A Novel L-Phenylalanine Dipeptide Inhibits the Growth and Metastasis of Prostate Cancer Cells via Targeting DUSP1 and TNFSF9
Abstract
1. Introduction
2. Results
2.1. Evaluation of the Toxic Effects of HXL131 on Human Regular Hepatic Cell Line LO2
2.2. Effect of HXL131 on the Growth of PC3 Cells
2.2.1. Effect of Different Concentrations of HXL131 on the Proliferation of PC3 Cells
2.2.2. Effect of Different Concentrations of HXL131 on Apoptosis of PC3 Cells
2.2.3. Effect of Different Concentrations of HXL131 on the Cycle of PC3 Cells
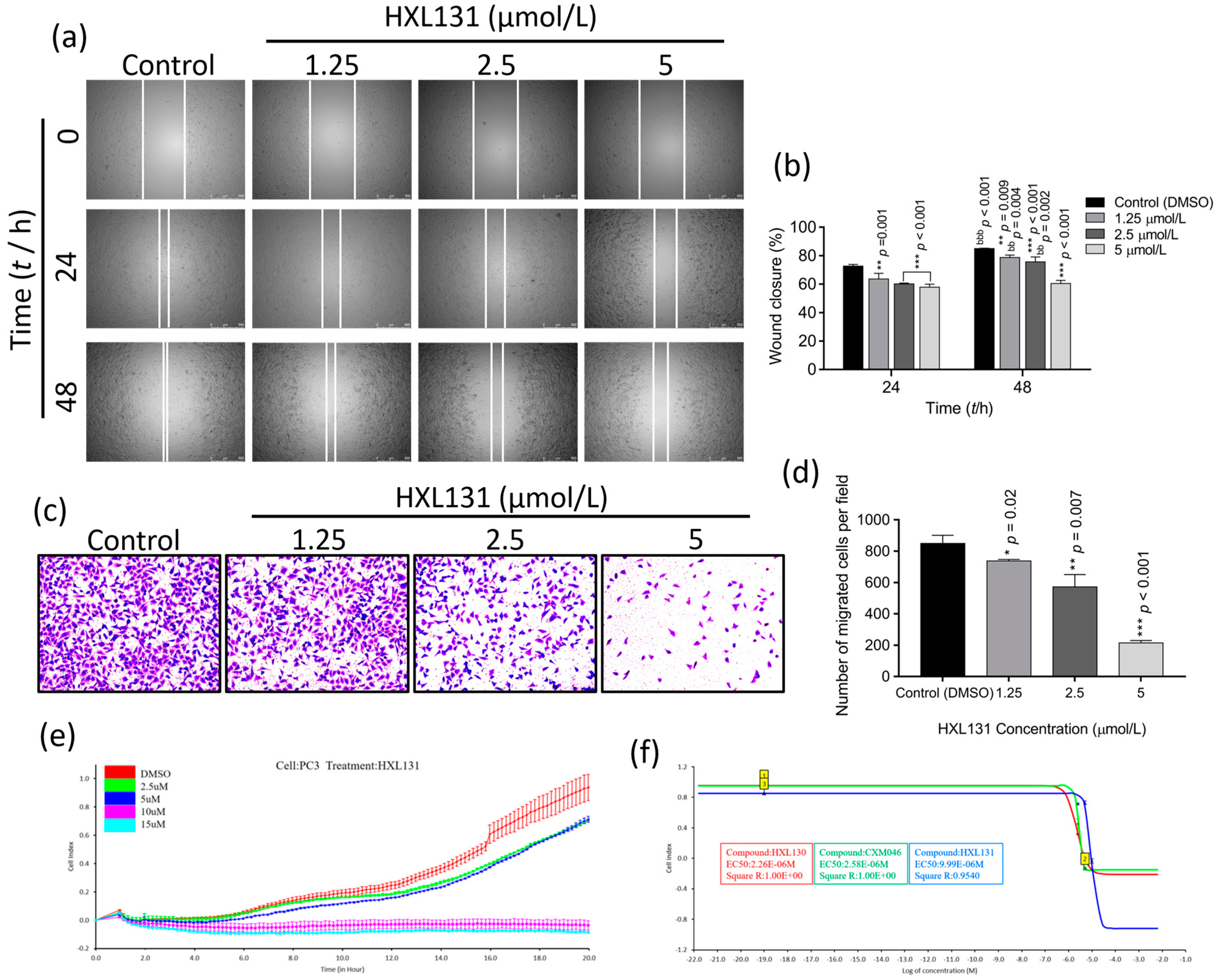
2.3. Effect of HXL131 on PC3 Cell Metastasis
2.3.1. Effect of Different Concentrations of HXL131 on the Wound-Healing Ability of PC3 Cells
2.3.2. Effect of Different Concentrations of HXL131 on the Migration of PC3 Cells
2.3.3. Effect of Different Concentrations of HXL131 on Real-Time Migration of PC3 Cells
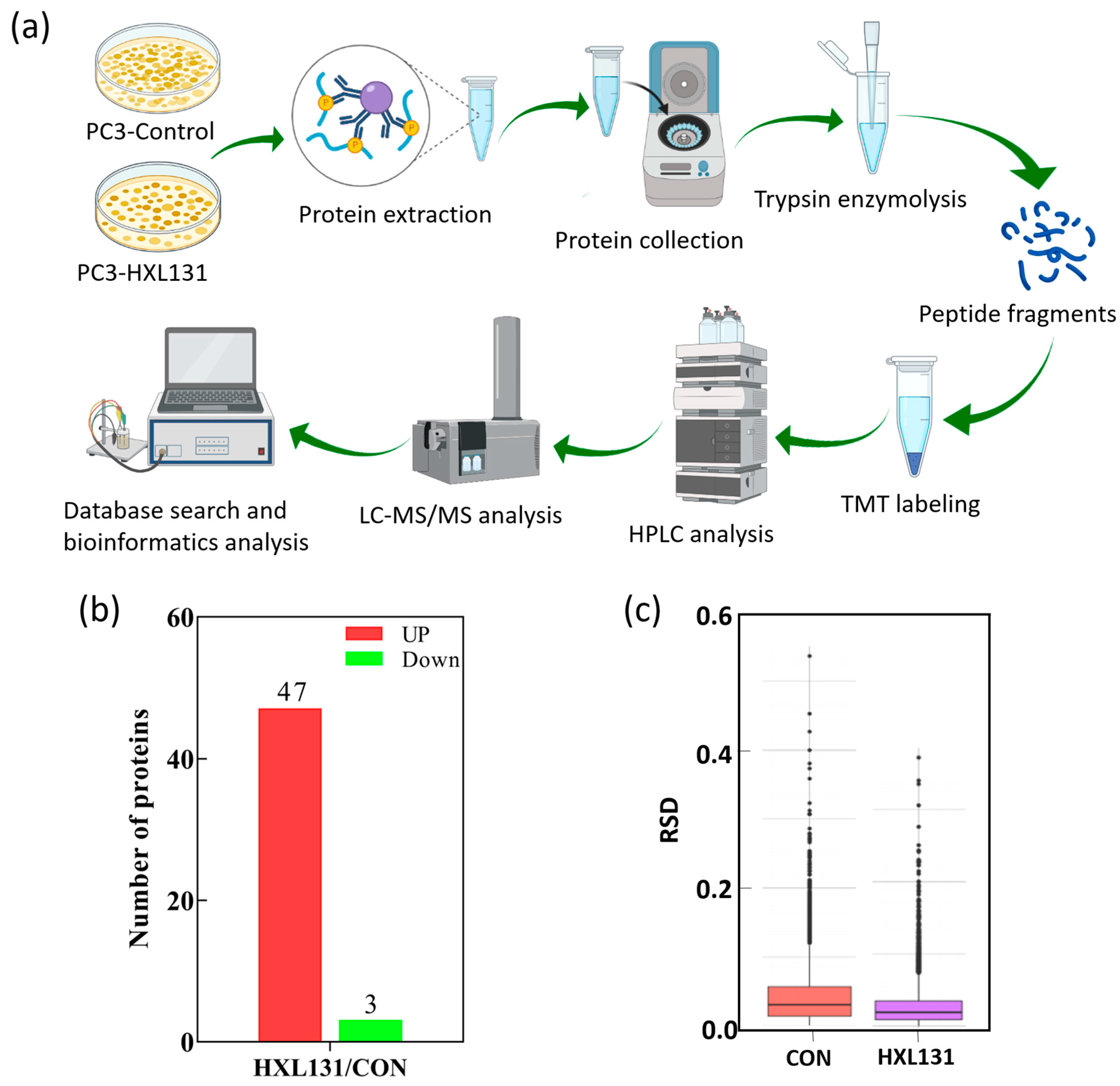
2.4. Effect of HXL131 on Protein Expression of PC3 Cells Was Analyzed by TMT Quantitative Proteomics
2.4.1. Expression of Differential Proteins in the HXL131 Treatment Group
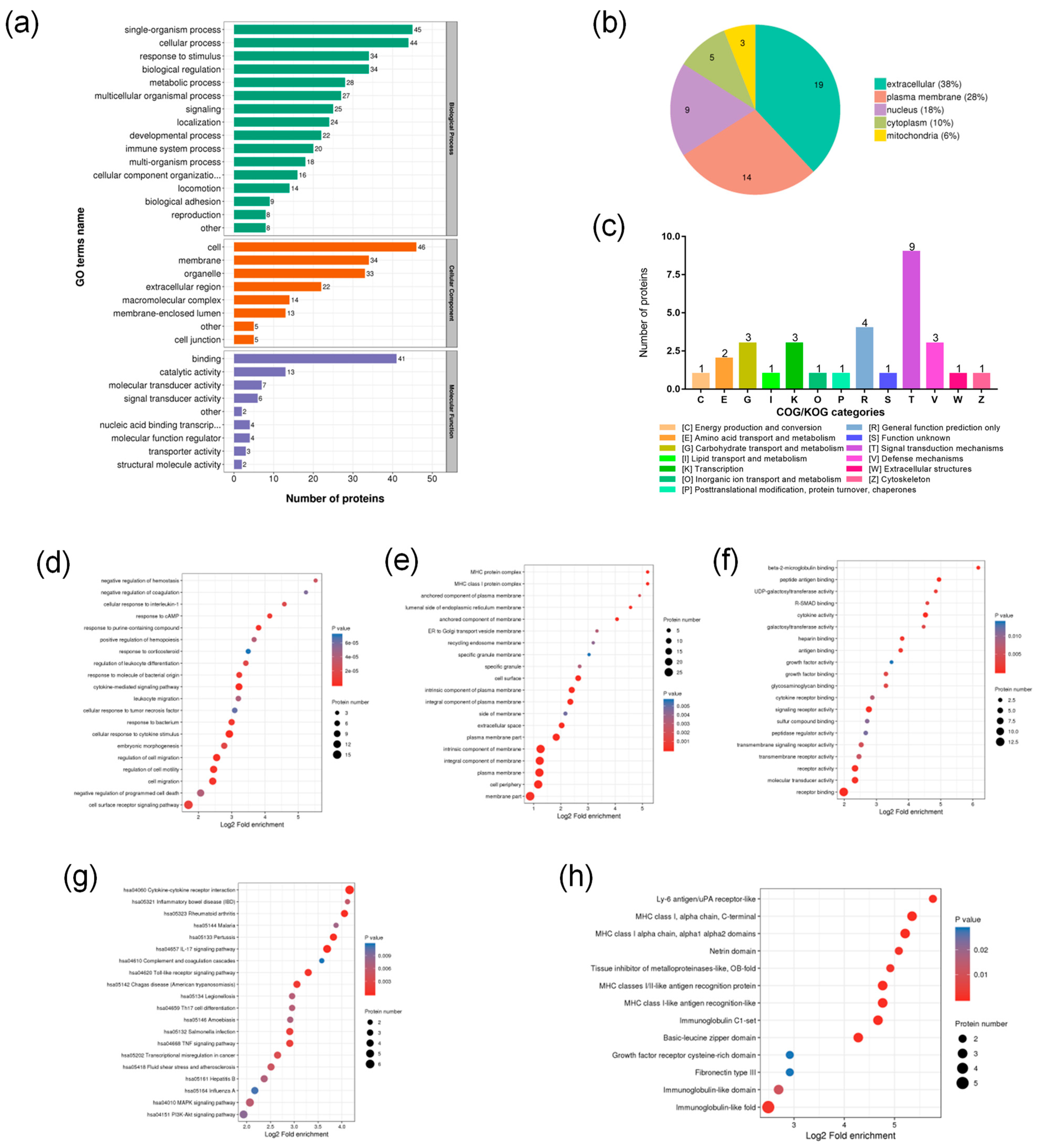
2.4.2. Functional Classification of DEPs
2.4.3. Functional Enrichment Analysis of DEPs
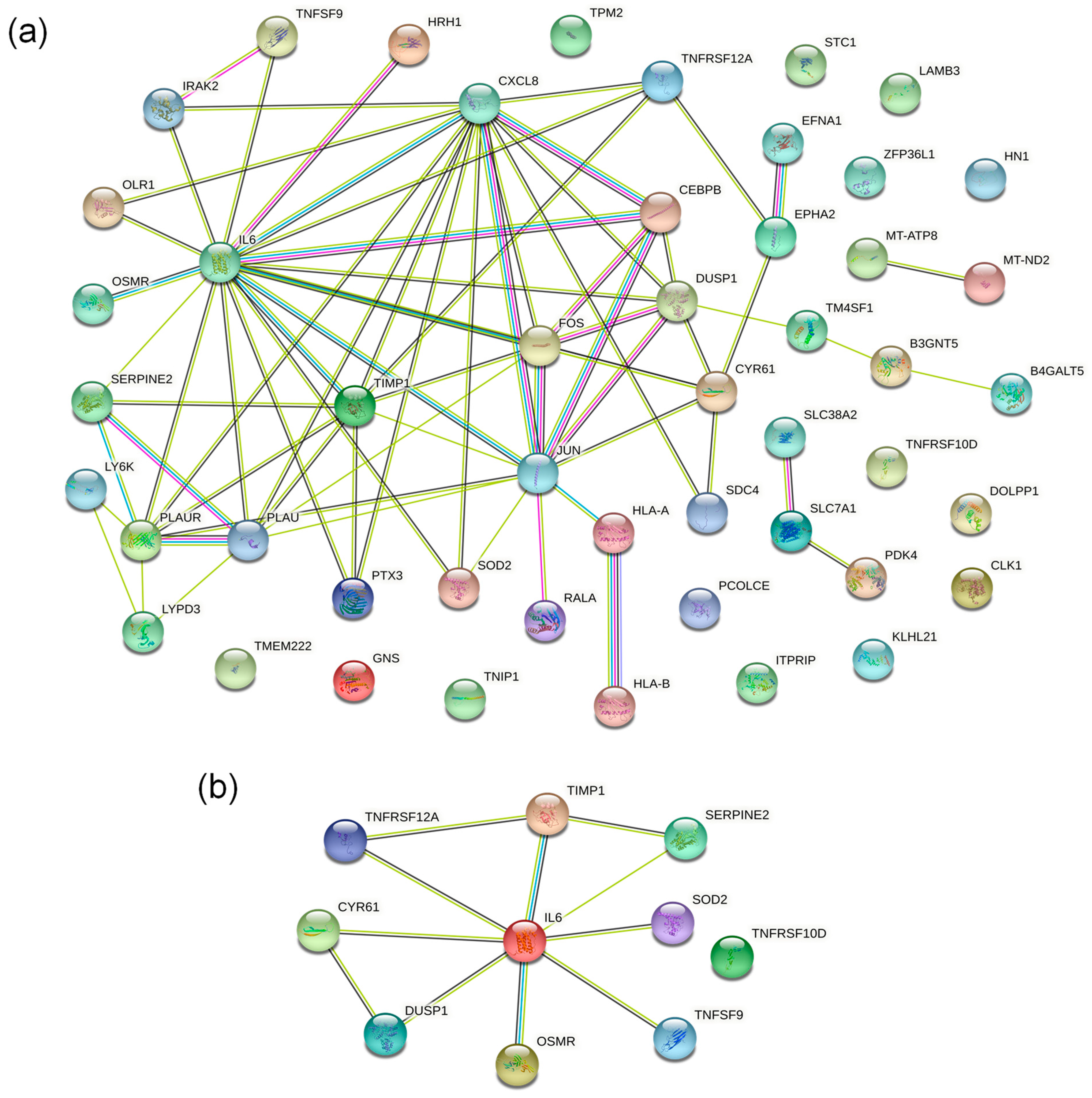
2.4.4. PPI Network Analysis of DEPs
2.5. Screening of Essential Proteins Regulated by HXL131
2.6. Validation of 10 Key DEPs
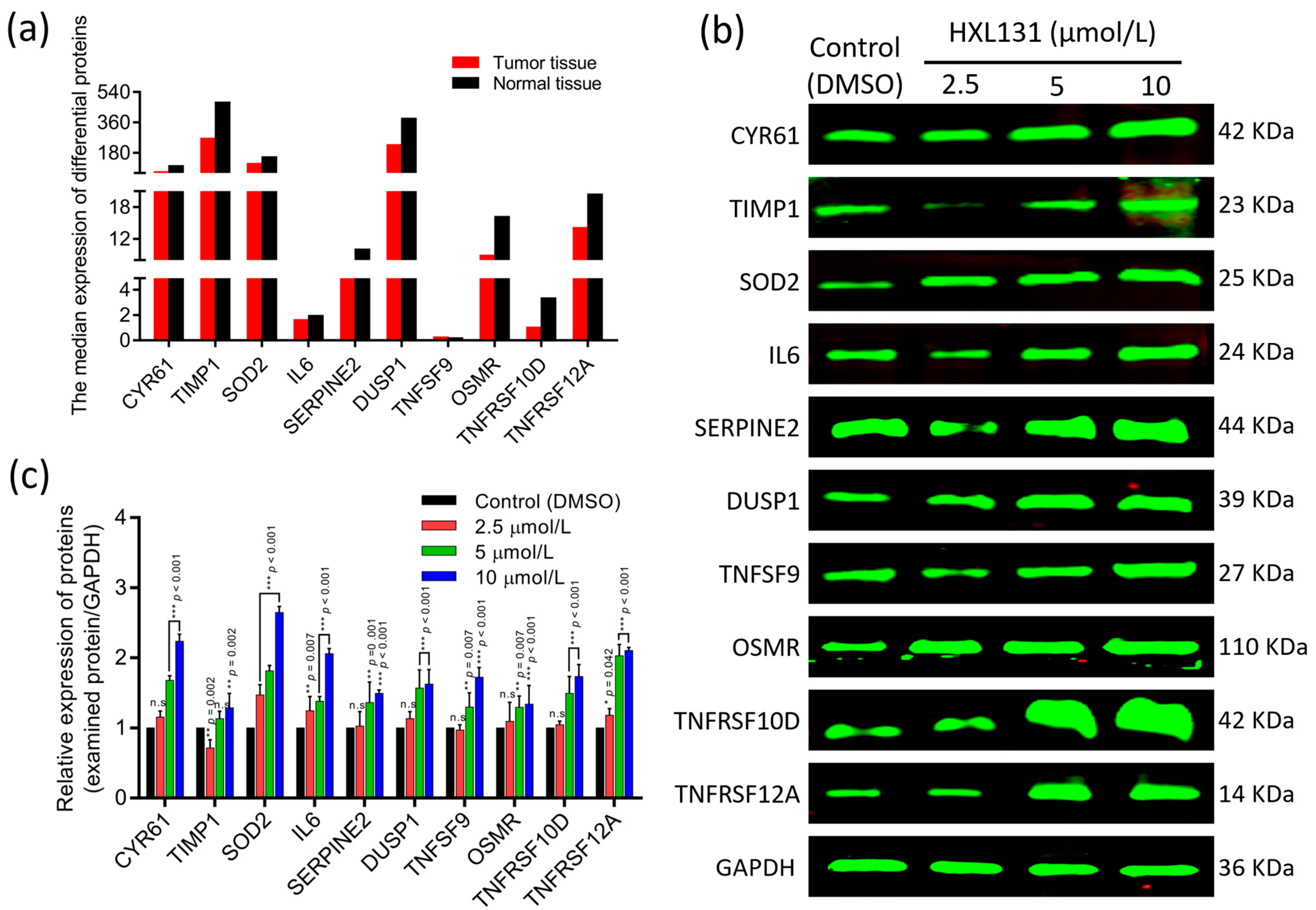
2.6.1. The GEPIA Database Verifies the Expression of 10 Key DEPs
2.6.2. Western Blot Verified the Expression of 10 Key DEPs
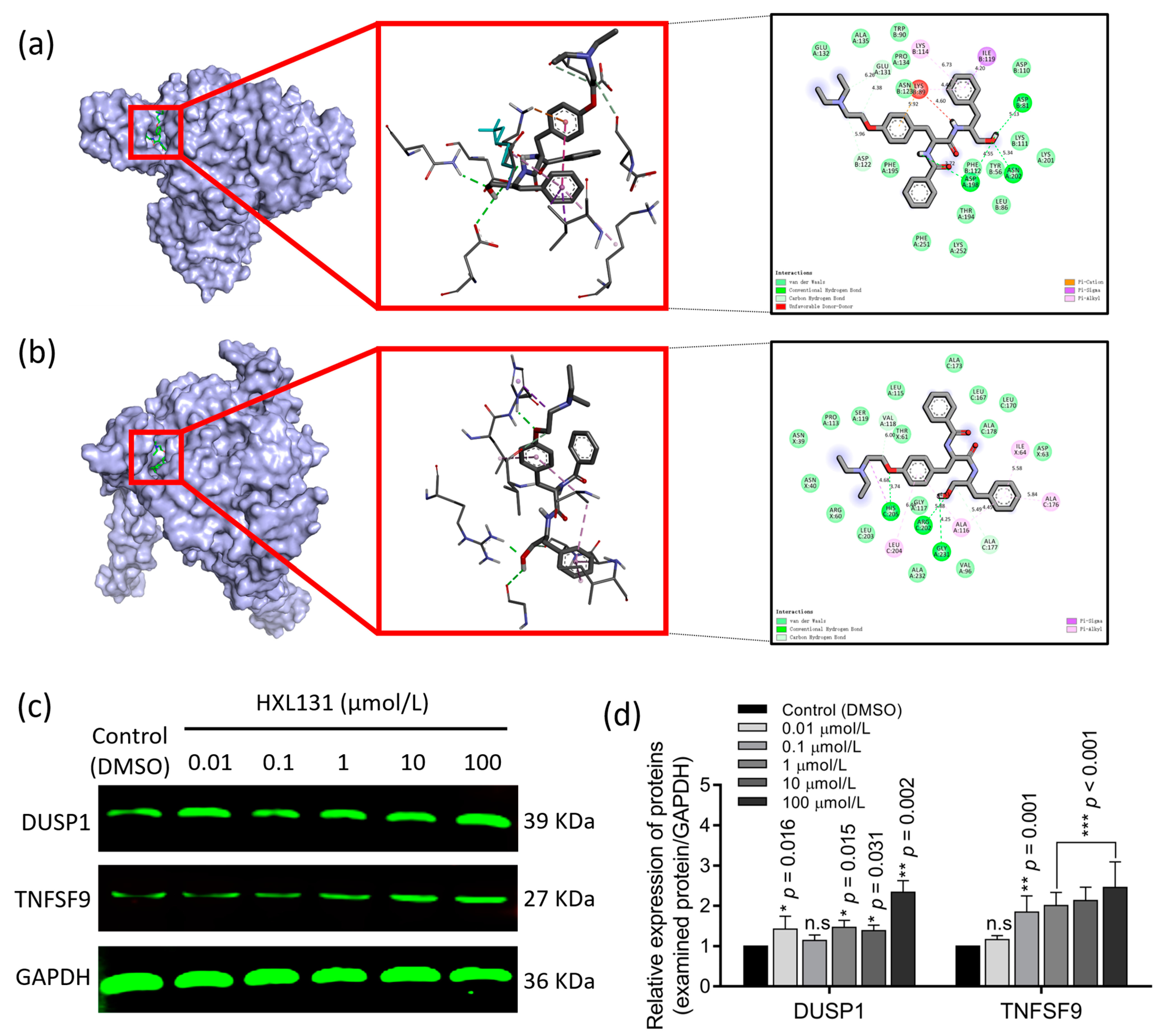
2.6.3. Molecular Docking Verified the Targeted Binding between the Target Protein and Small-Molecule Compound HXL131
2.6.4. CETSA Verified the Targeted Binding of HXL131 to DUSP1 and TNFSF9 in Molecular Docking
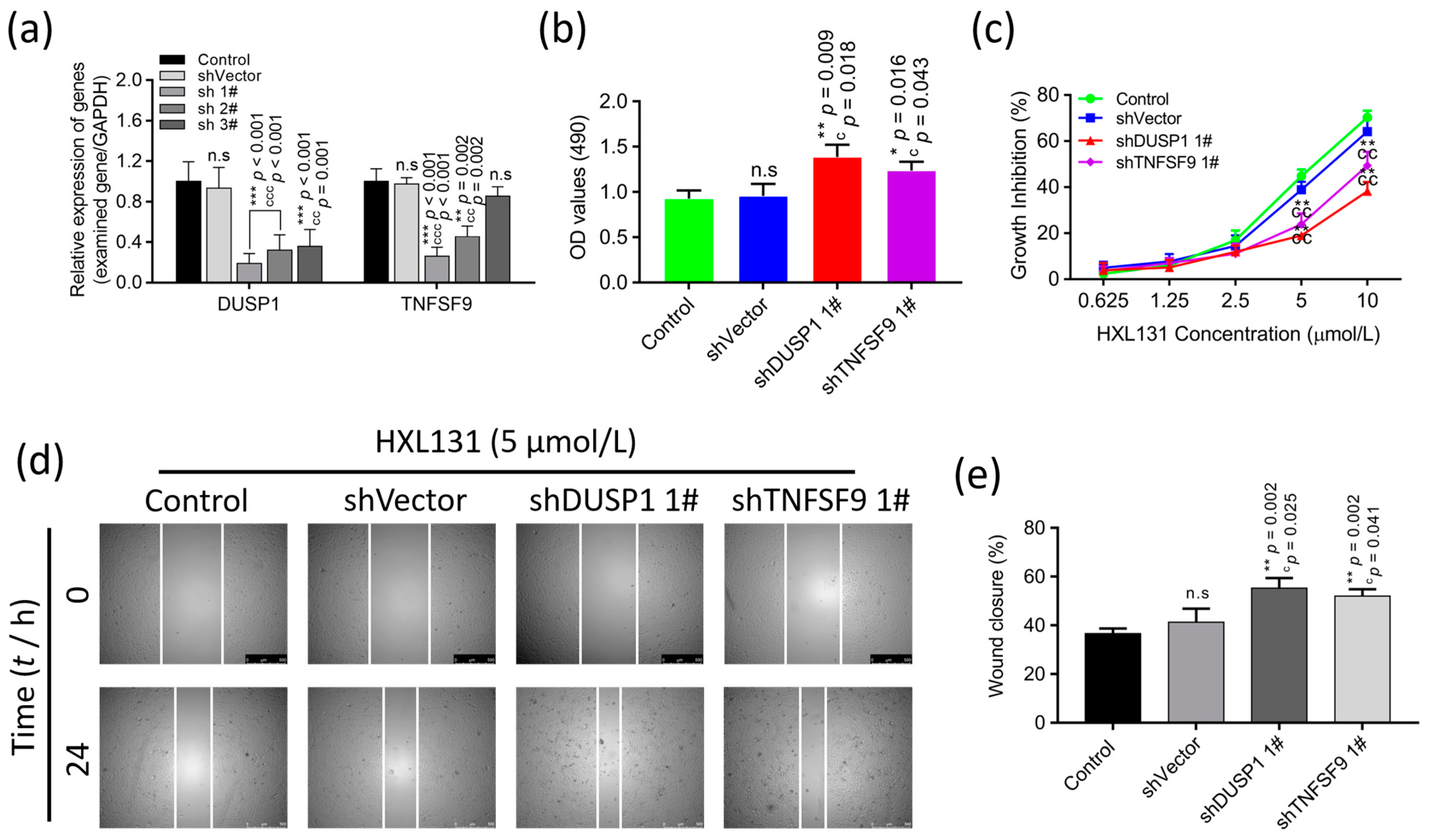
2.6.5. Gene Silencing Validated the Effect of Interfering with the Expression of DUSP1 and TNFSF9 on PC3 Cell Growth and Migration
3. Discussion
4. Materials and Methods
4.1. Chemicals, Reagents, and Instruments
4.2. Antibodies
4.3. Cell Culture and Compound Treatment
4.4. Cell Growth Assay
4.4.1. Cell Proliferation Assay
4.4.2. Cell Apoptosis Assay
4.4.3. Cell Cycle Assay
4.5. Cell Migration Assay
4.5.1. Wound-Healing Assay
4.5.2. Trans-Well Chamber Migration Assay
4.5.3. Real-Time Quantitative Cell Migration Assay
4.6. Proteomic Bioinformatics Analysis of TMT Labeling
4.7. DEP Validation Assay
4.7.1. GEPIA Assay
4.7.2. Western Blot Assay
4.7.3. Molecular Docking Assay
4.7.4. CETSA Assay
4.7.5. Gene Silencing Assay
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Song, Q.; Li, J.; Du, K.; Chen, Y.; Zou, C.; Mo, Z. Carcinogenic effect of adenylosuccinate lyase (ADSL) in prostate cancer development and progression through the cell cycle pathway. Cancer Cell Int. 2021, 21, 467. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Luo, L.; Xiang, Q.; Wang, J.; Liu, Y.; Deng, Y.; Zhao, Z. MiRNA-671-5p Promotes prostate cancer development and metastasis by targeting NFIA/CRYAB axis. Cell Death Dis. 2020, 11, 949. [Google Scholar] [CrossRef] [PubMed]
- Raspantini, G.L.; Luiz, M.T.; Abriata, J.P.; de Oliveira Eloy, J.; Vaidergorn, M.M.; da Silva Emery, F.; Marchetti, J.M. PCL-TPGS polymeric nanoparticles for docetaxel delivery to prostate cancer: Development, physicochemical and biological characterization. Colloids Surfaces A Physicochem. Eng. Asp. 2021, 627, 127144. [Google Scholar] [CrossRef]
- Yap, T.A.; Smith, A.D.; Ferraldeschi, R.; Al-Lazikani, B.; Workman, P.; de Bono, J.S. Drug discovery in advanced prostate cancer: Translating biology into therapy. Nat. Rev. Drug Discov. 2016, 15, 699–718. [Google Scholar] [CrossRef]
- Barlow, S.K.; Oyekunle, T.; Janes, J.L.; De Hoedt, A.M.; Aronson, W.J.; Kane, C.J.; Amling, C.L.; Cooperberg, M.R.; Klaassen, Z.W.; Terris, M.K.; et al. Prostate weight and prostate cancer outcomes after radical prostatectomy: Results from the SEARCH cohort study. Prostate 2022, 82, 366–372. [Google Scholar] [CrossRef]
- Cellini, F.; Tagliaferri, L.; Frascino, V.; Alitto, A.R.; Fionda, B.; Boldrini, L.; Romano, A.; Casà, C.; Catucci, F.; Mattiucci, G.C.; et al. Radiation therapy for prostate cancer: What’s the best in 2021. Urologia 2022, 89, 5–15. [Google Scholar] [CrossRef]
- Kamran, S.C.; D’Amico, A.V. Radiation Therapy for Prostate Cancer. Hematol. Oncol. Clin. N. Am. 2020, 34, 45–69. [Google Scholar] [CrossRef]
- Lopez, W.; Nguyen, N.; Cao, J.; Eddow, C.; Shung, K.K.; Lee, N.S.; Chow, M. Ultrasound Therapy, Chemotherapy and Their Combination for Prostate Cancer. Technol. Cancer Res. Treat. 2021, 20, 15330338211011965. [Google Scholar] [CrossRef] [PubMed]
- Schröder, F.H. Early versus delayed endocrine therapy for prostate cancer. Endocr. Relat. Cancer 2007, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Wang, Y.; Dong, Z.; Lai, C.; Chang, B.; Gong, Q.; Ren, S.; Sun, D.; Lu, J.; Gao, Y. Dichondra repens J.R.Forst. and G.Forst.: A Review of Its Traditional Uses, Chemistry, Pharmacology, Toxicology and Applications. Front. Pharmacol. 2020, 11, 608199. [Google Scholar] [CrossRef]
- Liu, X.; Xue, L.; Zhang, H.; Xu, Q.; Zhang, S.; Ma, S.; Ding, X.; Liu, L.; Dong, J.; Qian, L.; et al. Phase I, First-in-Human, Single and Multiple Ascending Dose- and Food-Effect Studies to Assess the Safety, Tolerability and Pharmacokinetics of a Novel Anti-hepatitis B Virus Drug, Bentysrepinine (Y101), in Healthy Chinese Subjects. Clin. Drug Investig. 2020, 40, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ma, C.; Liu, Z.; Kang, W. Natural Products: Review for Their Effects of Anti-HBV. BioMed Res. Int. 2020, 2020, 3972390. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Huang, Z.; Liu, C.; Cai, Z.; Pan, W.; Cao, P.; Hao, X.; Liang, G. Synthesis and anti-hepatitis B virus activities of Matijing-Su derivatives. Bioorganic Med. Chem. 2009, 17, 3118–3125. [Google Scholar] [CrossRef] [PubMed]
- Cui, J. Synthesis and anti-HBV activity evaluation of Matijin-Su derivatives containing trifluoromethyl. Chin. Pharmacol. J. 2019, 24, 1045–1053. [Google Scholar]
- Guo, D.; Zhang, Y.; Zhao, J.; He, H.; Hou, T. Selenium-biofortified corn peptides: Attenuating concanavalin A-Induced liver injury and structure characterization. J. Trace Elem. Med. Biol. 2019, 51, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Wang, N.; Pan, W.; Qiu, J.; Cao, P.; Zhu, M.; Feng, Y.; Liang, G. Synthesis and anti-tumor activity evaluation of Matijin-Su derivatives. Bioorganic Chem. 2014, 56, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Xu, B.; Gong, Q.; Pan, W.; Liu, C.; Huang, Z.; Gu, X.; Liang, G. Synthesis and Biological Evaluation of Matijin-Su Derivatives as Potential Antihepatitis B Virus and Anticancer Agents. Chem. Biodivers. 2016, 13, 1584–1592. [Google Scholar] [CrossRef]
- Qiu, J.; Xu, B.; Huang, Z.; Pan, W.; Cao, P.; Liu, C.; Hao, X.; Song, B.; Liang, G. Synthesis and biological evaluation of Matijing-Su derivatives as potent anti-HBV agents. Bioorganic Med. Chem. 2011, 19, 5352–5360. [Google Scholar] [CrossRef]
- Kuang, A.; Lu, W.; Zeng, X.; Liang, G.; Xu, B. Synthesis and anti-HBV activity evaluation of Matijin-Su derivatives containing veratric acid. Chin. J. New Drug 2019, 28, 1523–1530. [Google Scholar]
- Li, W.; Xu, F.; Shuai, W.; Sun, H.; Yao, H.; Ma, C.; Xu, S.; Yao, H.; Zhu, Z.; Yang, D.H.; et al. Discovery of Novel Quinoline-Chalcone Derivatives as Potent Antitumor Agents with Microtubule Polymerization Inhibitory Activity. J. Med. Chem. 2019, 62, 993–1013. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hung, A.; Yang, A. Herb-target virtual screening and network pharmacology for prediction of molecular mechanism of Danggui Beimu Kushen Wan for prostate cancer. Sci. Rep. 2021, 11, 6656. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, Y.; Yu, H.; Shen, B.; Liang, Y.; Jin, R.; Liu, X.; Shi, L.; Cai, X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016, 5, 2061–2068. [Google Scholar] [CrossRef]
- Martínez-Martínez, D.; Soto, A.; Gil-Araujo, B.; Gallego, B.; Chiloeches, A.; Lasa, M. Resveratrol promotes apoptosis through the induction of dual specificity phosphatase 1 and sensitizes prostate cancer cells to cisplatin. Food Chem. Toxicol. 2019, 124, 273–279. [Google Scholar] [CrossRef]
- Shen, J.; Zhou, S.; Shi, L.; Liu, X.; Lin, H.; Yu, H.; Xiaoliang; Tang, J.; Yu, T.; Cai, X. DUSP1 inhibits cell proliferation, metastasis and invasion and angiogenesis in gallbladder cancer. Oncotarget 2017, 8, 12133–12144. [Google Scholar] [CrossRef]
- Zhang, X.; Hyer, J.M.; Yu, H.; D’Silva, N.J.; Kirkwood, K.L. DUSP1 phosphatase regulates the proinflammatory milieu in head and neck squamous cell carcinoma. Cancer Res. 2014, 74, 7191–7197. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Shen, M.; Zhou, M.; Shi, X.; He, R.; Yin, T.; Wang, M.; Guo, X.; Qin, R. Long noncoding RNA LINC01111 suppresses pancreatic cancer aggressiveness by regulating DUSP1 expression via microRNA-3924. Cell Death Dis. 2019, 10, 883. [Google Scholar] [CrossRef] [PubMed]
- Kho, D.H.; Uddin, M.H.; Chatterjee, M.; Vogt, A.; Raz, A.; Wu, G.S. GP78 Cooperates with Dual-Specificity Phosphatase 1 to Stimulate Epidermal Growth Factor Receptor-Mediated Extracellular Signal-Regulated Kinase Signaling. Mol. Cell. Biol. 2019, 39, e00485-18. [Google Scholar] [CrossRef]
- Gil-Araujo, B.; Toledo Lobo, M.V.; Gutiérrez-Salmerón, M.; Gutiérrez-Pitalúa, J.; Ropero, S.; Angulo, J.C.; Chiloeches, A.; Lasa, M. Dual specificity phosphatase 1 expression inversely correlates with NF-κB activity and expression in prostate cancer and promotes apoptosis through a p38 MAPK dependent mechanism. Mol. Oncol. 2014, 8, 27–38. [Google Scholar] [CrossRef]
- Martínez-Martínez, D.; Toledo Lobo, M.V.; Baquero, P.; Ropero, S.; Angulo, J.C.; Chiloeches, A.; Lasa, M. Downregulation of Snail by DUSP1 Impairs Cell Migration and Invasion through the Inactivation of JNK and ERK and Is Useful as a Predictive Factor in the Prognosis of Prostate Cancer. Cancers 2021, 13, 1158. [Google Scholar] [CrossRef]
- Shen, Y.L.; Gan, Y.; Gao, H.F.; Fan, Y.C.; Wang, Q.; Yuan, H.; Song, Y.F.; Wang, J.D.; Tu, H. TNFSF9 exerts an inhibitory effect on hepatocellular carcinoma. J. Dig. Dis. 2017, 18, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Y.; Jiang, Z. TNFSF9 is a prognostic biomarker and correlated with immune infiltrates in pancreatic cancer. J. Gastrointest. Cancer 2021, 52, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Shen, Y.L.; Lan, H.Y.; Jin, J.M.; An, P.; Zhang, L.J.; Chen, L.L.; Peng, W.; Luan, X.; Zhang, H. The Cyr61 Is a Potential Target for Rotundifuran, a Natural Labdane-Type Diterpene from Vitex trifolia L., to Trigger Apoptosis of Cervical Cancer Cells. Oxidative Med. Cell. Longev. 2021, 2021, 6677687. [Google Scholar] [CrossRef] [PubMed]
- Chien, W.; Kumagai, T.; Miller, C.W.; Desmond, J.C.; Frank, J.M.; Said, J.W.; Koeffler, H.P. Cyr61 suppresses growth of human endometrial cancer cells. J. Biol. Chem. 2004, 279, 53087–53096. [Google Scholar] [CrossRef]
- Tanaka, F.; Rizqiawan, A.; Higashikawa, K.; Tobiume, K.; Okui, G.; Shigeishi, H.; Ono, S.; Shimasue, H.; Kamata, N. Snail promotes Cyr61 secretion to prime collective cell migration and form invasive tumor nests in squamous cell carcinoma. Cancer Lett. 2013, 329, 243–252. [Google Scholar] [CrossRef]
- Tong, X.; O’Kelly, J.; Xie, D.; Mori, A.; Lemp, N.; McKenna, R.; Miller, C.W.; Koeffler, H.P. Cyr61 suppresses the growth of non-small-cell lung cancer cells via the beta-catenin-c-myc-p53 pathway. Oncogene 2004, 23, 4847–4855. [Google Scholar] [CrossRef]
- Kim, H.; Son, S.; Ko, Y.; Lee, J.E.; Kim, S.; Shin, I. YAP, CTGF and Cyr61 are overexpressed in tamoxifen-resistant breast cancer and induce transcriptional repression of ERα. J. Cell Sci. 2021, 134, jcs256503. [Google Scholar] [CrossRef]
- Da Silva-Álvarez, S.; Collado, M. The Jekyll and Hyde of Senescence in Cancer: TIMP1 Controls the Switch from Tumor-Controlling to Tumor-Promoting Senescence. Cancer Cell 2021, 39, 13–15. [Google Scholar] [CrossRef]
- Rabelo-Santos, S.H.; Termini, L.; Boccardo, E.; Derchain, S.; Longatto-Filho, A.; Andreoli, M.A.; Costa, M.C.; Lima Nunes, R.A.; Lucci Ângelo-Andrade, L.A.; Villa, L.L.; et al. Strong SOD2 expression and HPV-16/18 positivity are independent events in cervical cancer. Oncotarget 2018, 9, 21630–21640. [Google Scholar] [CrossRef]
- Mumyatova, V.A.; Balakina, A.A.; Lapshina, M.A.; Sen’, V.D.; Kornev, A.B.; Terent’ev, A.A. Influence of Tumor Suppressor p53 Functioning on the Expression of Antioxidant System Genes under the Action of Cytotoxic Compounds. Bull. Exp. Biol. Med. 2020, 169, 169–175. [Google Scholar] [CrossRef]
- Kim, J.W.; Gautam, J.; Kim, J.E.; Kim, J.A.; Kang, K.W. Inhibition of tumor growth and angiogenesis of tamoxifen-resistant breast cancer cells by ruxolitinib, a selective JAK2 inhibitor. Oncol. Lett. 2019, 17, 3981–3989. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, S.; Moradi, S.Z.; Ash-Rafzadeh, A.; Bishayee, A. Targeting cellular senescence in cancer by plant secondary metabolites: A systematic review. Pharmacol. Res. 2022, 177, 105961. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.L.; Lu, C.; Klement, J.D.; Redd, P.S.; Yang, D.; Smith, A.D.; Liu, K. Expression profiles and function of IL6 in polymorphonuclear myeloid-derived suppressor cells. Cancer Immunol. Immunother. 2020, 69, 2233–2245. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, J.; Cheng, S.; Peng, Z.; Luo, H. Transcription factor Fli-1 as a new target for antitumor drug development. Int. J. Biol. Macromol. 2022, 209, 1155–1168. [Google Scholar] [CrossRef]
- Xia, T.; Li, J.; Ren, X.; Liu, C.; Sun, C. Research progress of phenolic compounds regulating IL-6 to exert antitumor effects. Phytother. Res. 2021, 35, 6720–6734. [Google Scholar] [CrossRef]
- Fisher, D.T.; Appenheimer, M.M.; Evans, S.S. The two faces of IL-6 in the tumor microenvironment. Semin. Immunol. 2014, 26, 38–47. [Google Scholar] [CrossRef]
- Pagliara, V.; Adornetto, A.; Mammì, M.; Masullo, M.; Sarnataro, D.; Pietropaolo, C.; Arcone, R. Protease Nexin-1 affects the migration and invasion of C6 glioma cells through the regulation of urokinase Plasminogen Activator and Matrix Metalloproteinase-9/2. Biochim. Biophys. Acta 2014, 1843, 2631–2644. [Google Scholar] [CrossRef]
- Xu, D.; McKee, C.M.; Cao, Y.; Ding, Y.; Kessler, B.M.; Muschel, R.J. Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Cancer Res. 2010, 70, 6988–6998. [Google Scholar] [CrossRef] [PubMed]
- Hibi, K.; Goto, T.; Sakuraba, K.; Shirahata, A.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; Sanada, Y. Methylation of OSMR gene is frequently observed in non-invasive colorectal cancer. Anticancer Res. 2011, 31, 1293–1295. [Google Scholar]
- Jahani-Asl, A.; Yin, H.; Soleimani, V.D.; Haque, T.; Luchman, H.A.; Chang, N.C.; Sincennes, M.C.; Puram, S.V.; Scott, A.M.; Lorimer, I.A.; et al. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat. Neurosci. 2016, 19, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhong, J.; Cai, C.; Lu, J.; Wu, W.; Zeng, G. Immune-related biomarker risk score predicts prognosis in prostate cancer. Aging 2020, 12, 22776–22793. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.H.; Niu, X.; Wu, G.H.; Cheng, Q. Decreased expression of TNFRSF12A in thyroid gland cancer predicts poor prognosis: A study based on TCGA data. Medicine 2020, 99, e21882. [Google Scholar] [CrossRef] [PubMed]
- Naimi, A.; Movassaghpour, A.A.; Hagh, M.F.; Talebi, M.; Entezari, A.; Jadidi-Niaragh, F.; Solali, S. TNF-related apoptosis-inducing ligand (TRAIL) as the potential therapeutic target in hematological malignancies. Biomed. Pharmacother. 2018, 98, 566–576. [Google Scholar] [CrossRef]
- Gamie, Z.; Kapriniotis, K.; Papanikolaou, D.; Haagensen, E.; Da Conceicao Ribeiro, R.; Dalgarno, K.; Krippner-Heidenreich, A.; Gerrand, C.; Tsiridis, E.; Rankin, K.S. TNF-related apoptosis-inducing ligand (TRAIL) for bone sarcoma treatment: Pre-clinical and clinical data. Cancer Lett. 2017, 409, 66–80. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Wen, Z.H.; Wan, K.; Yuan, D.; Zeng, X.; Liang, G.; Zhu, J.; Xu, B.; Luo, H. A novel synthesized 3’, 5’-diprenylated chalcone mediates the proliferation of human leukemia cells by regulating apoptosis and autophagy pathways. Biomed. Pharmacother. 2018, 106, 794–804. [Google Scholar] [CrossRef]
- Karim, S.; Burzangi, A.S.; Ahmad, A.; Siddiqui, N.A.; Ibrahim, I.M.; Sharma, P.; Abualsunun, W.A.; Gabr, G.A. PI3K-AKT Pathway Modulation by Thymoquinone Limits Tumor Growth and Glycolytic Metabolism in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 2305. [Google Scholar] [CrossRef]
- Fu, J.; Qin, L.; He, T.; Qin, J.; Hong, J.; Wong, J.; Liao, L.; Xu, J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011, 21, 275–289. [Google Scholar] [CrossRef]
- DU, X.; Xiao, J.; Fu, X.; Xu, B.; Han, H.; Wang, Y.; Pei, X. A proteomic analysis of Bcl-2 regulation of cell cycle arrest: Insight into the mechanisms. J. Zhejiang Univ. Sci. B 2021, 22, 839–855. [Google Scholar] [CrossRef] [PubMed]

| Type of Regulation | Protein Symbol |
|---|---|
| Up | CYR61, IRAK2, B4GALT5, LYPD3, PLAU, TIMP1, FOS, HLA-A(68), MT-ND2, MT-ATP8, SOD2, IL6, JUN, HLA-A(24), SERPINE2, CXCL8, RALA, CEBPB, EFNA1, PTX3, DUSP1, EPHA2, TM4SF1, SLC7A1, SDC4, HRH1, TNFSF9, CLK1, STC1, OLR1, PLAUR, ZFP36L1, LAMB3, TNIP1, PCOLCE, PDK4, LY6K, DOLPP1, ITPRIP, HLA-B, SLC38A2, OSMR, B3GNT5, TMEM222, TNFRSF10D, TNFRSF12A, KLHL21 |
| Down | TPM2, GNS, JPT1 |
| Target Symbol | Entry | PDB ID | Binding Affinity (kcal/mol) |
|---|---|---|---|
| CYR61 | O00622 | 5WTT | −6.9 |
| TIMP1 | P01033 | 3V96 | −7.8 |
| SOD2 | P04179 | 1PL4 | −7.2 |
| IL6 | P05231 | 7NXZ | −6.0 |
| SERPINE2 | P07093 | 4DY0 | −7.3 |
| DUSP1 | P28562 | 6D66 | −8.7 |
| TNFSF9 | P41273 | 6MGP | −8.4 |
| OSMR | Q99650 | NA | NA |
| TNFRSF10D | Q9UBN6 | NA | NA |
| TNFRSF12A | Q9NP84 | 2KMZ | −5.6 |
| Target Name | Target Sequence |
|---|---|
| ShDUSP1 1# | GCTCTGTCAACGTGCGCTTCA |
| ShDUSP1 2# | CAAAGGAGGATACGAAGCGTT |
| ShDUSP1 3# | AGTTTGTGAAGCAGAGGCGAA |
| ShTNFSF9 1# | TGAGCTACAAAGAGGACACGA |
| ShTNFSF9 2# | CGAGGCTCGGAACTCGGCCTT |
| ShTNFSF9 3# | CCCTTCACCGAGGTCGGAATA |
| shVector | TTCTCCGAACGTGTCACGTAA |
| Gene Name | Primer Direction | Primer Sequence |
|---|---|---|
| DUSP1 | Forward | 5′-AGAGCCCCATTACGACCTCT-3′ |
| Reverse | 5′-CCAGAGGAACTCGGGTGAAG-3′ | |
| TNFSF9 | Forward | 5′-AAATGTTCTGATCGATGGG-3′ |
| Reverse | 5′-CCGCAGCTCTAGTTGAAAGAAGA-3′ | |
| GAPDH | Forward | 5′-GGAGCGAGATCCCTCCAAAAT-3′ |
| Reverse | 5′-GGCTGTTGTCATACTTCTCATGG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Yang, M.; Yu, J.; Cheng, S.; Ahmad, M.; Wu, C.; Wan, X.; Xu, B.; Ben-David, Y.; Luo, H. A Novel L-Phenylalanine Dipeptide Inhibits the Growth and Metastasis of Prostate Cancer Cells via Targeting DUSP1 and TNFSF9. Int. J. Mol. Sci. 2022, 23, 10916. https://doi.org/10.3390/ijms231810916
Li L, Yang M, Yu J, Cheng S, Ahmad M, Wu C, Wan X, Xu B, Ben-David Y, Luo H. A Novel L-Phenylalanine Dipeptide Inhibits the Growth and Metastasis of Prostate Cancer Cells via Targeting DUSP1 and TNFSF9. International Journal of Molecular Sciences. 2022; 23(18):10916. https://doi.org/10.3390/ijms231810916
Chicago/Turabian StyleLi, Lanlan, Mingfei Yang, Jia Yu, Sha Cheng, Mashaal Ahmad, Caihong Wu, Xinwei Wan, Bixue Xu, Yaacov Ben-David, and Heng Luo. 2022. "A Novel L-Phenylalanine Dipeptide Inhibits the Growth and Metastasis of Prostate Cancer Cells via Targeting DUSP1 and TNFSF9" International Journal of Molecular Sciences 23, no. 18: 10916. https://doi.org/10.3390/ijms231810916
APA StyleLi, L., Yang, M., Yu, J., Cheng, S., Ahmad, M., Wu, C., Wan, X., Xu, B., Ben-David, Y., & Luo, H. (2022). A Novel L-Phenylalanine Dipeptide Inhibits the Growth and Metastasis of Prostate Cancer Cells via Targeting DUSP1 and TNFSF9. International Journal of Molecular Sciences, 23(18), 10916. https://doi.org/10.3390/ijms231810916






