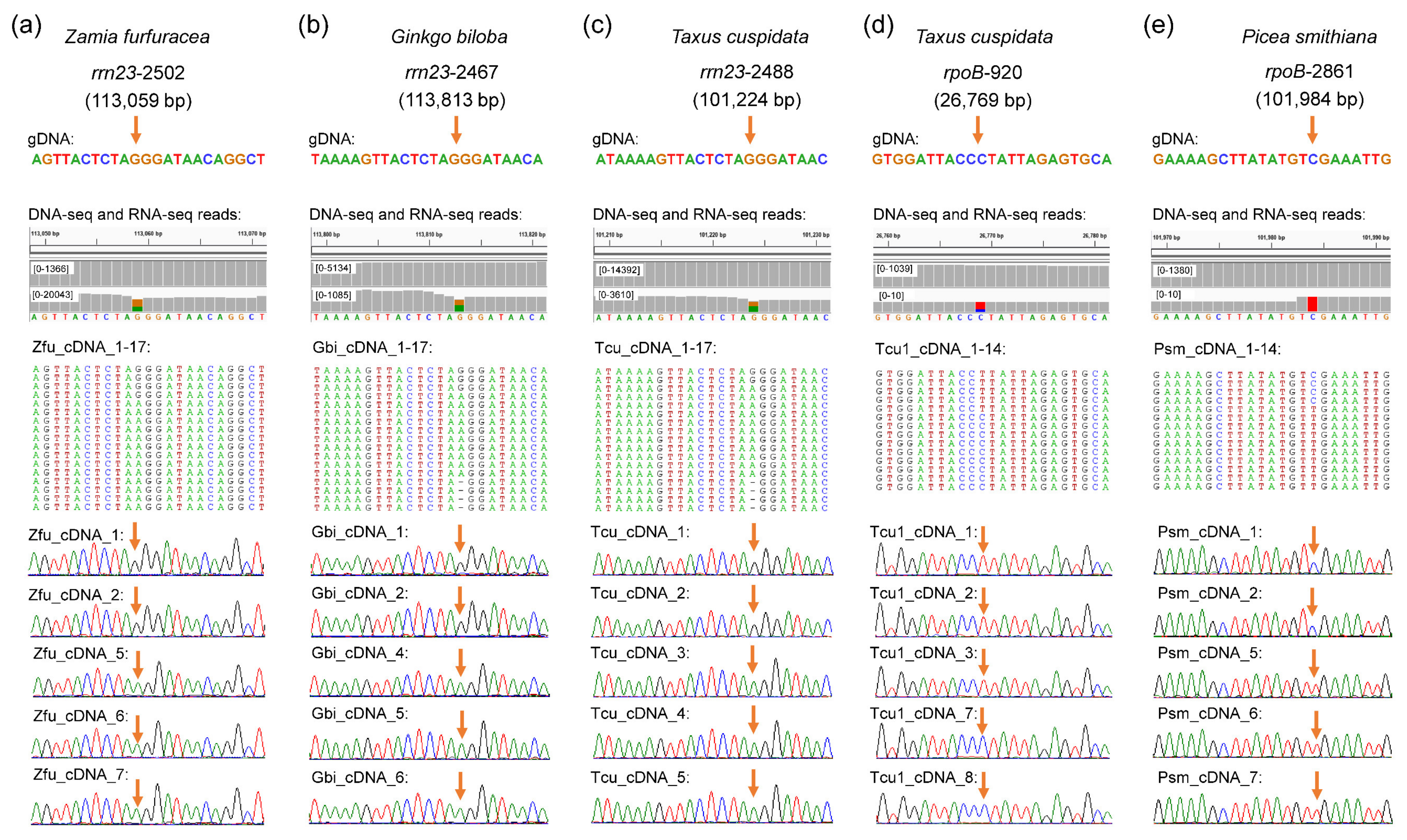
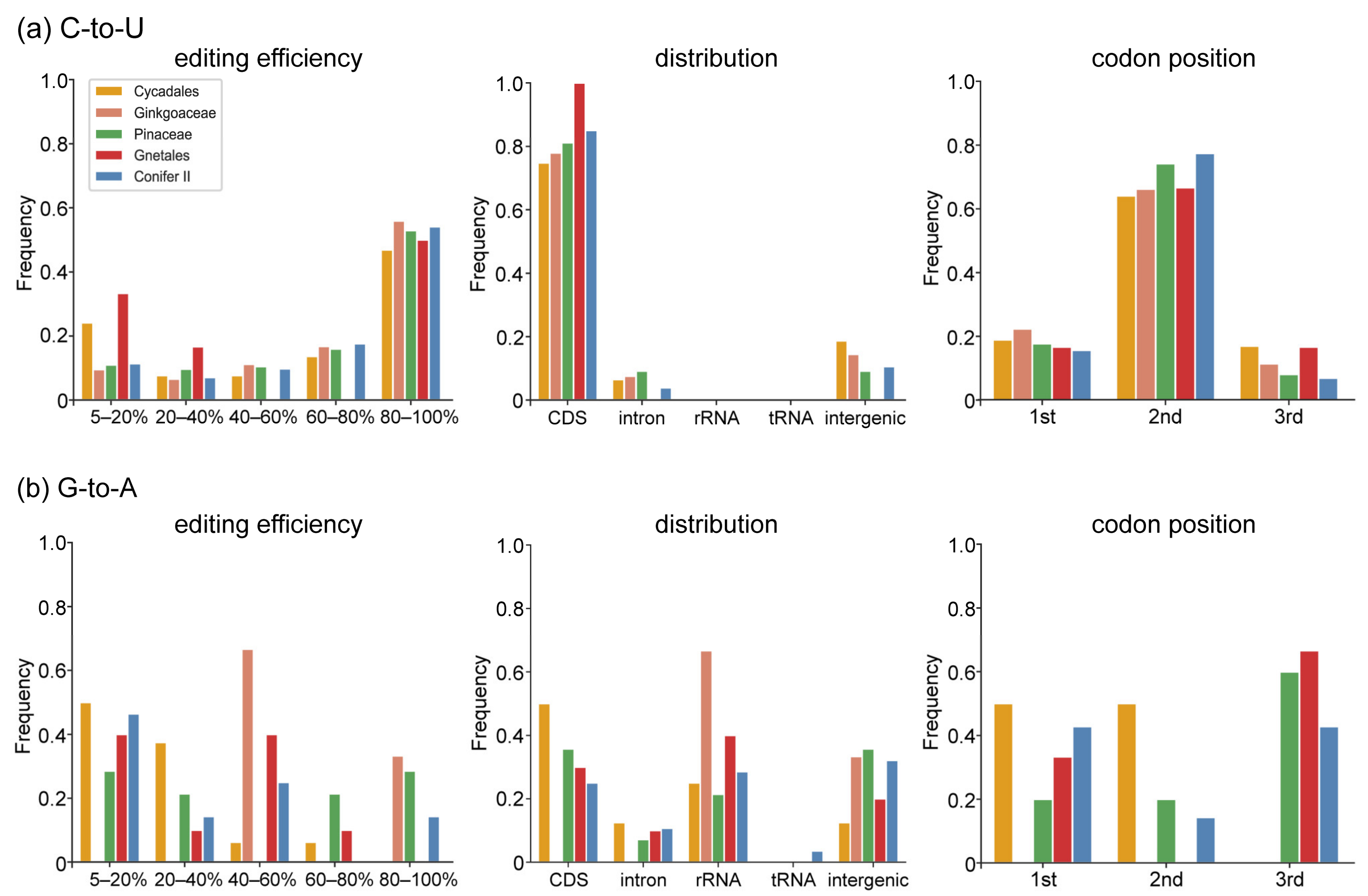
2.1. Characteristics of Chloroplast RNA Editing in Gymnosperms
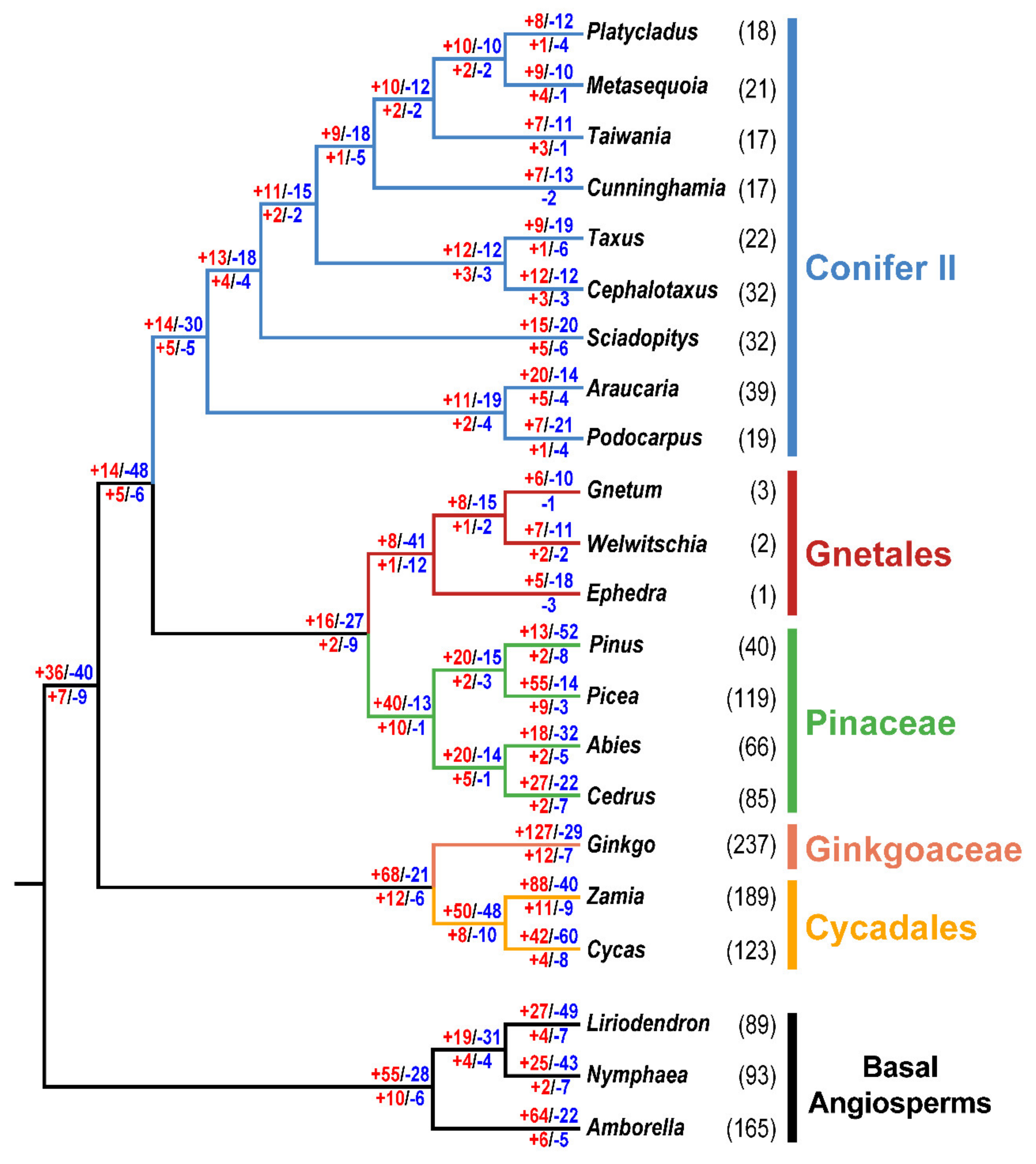
2.2. Variability of Chloroplast RNA Editing Sites in Gymnosperms
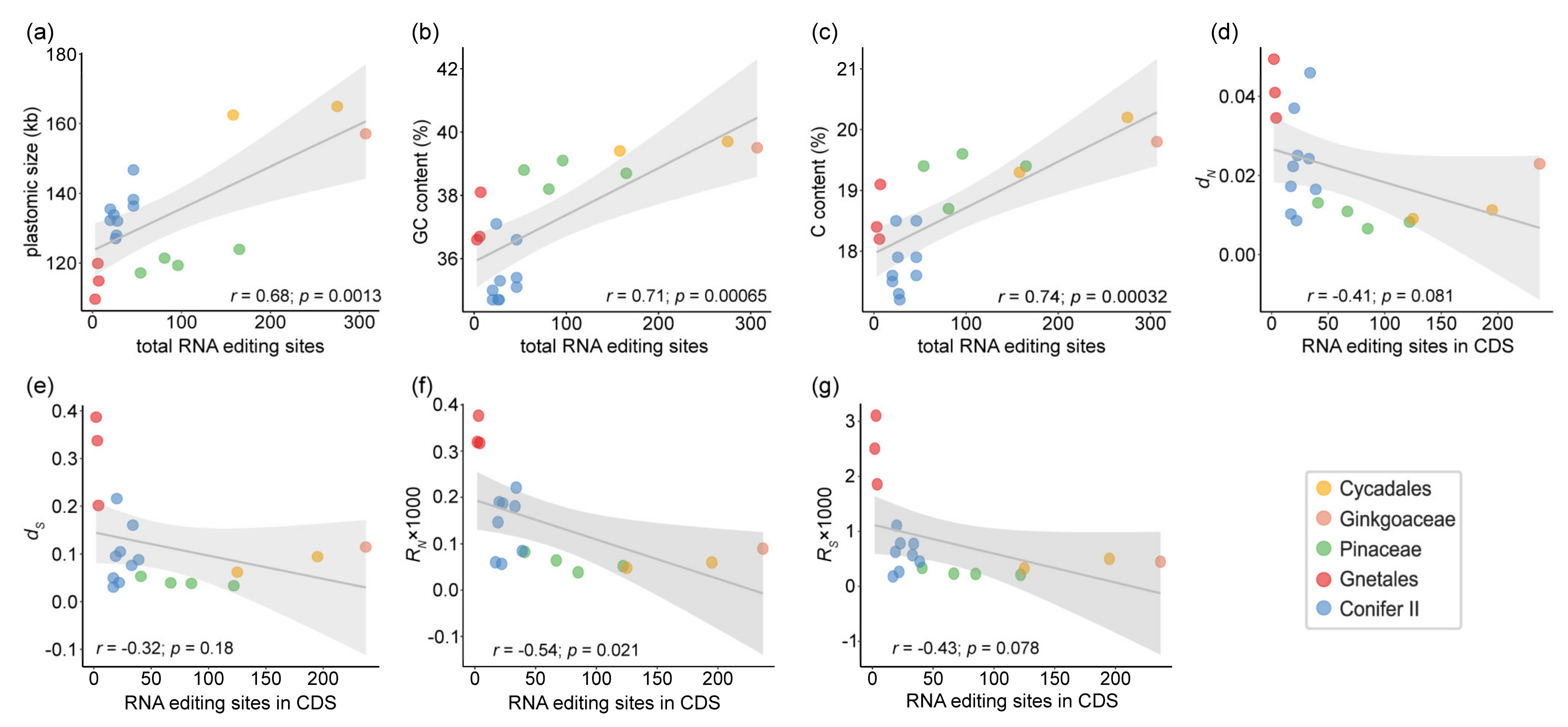
2.3. Factors Correlated with the Number of Chloroplast RNA Editing Sites in Gymnosperms
3. Discussion
3.1. G-to-A, a Novel Type of RNA Editing Site, Is Common in Gymnosperm Plastomes
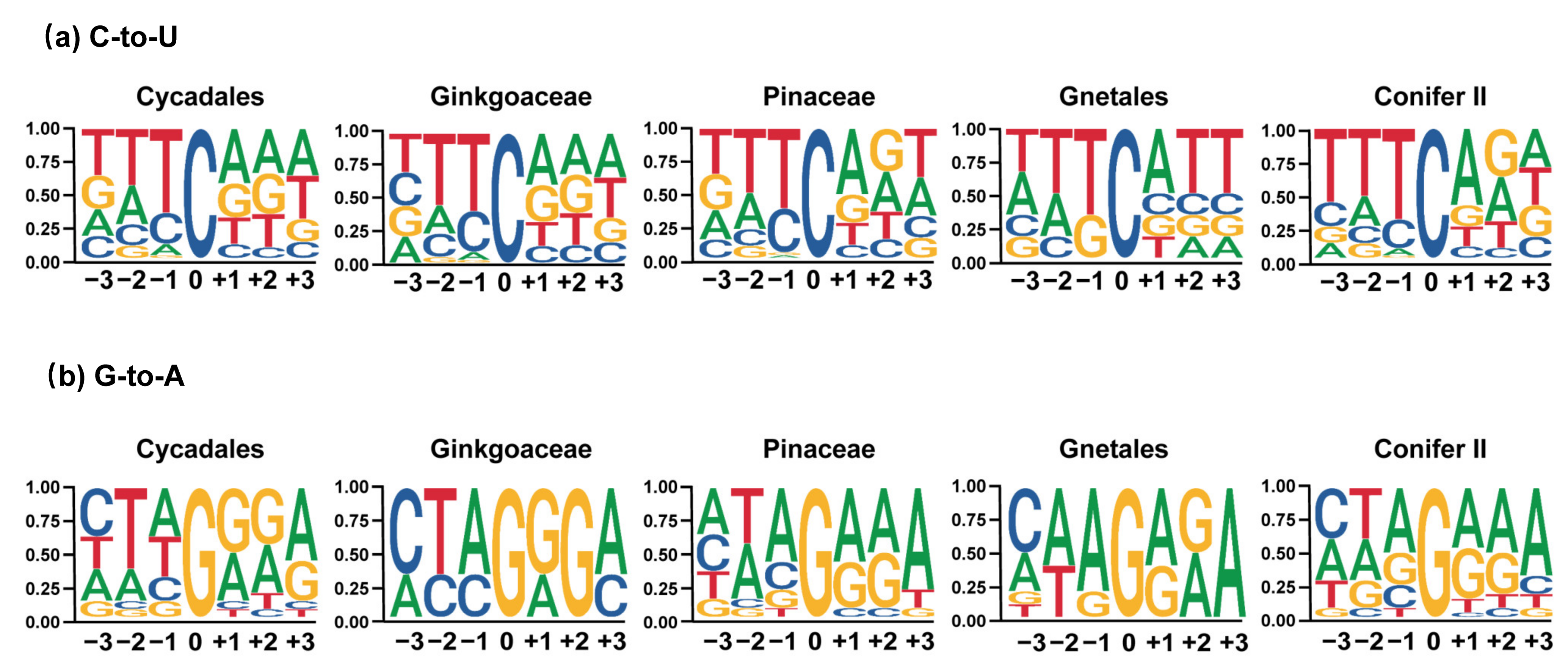
3.2. Chloroplast C-to-U RNA Editing Sites of Gymnosperms Share Many Common Characteristics with Other Land Plants but Also Have Some Unique Characteristics
3.3. Several Factors Could Be Related to Variation in Chloroplast RNA Editing Sites in Gymnosperms
4. Conclusions
5. Materials and Methods
5.1. Taxon Sampling and Sequencing Data Processing
5.2. Plastomic Assembly and Annotation
5.3. RNA Editing Site Identification and Analysis
5.4. Correlation Analyses between Some Important Factors and the Number of RNA Editing Sites
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knoop, V. When you can’t trust the DNA: RNA editing changes transcript sequences. Experientia 2010, 68, 567–586. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Shikanai, T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA 2007, 104, 8178–8183. [Google Scholar] [CrossRef] [PubMed]
- Bégu, D.; Graves, P.V.; Domec, C.; Arselin, G.; Litvak, S.; Araya, A. RNA editing of wheat mitochondrial ATP synthase subunit 9: Direct protein and cDNA sequencing. Plant Cell 1990, 2, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Kotera, E.; Tasaka, M.; Shikanai, T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 2005, 433, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.M.; Zeitz, P.; Kössel, H.; Bonnard, G.; Gualberto, J.M.; Grienenberger, J.M. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996, 32, 343–365. [Google Scholar] [CrossRef]
- Rüdinger, M.; Polsakiewicz, M.; Knoop, V. Organellar RNA Editing and Plant-Specific Extensions of Pentatricopeptide Repeat Proteins in Jungermanniid but not in Marchantiid Liverworts. Mol. Biol. Evol. 2008, 25, 1405–1414. [Google Scholar] [CrossRef]
- Schallenberg-Rüdinger, M.; Knoop, V. Coevolution of Organelle RNA Editing and Nuclear Specificity Factors in Early Land Plants. Adv. Bot. Res. 2016, 78, 37–93. [Google Scholar] [CrossRef]
- Fauskee, B.D.; Sigel, E.M.; Pryer, K.M.; Grusz, A.L. Variation in frequency of plastid RNA editing within Adiantum implies rapid evolution in fern plastomes. Am. J. Bot. 2021, 108, 820–827. [Google Scholar] [CrossRef]
- Knie, N.; Grewe, F.; Fischer, S.; Knoop, V. Reverse U-to-C editing exceeds C-to-U RNA editing in some ferns—A monilophyte-wide comparison of chloroplast and mitochondrial RNA editing suggests independent evolution of the two processes in both organelles. BMC Evol. Biol. 2016, 16, 134. [Google Scholar] [CrossRef]
- Grewe, F.; Herres, S.; Viehöver, P.; Polsakiewicz, M.; Weisshaar, B.; Knoop, V. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2011, 39, 2890–2902. [Google Scholar] [CrossRef]
- Duff, R.J. Divergent RNA editing frequencies in hornwort mitochondrial nad5 sequences. Gene 2006, 366, 285–291. [Google Scholar] [CrossRef]
- Wolf, P.G.; Rowe, C.A.; Hasebe, M. High levels of RNA editing in a vascular plant chloroplast genome: Analysis of transcripts from the fern Adiantum capillus-veneris. Gene 2004, 339, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Kugita, M.; Yamamoto, Y.; Fujikawa, T.; Matsumoto, T.; Yoshinaga, K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 2003, 31, 2417–2423. [Google Scholar] [CrossRef]
- Li, M.; Xia, L.; Zhang, Y.; Niu, G.; Li, M.; Wang, P.; Zhang, Y.; Sang, J.; Zou, D.; Hu, S.; et al. Plant editosome database: A curated database of RNA editosome in plants. Nucleic Acids Res. 2019, 47, D170–D174. [Google Scholar] [CrossRef]
- Members, B.D.C.; Zhang, Z.; Zhao, W.; Xiao, J.; Bao, Y.; Wang, F.; Hao, L.; Zhu, J.; Chen, T.; Zhang, S.; et al. Database Resources of the BIG Data Center in 2019. Nucleic Acids Res. 2018, 47, D8–D14. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Morton, B.R.; Maier, U.G. The Evolution of Chloroplast RNA Editing. Mol. Biol. Evol. 2006, 23, 1912–1921. [Google Scholar] [CrossRef]
- He, P.; Huang, S.; Xiao, G.; Zhang, Y.; Yu, J. Abundant RNA editing sites of chloroplast protein-coding genes in Ginkgo biloba and an evolutionary pattern analysis. BMC Plant Biol. 2016, 16, 257. [Google Scholar] [CrossRef] [PubMed]
- Hein, A.; Polsakiewicz, M.; Knoop, V. Frequent chloroplast RNA editing in early-branching flowering plants: Pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors. BMC Evol. Biol. 2016, 16, 23. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Small, I.; Shikanai, T. Evolutionary Model of Plastidial RNA Editing in Angiosperms Presumed from Genome-Wide Analysis of Amborella trichopoda. Plant Cell Physiol. 2019, 60, 2141–2151. [Google Scholar] [CrossRef]
- Oldenkott, B.; Yamaguchi, K.; Tsuji-Tsukinoki, S.; Knie, N.; Knoop, V. Chloroplast RNA editing going extreme: More than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 2014, 20, 1499–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Fang, J.; Jiang, X.; Wang, T.; Zhang, X. Gaining comprehensive biological insight into chloroplast RNA editing by performing a broad-spectrum RNA-seq analysis. bioRxiv 2020. [CrossRef]
- Binder, S.; Marchfelder, A.; Brennicke, A. RNA editing of tRNAPhe and tRNACys in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet. 1994, 244, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.; Grewe, F.; Knoop, V. Extreme RNA Editing in Coding Islands and Abundant Microsatellites in Repeat Sequences of Selaginella moellendorffii Mitochondria: The Root of Frequent Plant mtDNA Recombination in Early Tracheophytes. Genome Biol. Evol. 2011, 3, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Börner, G.V.; Mörl, M.; Wissinger, B.; Brennicke, A.; Schmelzer, C. RNA editing of a group II intron in Oenothera as a prerequisite for splicing. Mol. Gen. Genet. 1995, 246, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, E.; Le Ret, M.; Faivre-Nitschke, E.; Estavillo, G.M.; Bergdoll, M.; Taylor, N.L.; Pogson, B.J.; Small, I.; Imbault, P.; Gualberto, J.M. Arabidopsis tRNA Adenosine Deaminase Arginine Edits the Wobble Nucleotide of Chloroplast tRNAArg(ACG) and Is Essential for Efficient Chloroplast Translation. Plant Cell 2009, 21, 2058–2071. [Google Scholar] [CrossRef]
- Lu, Y.; Ran, J.-H.; Guo, D.-M.; Yang, Z.-Y.; Wang, X.-Q. Phylogeny and Divergence Times of Gymnosperms Inferred from Single-Copy Nuclear Genes. PLoS ONE 2014, 9, e107679. [Google Scholar] [CrossRef]
- Ran, J.-H.; Shen, T.-T.; Wang, M.-M.; Wang, X.-Q. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc. R. Soc. B: Boil. Sci. 2018, 285, 20181012. [Google Scholar] [CrossRef]
- Fan, W.; Guo, W.; Funk, L.; Mower, J.P.; Zhu, A. Complete loss of RNA editing from the plastid genome and most highly expressed mitochondrial genes of Welwitschia mirabilis. Sci. China Life Sci. 2019, 62, 498–506. [Google Scholar] [CrossRef]
- Chen, H.; Deng, L.; Jiang, Y.; Lu, P.; Yu, J. RNA Editing Sites Exist in Protein-coding Genes in the Chloroplast Genome of Cycas taitungensis. J. Integr. Plant Biol. 2011, 53, 961–970. [Google Scholar] [CrossRef]
- Wakasugi, T.; Hirose, T.; Horihata, M.; Tsudzuki, T.; Kössel, H.; Sugiura, M. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: The pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl. Acad. Sci. USA 1996, 93, 8766–8770. [Google Scholar] [CrossRef] [Green Version]
- Saina, J.K.; Li, Z.-Z.; Gichira, A.W.; Avoga, S.; Wang, Q.-F.; Kuo, L. The complete plastome of real yellow wood (Podocarpus latifolius): Gene organization and comparison with related species. Holzforschung 2019, 73, 525–536. [Google Scholar] [CrossRef]
- Wu, C.-S.; Sudianto, E.; Chaw, S.-M. Tight association of genome rearrangements with gene expression in conifer plastomes. BMC Plant Biol. 2021, 21, 33. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-S.; Chaw, S.-M. Evolution of mitochondrial RNA editing in extant gymnosperms. Plant J. 2022. [CrossRef] [PubMed]
- Ran, J.-H.; Shen, T.-T.; Wu, H.; Gong, X.; Wang, X.-Q. Phylogeny and evolutionary history of Pinaceae updated by transcriptomic analysis. Mol. Phylogenetics Evol. 2018, 129, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Wagih, O. ggseqlogo: A versatile R package for drawing sequence logos. Bioinformatics 2017, 33, 3645–3647. [Google Scholar] [CrossRef]
- Wang, S.; Yang, C.; Zhao, X.; Chen, S.; Qu, G.-Z. Complete chloroplast genome sequence of Betula platyphylla: Gene organization, RNA editing, and comparative and phylogenetic analyses. BMC Genom. 2018, 19, 950. [Google Scholar] [CrossRef]
- Nawae, W.; Yundaeng, C.; Naktang, C.; Kongkachana, W.; Yoocha, T.; Sonthirod, C.; Narong, N.; Somta, P.; Laosatit, K.; Tangphatsornruang, S.; et al. The Genome and Transcriptome Analysis of the Vigna mungo Chloroplast. Plants 2020, 9, 1247. [Google Scholar] [CrossRef]
- Sander, I.M.; Chaney, J.L.; Clark, P.L. Expanding Anfinsen’s Principle: Contributions of Synonymous Codon Selection to Rational Protein Design. J. Am. Chem. Soc. 2014, 136, 858–861. [Google Scholar] [CrossRef]
- Buhr, F.; Jha, S.; Thommen, M.; Mittelstaet, J.; Kutz, F.; Schwalbe, H.; Rodnina, M.V.; Komar, A.A. Synonymous Codons Direct Cotranslational Folding toward Different Protein Conformations. Mol. Cell 2016, 61, 341–351. [Google Scholar] [CrossRef]
- Yu, C.-H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Mol. Cell 2015, 59, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Walsh, I.M.; Bowman, M.A.; Santarriaga, I.F.S.; Rodriguez, A.; Clark, P.L. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc. Natl. Acad. Sci. USA 2020, 117, 3528–3534. [Google Scholar] [CrossRef] [PubMed]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.-W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “Silent” Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science 2007, 315, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Sugita, M. RNA Editing and Its Molecular Mechanism in Plant Organelles. Genes 2017, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Funk, H.T.; Schmitz-Linneweber, C.; Poltnigg, P.; Sabater, B.; Martin, M.; Maier, R.M. Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 2005, 43, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Niavarani, A.; Currie, E.; Reyal, Y.; Afonso, F.D.A.; Horswell, S.; Griessinger, E.; Sardina, J.L.; Bonnet, D. APOBEC3A Is Implicated in a Novel Class of G-to-A mRNA Editing in WT1 Transcripts. PLoS ONE 2015, 10, e0120089. [Google Scholar] [CrossRef] [PubMed]
- Lopdell, T.J.; Hawkins, V.; Couldrey, C.; Tiplady, K.; Davis, S.R.; Harris, B.L.; Snell, R.G.; Littlejohn, M.D. Widespread cis-regulation of RNA editing in a large mammal. RNA 2019, 25, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Bentolila, S.; Hanson, M.R. The Unexpected Diversity of Plant Organelle RNA Editosomes. Trends Plant Sci. 2016, 21, 962–973. [Google Scholar] [CrossRef]
- Hayes, M.L.; Giang, K.; Berhane, B.; Mulligan, R.M. Identification of Two Pentatricopeptide Repeat Genes Required for RNA Editing and Zinc Binding by C-terminal Cytidine Deaminase-like Domains. J. Biol. Chem. 2013, 288, 36519–36529. [Google Scholar] [CrossRef]
- Oldenkott, B.; Yang, Y.; Lesch, E.; Knoop, V.; Schallenberg-Rüdinger, M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019, 2, s42003–s42019. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Hayes, M.L.; Santibanez, P.I. A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J. Biol. Chem. 2020, 295, 3497–3505. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Takenaka, S.; Barthel, T.; Frink, B.; Haag, S.; Verbitskiy, D.; Oldenkott, B.; Schallenberg-Rüdinger, M.; Feiler, C.G.; Weiss, M.S.; et al. DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis. Nat. Catal. 2021, 4, 510–522. [Google Scholar] [CrossRef]
- Cheong, C.; Tinoco, I.; Chollet, A. Thermodynamic studies of base pairing involving 2,6-diaminopurine. Nucleic Acids Res. 1988, 16, 5115–5122. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, C.; Zhang, S.; Wu, H.; Mu, W.; Wei, T.; Li, N.; Wan, T.; Liu, H.; Cui, J.; et al. The Amount of RNA Editing Sites in Liverwort Organellar Genes Is Correlated with GC Content and Nuclear PPR Protein Diversity. Genome Biol. Evol. 2019, 11, 3233–3239. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Grewe, F.; Mower, J.P. Variable Frequency of Plastid RNA Editing among Ferns and Repeated Loss of Uridine-to-Cytidine Editing from Vascular Plants. PLoS ONE 2015, 10, e0117075. [Google Scholar] [CrossRef]
- He, Z.-S.; Zhu, A.; Yang, J.-B.; Fan, W.; Li, D.-Z. Organelle Genomes and Transcriptomes of Nymphaea Reveal the Interplay between Intron Splicing and RNA Editing. Int. J. Mol. Sci. 2021, 22, 9842. [Google Scholar] [CrossRef]
- Yura, K.; Go, M. Correlation between amino acid residues converted by RNA editing and functional residues in protein three-dimensional structures in plant organelles. BMC Plant Biol. 2008, 8, 79. [Google Scholar] [CrossRef]
- Chou, P.Y.; Fasman, G.D. Prediction of the Secondary Structure of Proteins from their Amino Acid Sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978, 47, 45–148. [Google Scholar] [CrossRef]
- Covello, P.; Gray, M. On the evolution of RNA editing. Trends Genet. 1993, 9, 265–268. [Google Scholar] [CrossRef]
- Stern, D.B.; Goldschmidt-Clermont, M.; Hanson, M.R. Chloroplast RNA Metabolism. Annu. Rev. Plant Biol. 2010, 61, 125–155. [Google Scholar] [CrossRef]
- Bock, R.; Hermann, M.; Kössel, H. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 1996, 15, 5052–5059. [Google Scholar] [CrossRef] [PubMed]
- Kahlau, S.; Aspinall, S.; Gray, J.C.; Bock, R. Sequence of the Tomato Chloroplast DNA and Evolutionary Comparison of Solanaceous Plastid Genomes. J. Mol. Evol. 2006, 63, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Chateigner-Boutin, A.-L.; Small, I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007, 35, e114. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, W.; Wu, Y.; Maliga, P.; Messing, J. RNA Editing in Chloroplasts of Spirodela polyrhiza, an Aquatic Monocotelydonous Species. PLoS ONE 2015, 10, e0140285. [Google Scholar] [CrossRef]
- Wu, C.-S.; Lai, Y.-T.; Lin, C.-P.; Wang, Y.-N.; Chaw, S.-M. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: Selection toward a lower-cost strategy. Mol. Phylogenetics Evol. 2009, 52, 115–124. [Google Scholar] [CrossRef]
- Braukmann, T.W.A.; Kuzmina, M.; Stefanović, S. Loss of all plastid ndh genes in Gnetales and conifers: Extent and evolutionary significance for the seed plant phylogeny. Curr. Genet. 2009, 55, 323–337. [Google Scholar] [CrossRef]
- McCoy, S.R.; Kuehl, J.V.; Boore, J.L.; Raubeson, L.A. The complete plastid genome sequence of Welwitschia mirabilis: An unusually compact plastome with accelerated divergence rates. BMC Evol. Biol. 2008, 8, 130. [Google Scholar] [CrossRef]
- Wakasugi, T.; Tsudzuki, J.; Ito, S.; Nakashima, K.; Tsudzuki, T.; Sugiura, M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 1994, 91, 9794–9798. [Google Scholar] [CrossRef]
- Smith, D.R. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol. Biol. 2009, 71, 627–639. [Google Scholar] [CrossRef]
- Jobson, R.W.; Qiu, Y.-L. Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol. Direct 2008, 3, 43. [Google Scholar] [CrossRef] [Green Version]
- Malek, O.; Lättig, K.; Hiesel, R.; Brennicke, A.; Knoop, V. RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J. 1996, 15, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Chaw, S.-M.; Wu, C.-S.; Sudianto, E. Evolution of Gymnosperm Plastid Genomes. Adv. Bot. Res. 2018, 85, 195–222. [Google Scholar] [CrossRef]
- Sloan, D.B.; MacQueen, A.H.; Alverson, A.J.; Palmer, J.D.; Taylor, D.R. Extensive Loss of RNA Editing Sites in Rapidly Evolving Silene Mitochondrial Genomes: Selection vs. Retroprocessing as the Driving Force. Genetics 2010, 185, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Cuenca, A.; Petersen, G.; Seberg, O.; Davis, J.I.; Stevenson, D.W. Are substitution rates and RNA editing correlated? BMC Evol. Biol. 2010, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- Rüdinger, M.; Volkmar, U.; Lenz, H.; Groth-Malonek, M.; Knoop, V. Nuclear DYW-Type PPR Gene Families Diversify with Increasing RNA Editing Frequencies in Liverwort and Moss Mitochondria. J. Mol. Evol. 2012, 74, 37–51. [Google Scholar] [CrossRef]
- Salone, V.; Rüdinger, M.; Polsakiewicz, M.; Hoffmann, B.; Groth-Malonek, M.; Szurek, B.; Small, I.; Knoop, V.; Lurin, C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007, 581, 4132–4138. [Google Scholar] [CrossRef]
- Guillaumot, D.; Lopez-Obando, M.; Baudry, K.; Avon, A.; Rigaill, G.; de Longevialle, A.F.; Broche, B.; Takenaka, M.; Berthomé, R.; De Jaeger, G.; et al. Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc. Natl. Acad. Sci. USA 2017, 114, 8877–8882. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.-L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Takenaka, M.; Kühn, K.; Craddock, C.; Smalle, J.; Karampelias, M.; Denecke, J.; Peters, J.; et al. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. Plant J. 2012, 71, 836–849. [Google Scholar] [CrossRef]
- Kim, S.-R.; Yang, J.-I.; Moon, S.; Ryu, C.-H.; An, K.; Kim, K.-M.; Yim, J.; An, G. Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 2009, 59, 738–749. [Google Scholar] [CrossRef]
- Glass, F.; Härtel, B.; Zehrmann, A.; Verbitskiy, D.; Takenaka, M. MEF13 Requires MORF3 and MORF8 for RNA Editing at Eight Targets in Mitochondrial mRNAs in Arabidopsis thaliana. Mol. Plant 2015, 8, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Yap, A.; Kindgren, P.; Francs-Small, C.C.D.; Kazama, T.; Tanz, S.K.; Toriyama, K.; Small, I. AEF1/MPR25 is implicated in RNA editing of plastid atpFand mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 2015, 81, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Royan, S.; Schallenberg-Rüdinger, M.; Lenz, H.; Castleden, I.R.; McDowell, R.; Vacher, M.A.; Tonti-Filippini, J.; Bond, C.S.; Knoop, V.; et al. The Expansion and Diversification of Pentatricopeptide Repeat RNA-Editing Factors in Plants. Mol. Plant 2020, 13, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Kan, S.-L.; Shen, T.-T.; Ran, J.-H.; Wang, X.-Q. Both Conifer II and Gnetales are characterized by a high frequency of ancient mitochondrial gene transfer to the nuclear genome. BMC Biol. 2021, 19, 146. [Google Scholar] [CrossRef] [PubMed]
- Kan, S.-L.; Shen, T.-T.; Gong, P.; Ran, J.-H.; Wang, X.-Q. The complete mitochondrial genome of Taxus cuspidata (Taxaceae): Eight protein-coding genes have transferred to the nuclear genome. BMC Evol. Biol. 2020, 20, 10. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.-J.; Moore, M.J.; Li, D.-Z.; Yi, T.-S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lian, J.; Li, Q.; Zhang, P.; Zhou, Y.; Zhan, X.; Zhang, G. RES-Scanner: A software package for genome-wide identification of RNA-editing sites. GigaScience 2016, 5, 37. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Cohen, O.; Ashkenazy, H.; Belinky, F.; Huchon, D.; Pupko, T. GLOOME: Gain loss mapping engine. Bioinformatics 2010, 26, 2914–2915. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0 Contributors. SciPy 1.0 Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Smith, D.R. Unparalleled variation in RNA editing among Selaginella plastomes. Plant Physiol. 2020, 182, 12–14. [Google Scholar] [CrossRef]
- Ren, T.; Li, Z.-X.; Xie, D.-F.; Gui, L.-J.; Peng, C.; Wen, J.; He, X.-J. Plastomes of eight Ligusticum species: Characterization, genome evolution, and phylogenetic relationships. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Xu, Y.; Wang, L.; Liu, J.; Yu, J.; Chen, H. High level of intraspecific divergence and low frequency of RNA editing in the chloroplast genome sequence of Tagetes erecta. Mitochondrial DNA B Resour. 2020, 5, 2948–2953. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, A.; Furihata, H.Y.; Tsujino, Y.; Kawanabe, T.; Fujii, S.; Yoshida, T. Divergence of RNA editing among Arabidopsis species. Plant Sci. 2019, 280, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Wang, W.; Huang, Q.; Fan, R.; Wang, X.; Feng, P.; Zhao, G.; Bian, S.; Ren, H.; Chang, Y. Complete chloroplast genome sequence of Dryopteris fragrans (L.) Schott and the repeat structures against the thermal environment. Sci. Rep. 2018, 8, 16635. [Google Scholar] [CrossRef]
- De Santana Lopes, A.; Pacheco, T.G.; Santos, K.G.d.; Vieira, L.d.N.; Guerra, M.P.; Nodari, R.O.; de Souza, E.M.; de Oliveira Pedrosa, F.; Rogalski, M. The Linum usitatissimum L. plastome reveals atypical structural evolution, new editing sites, and the phylogenetic position of Linaceae within Malpighiales. Plant Cell Rep. 2018, 37, 307–328. [Google Scholar] [CrossRef]
- Wang, W.; Yu, H.; Wang, J.; Lei, W.; Gao, J.; Qiu, X.; Wang, J. The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa (Oleaceae). Int. J. Mol. Sci. 2017, 18, 2288. [Google Scholar] [CrossRef]
- Labiak, P.H.; Karol, K.G. Plastome sequences of an ancient fern lineage reveal remarkable changes in gene content and architecture. Am. J. Bot. 2017, 104, 1008–1018. [Google Scholar] [CrossRef]
- Wang, M.; Liu, H.; Ge, L.; Xing, G.; Wang, M.; Weining, S.; Nie, X. Identification and analysis of RNA editing sites in the chloroplast transcripts of Aegilops tauschii L. Genes 2016, 8, 13. [Google Scholar] [CrossRef]
- Yan, L.; Lai, X.; Li, X.; Wei, C.; Tan, X.; Zhang, Y. Analyses of the complete genome and gene expression of chloroplast of sweet potato [Ipomoea batata]. PLoS ONE 2015, 10, e0124083. [Google Scholar] [CrossRef]
- Lin, C.-P.; Ko, C.-Y.; Kuo, C.-I.; Liu, M.-S.; Schafleitner, R.; Chen, L.-F.O. Transcriptional slippage and RNA editing increase the diversity of transcripts in chloroplasts: Insight from deep sequencing of Vigna radiata genome and transcriptome. PLoS ONE 2015, 10, e0129396. [Google Scholar] [CrossRef]
- Weng, M.-L.; Blazier, J.C.; Govindu, M.; Jansen, R.K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 2014, 31, 645–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Hou, B.-W.; Niu, Z.-T.; Liu, W.; Xue, Q.-Y.; Ding, X.-Y. Comparative chloroplast genomes of photosynthetic orchids: Insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS ONE 2014, 9, e99016. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kang, Y.; Shin, S.C.; Park, H.; Lee, H. Combined analysis of the chloroplast genome and transcriptome of the Antarctic vascular plant Deschampsia antarctica Desv. PLoS ONE 2014, 9, e92501. [Google Scholar] [CrossRef] [PubMed]
- Ruwe, H.; Castandet, B.; Schmitz-Linneweber, C.; Stern, D.B. Arabidopsis chloroplast quantitative editotype. FEBS Lett. 2013, 587, 1429–1433. [Google Scholar] [CrossRef]
- Huang, Y.-Y.; Matzke, A.J.; Matzke, M. Complete sequence and comparative analysis of the chloroplast genome of coconut palm (Cocos nucifera). PLoS ONE 2013, 8, e74736. [Google Scholar] [CrossRef]
- Uthaipaisanwong, P.; Chanprasert, J.; Shearman, J.; Sangsrakru, D.; Yoocha, T.; Jomchai, N.; Jantasuriyarat, C.; Tragoonrung, S.; Tangphatsornruang, S. Characterization of the chloroplast genome sequence of oil palm (Elaeis guineensis Jacq.). Gene 2012, 500, 172–180. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Wu, M.; Palmer, J.D.; Taylor, D.R. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol. Evol. 2012, 4, 294–306. [Google Scholar] [CrossRef]
- Kazakoff, S.H.; Imelfort, M.; Edwards, D.; Koehorst, J.; Biswas, B.; Batley, J.; Scott, P.T.; Gresshoff, P.M. Capturing the biofuel wellhead and powerhouse: The chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. PLoS ONE 2012, 7, e51687. [Google Scholar] [CrossRef]
- Jiang, Y.; Fan, S.; Song, M.; Yu, J.; Yu, S. Identification of RNA editing sites in cotton (Gossypium hirsutum) chloroplasts and editing events that affect secondary and three-dimensional protein structures. Genet. Mol. Res. 2012, 11, 987–1001. [Google Scholar] [CrossRef]
- Jheng, C.-F.; Chen, T.-C.; Lin, J.-Y.; Chen, T.-C.; Wu, W.-L.; Chang, C.-C. The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis orchids. Plant Sci. 2012, 190, 62–73. [Google Scholar] [CrossRef]
- Grosche, C.; Funk, H.T.; Maier, U.G.; Zauner, S. The chloroplast genome of Pellia endiviifolia: Gene content, RNA-editing pattern, and the origin of chloroplast editing. Genome Biol. Evol. 2012, 4, 1349–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, H.A.; Lanzatella, C.L.; Sarath, G.; Tobias, C.M. Chloroplast genome variation in upland and lowland switchgrass. PLoS ONE 2011, 6, e23980. [Google Scholar] [CrossRef]
- Tangphatsornruang, S.; Uthaipaisanwong, P.; Sangsrakru, D.; Chanprasert, J.; Yoocha, T.; Jomchai, N.; Tragoonrung, S. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene 2011, 475, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Guzowska-Nowowiejska, M.; Fiedorowicz, E.; Pląder, W. Cucumber, melon, pumpkin, and squash: Are rules of editing in flowering plants chloroplast genes so well known indeed? Gene 2009, 434, 1–8. [Google Scholar] [CrossRef]
- Yura, K.; Miyata, Y.; Arikawa, T.; Higuchi, M.; Sugita, M. Characteristics and prediction of RNA editing sites in transcripts of the moss Takakia lepidozioides chloroplast. DNA Res. 2008, 15, 309–321. [Google Scholar] [CrossRef]
- Zeng, W.-H.; Liao, S.-C.; Chang, C.-C. Identification of RNA editing sites in chloroplast transcripts of Phalaenopsis aphrodite and comparative analysis with those of other seed plants. Plant Cell Physiol. 2007, 48, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Miyata, Y.; Sugita, M. Tissue- and stage-specific RNA editing of rps14 transcripts in moss (Physcomitrella patens) chloroplasts. J. Plant Physiol. 2004, 161, 113–115. [Google Scholar] [CrossRef] [PubMed]
- Inada, M.; Sasaki, T.; Yukawa, M.; Tsudzuki, T.; Sugiura, M. A systematic search for RNA editing sites in pea chloroplasts: An editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol. 2004, 45, 1615–1622. [Google Scholar] [CrossRef]
- Calsa Júnior, T.; Carraro, D.M.; Benatti, M.R.; Barbosa, A.C.; Kitajima, J.P.; Carrer, H. Structural features and transcript-editing analysis of sugarcane (Saccharum officinarum L.) chloroplast genome. Curr. Genet. 2004, 46, 366–373. [Google Scholar] [CrossRef]
- Sasaki, T.; Yukawa, Y.; Miyamoto, T.; Obokata, J.; Sugiura, M. Identification of RNA editing sites in chloroplast transcripts from the maternal and paternal progenitors of tobacco (Nicotiana tabacum): Comparative analysis shows the involvement of distinct trans-factors for ndhB editing. Mol. Biol. Evol. 2003, 20, 1028–1035. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Regel, R.; Du, T.G.; Hupfer, H.; Herrmann, R.G.; Maier, R.M. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: The role of RNA editing in generating divergence in the process of plant speciation. Mol. Biol. Evol. 2002, 19, 1602–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsudzuki, T.; Wakasugi, T.; Sugiura, M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 2001, 53, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Corneille, S.; Lutz, K.; Maliga, P. Conservation of RNA editing between rice and maize plastids: Are most editing events dispensable? Mol. Gen. Genet. 2000, 264, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Kusumegi, T.; Tsudzuki, T.; Sugiura, M. RNA editing sites in tobacco chloroplast transcripts: Editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet. 1999, 262, 462–467. [Google Scholar] [CrossRef] [PubMed]



| Species | Editing Type | Number of Editing Sites | RNA Editing Sites in Coding Regions | RNA Editing Sites in Non-Coding Regions | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st Codon | 2nd Codon | 3rd Codon | Silent Editing | Non-Synonymous Editing | tRNA | rRNA | Intron | Intergenic Region | |||
| Cycas revoluta | C-to-U | 152 | 23 | 89 | 11 | 13 | 110 | 0 | 0 | 7 | 22 |
| G-to-A | 6 | 1 | 1 | 0 | 0 | 2 | 0 | 2 | 1 | 1 | |
| Zamia furfuracea | C-to-U | 265 | 36 | 111 | 42 | 41 | 148 | 0 | 0 | 20 | 56 |
| G-to-A | 10 | 3 | 3 | 0 | 0 | 6 | 0 | 2 | 1 | 1 | |
| Ginkgo biloba | C-to-U | 304 | 53 | 157 | 27 | 27 | 210 | 0 | 0 | 23 | 44 |
| G-to-A | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | |
| Cedrus deodara | C-to-U | 95 | 15 | 67 | 3 | 3 | 82 | 0 | 0 | 9 | 1 |
| G-to-A | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Abies firma | C-to-U | 77 | 10 | 51 | 5 | 5 | 61 | 0 | 1 | 8 | 2 |
| G-to-A | 4 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | |
| Picea smithiana | C-to-U | 161 | 23 | 84 | 12 | 12 | 107 | 0 | 1 | 14 | 27 |
| G-to-A | 4 | 1 | 0 | 2 | 2 | 1 | 0 | 1 | 0 | 0 | |
| Pinus armandii | C-to-U | 49 | 7 | 28 | 5 | 6 | 34 | 0 | 0 | 4 | 5 |
| G-to-A | 5 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | |
| Ephedra przewalskii | C-to-U | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| G-to-A | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | |
| Welwitschia mirabilis | C-to-U | 2 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| G-to-A | 4 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | |
| Gnetum montanum | C-to-U | 3 | 0 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| G-to-A | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | |
| Podocarpus macrophyllus | C-to-U | 20 | 2 | 17 | 0 | 0 | 19 | 0 | 0 | 0 | 1 |
| G-to-A | 4 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 2 | |
| Araucaria cunninghamii | C-to-U | 45 | 4 | 32 | 3 | 5 | 34 | 0 | 0 | 1 | 5 |
| G-to-A | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Sciadopitys verticillata | C-to-U | 41 | 7 | 20 | 5 | 5 | 27 | 0 | 0 | 4 | 5 |
| G-to-A | 5 | 2 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 2 | |
| Cephalotaxus sinensis | C-to-U | 40 | 6 | 25 | 1 | 1 | 31 | 0 | 1 | 3 | 4 |
| G-to-A | 6 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 4 | |
| Taxus cuspidata | C-to-U | 25 | 1 | 19 | 2 | 2 | 20 | 0 | 0 | 0 | 3 |
| G-to-A | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | |
| Cunninghamia lanceolata | C-to-U | 18 | 3 | 14 | 0 | 0 | 17 | 0 | 0 | 1 | 0 |
| G-to-A | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | |
| Taiwania cryptomerioides | C-to-U | 18 | 2 | 14 | 1 | 1 | 16 | 0 | 0 | 0 | 1 |
| G-to-A | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | |
| Metasequoia glyptostroboides | C-to-U | 24 | 4 | 14 | 3 | 4 | 17 | 0 | 0 | 0 | 3 |
| G-to-A | 4 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | |
| Platycladus orientalis | C-to-U | 24 | 5 | 13 | 0 | 1 | 17 | 0 | 0 | 1 | 5 |
| G-to-A | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | |
| Total | C-to-U | 1364 | 202 | 759 | 121 | 127 | 955 | 0 | 3 | 95 | 184 |
| G-to-A | 71 | 9 | 6 | 8 | 7 | 16 | 1 | 21 | 7 | 19 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, K.-Y.; Kan, S.-L.; Shen, T.-T.; Gong, P.; Feng, Y.-Y.; Du, H.; Zhao, Y.-P.; Wan, T.; Wang, X.-Q.; Ran, J.-H. A Comprehensive Evolutionary Study of Chloroplast RNA Editing in Gymnosperms: A Novel Type of G-to-A RNA Editing Is Common in Gymnosperms. Int. J. Mol. Sci. 2022, 23, 10844. https://doi.org/10.3390/ijms231810844
Huang K-Y, Kan S-L, Shen T-T, Gong P, Feng Y-Y, Du H, Zhao Y-P, Wan T, Wang X-Q, Ran J-H. A Comprehensive Evolutionary Study of Chloroplast RNA Editing in Gymnosperms: A Novel Type of G-to-A RNA Editing Is Common in Gymnosperms. International Journal of Molecular Sciences. 2022; 23(18):10844. https://doi.org/10.3390/ijms231810844
Chicago/Turabian StyleHuang, Kai-Yuan, Sheng-Long Kan, Ting-Ting Shen, Pin Gong, Yuan-Yuan Feng, Hong Du, Yun-Peng Zhao, Tao Wan, Xiao-Quan Wang, and Jin-Hua Ran. 2022. "A Comprehensive Evolutionary Study of Chloroplast RNA Editing in Gymnosperms: A Novel Type of G-to-A RNA Editing Is Common in Gymnosperms" International Journal of Molecular Sciences 23, no. 18: 10844. https://doi.org/10.3390/ijms231810844
APA StyleHuang, K.-Y., Kan, S.-L., Shen, T.-T., Gong, P., Feng, Y.-Y., Du, H., Zhao, Y.-P., Wan, T., Wang, X.-Q., & Ran, J.-H. (2022). A Comprehensive Evolutionary Study of Chloroplast RNA Editing in Gymnosperms: A Novel Type of G-to-A RNA Editing Is Common in Gymnosperms. International Journal of Molecular Sciences, 23(18), 10844. https://doi.org/10.3390/ijms231810844





