High-Resolution Conformational Analysis of RGDechi-Derived Peptides Based on a Combination of NMR Spectroscopy and MD Simulations
Abstract
1. Introduction
2. Results and Discussion
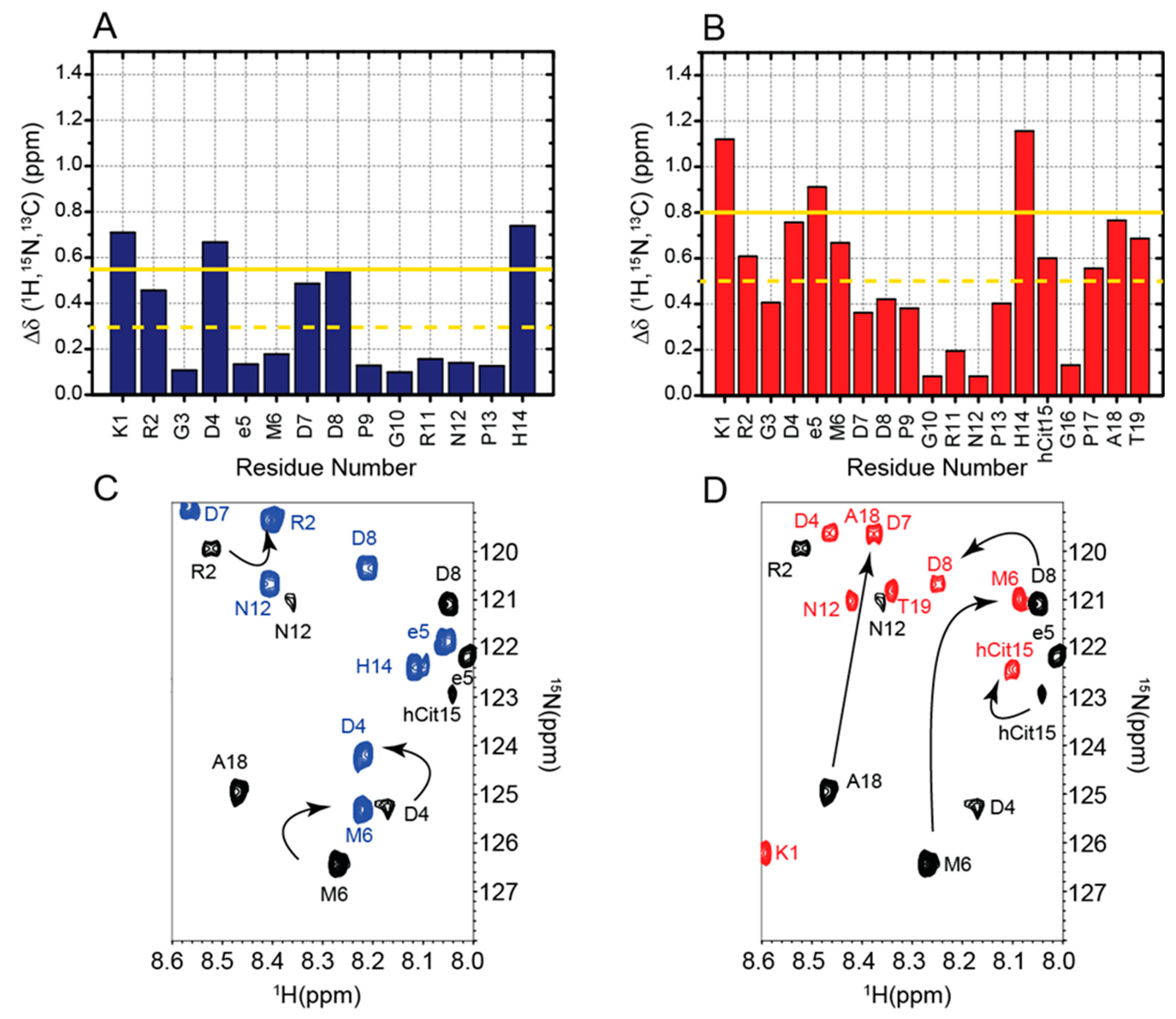
2.1. Chemical Shifts Assignment of RGDechi1-14 and ψRGDechi
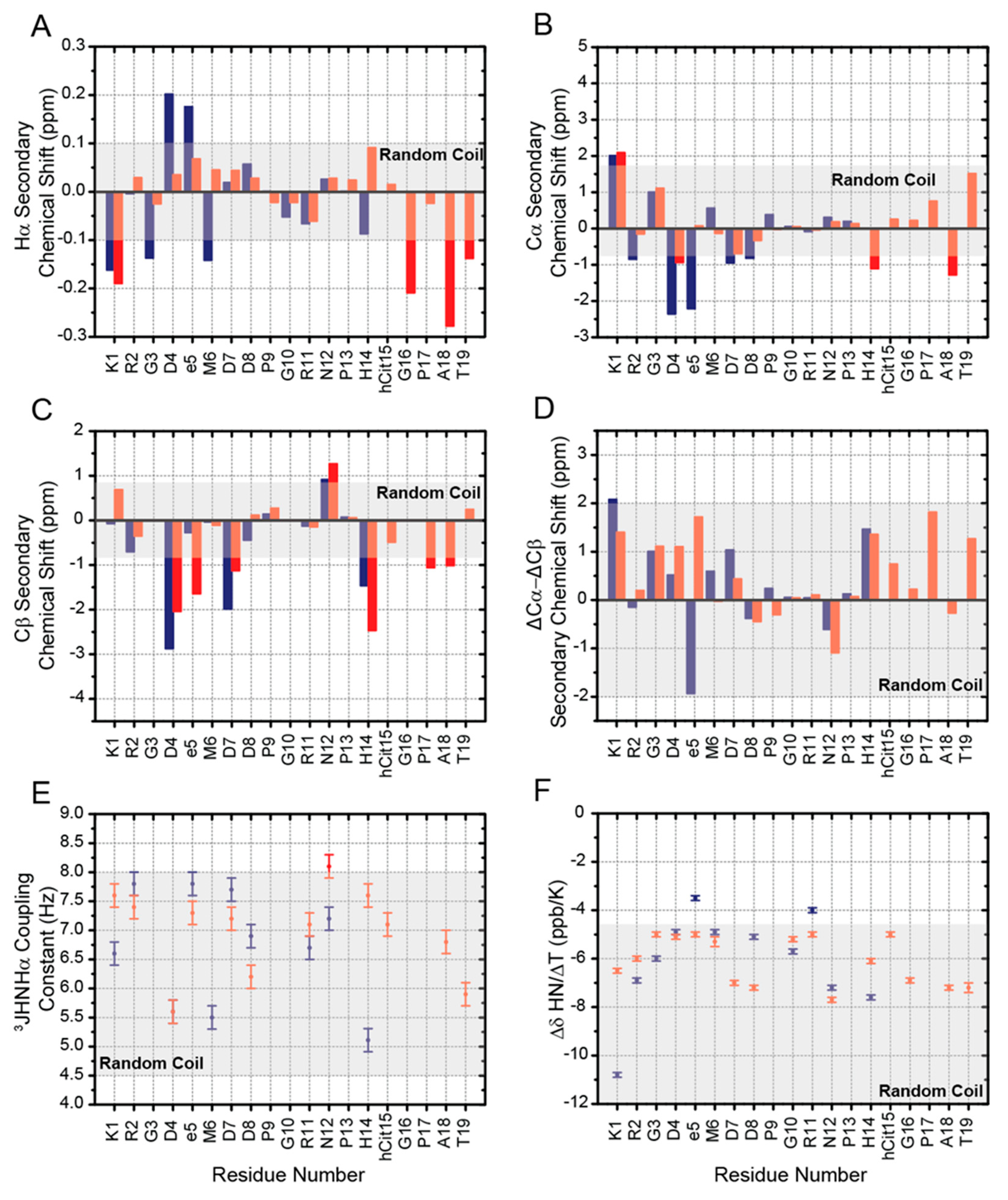
2.2. Chemical Shifts Analysis of RGDechi1-14 and ψRGDechi
2.3. 3JHNHα Coupling Constants, HN Temperature Coefficients and NOEs Evaluation
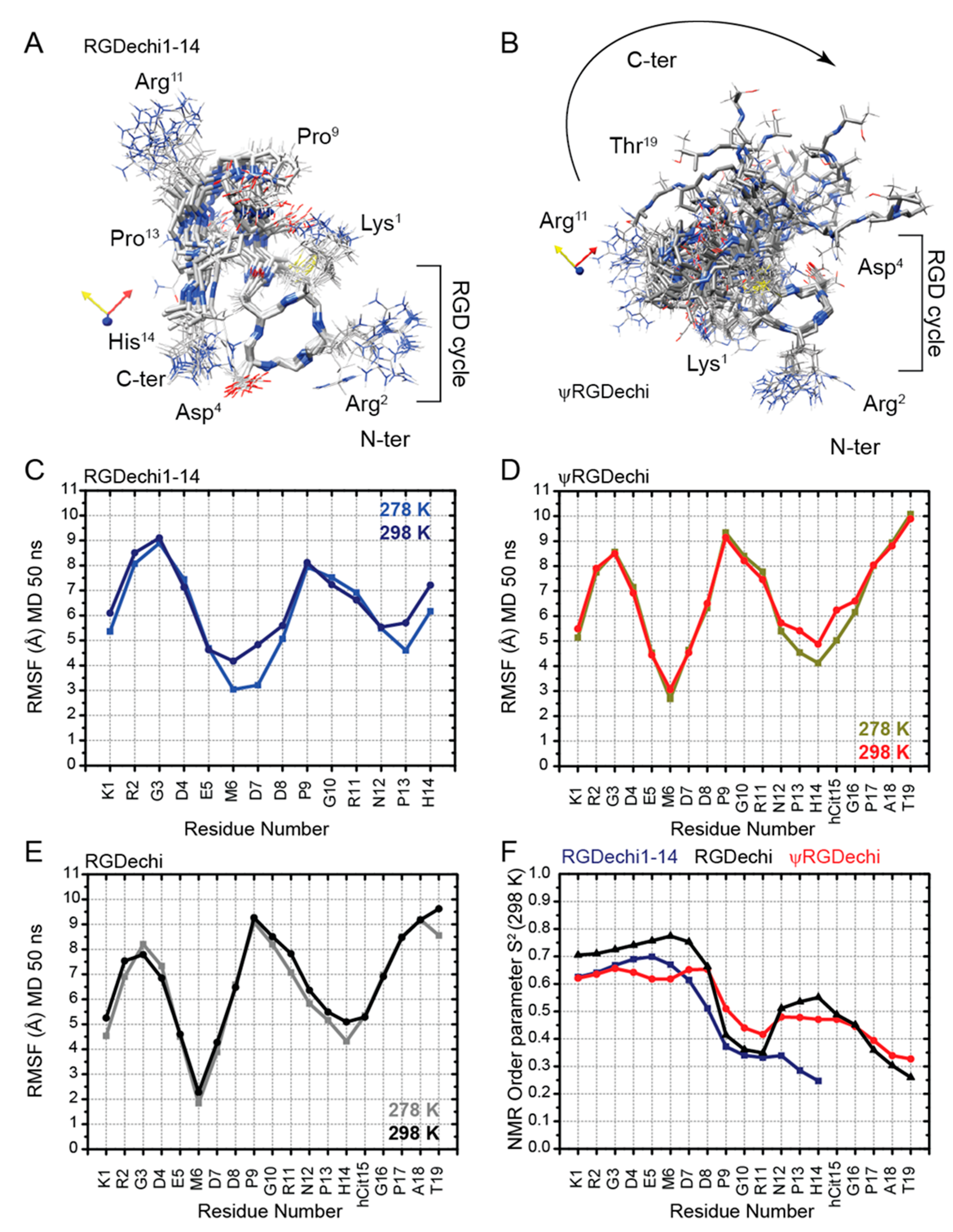
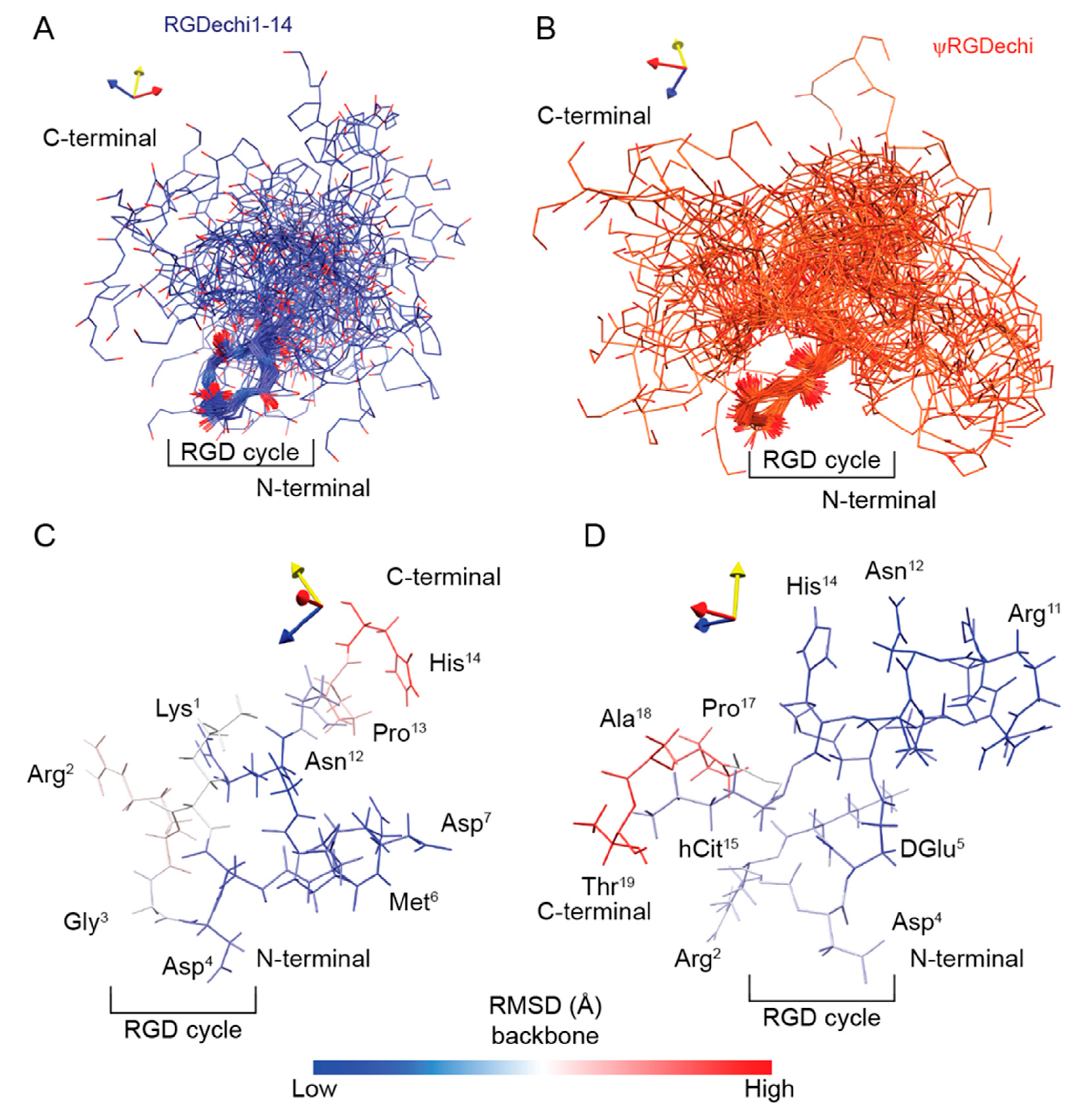
2.4. Structural Comparison of RGDechi1-14 and ψRGDechi versus RGDechi
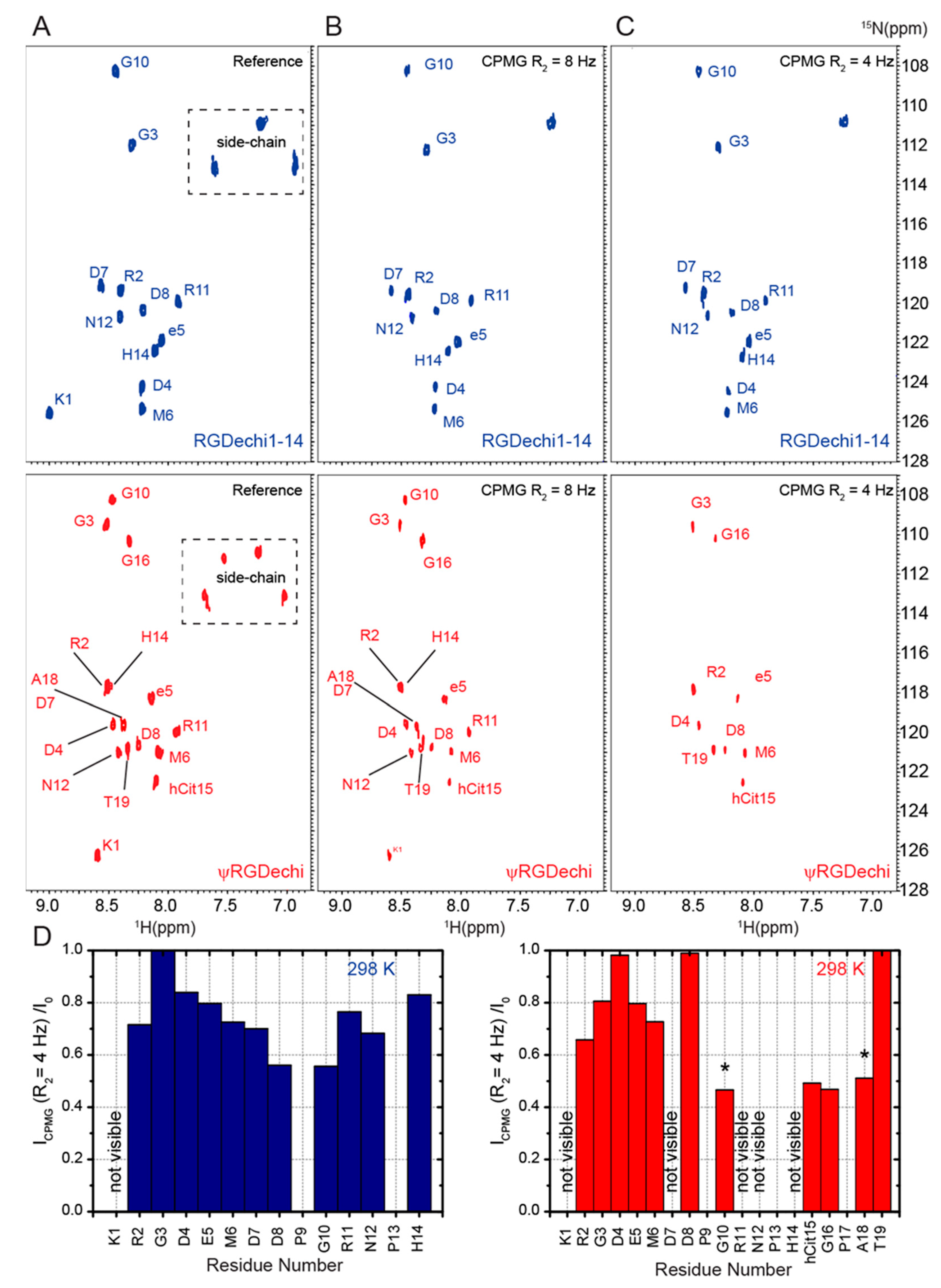
2.5. RGDechi1-14 and ψRGDechi Backbone Motions
2.6. Conformational Ensemble of RGDechi1-14 and ψRGDechi Peptides
2.7. Elucidation on the Recognition Mechanism of αvβ3 Integrin by ψRGDechi
3. Materials and Methods
3.1. Peptides Preparation
3.2. NMR Experiments
- (i)
- The 2D [1H-1H] TOCSY (total correlation spectroscopy) [55], 2D [1H-1H] NOESY (nuclear overhauser effect spectroscopy) [56], and 2D [1H-1H] ROESY (rotating frame overhauser effect spectroscopy) [57] data were acquired using 32 scans per t1 increment, a spectral width (SW) of 6001.30 Hz along both t1 and t2, 2048 × 256 complex points in t2 and t1, and a relaxation delay of 3.0 s. In all NMR experiments reported above, the water suppression was achieved using the Watergate pulse sequence with the application of gradients using double echo. The 2D [1H-1H] TOCSY experiments were acquired by homonuclear Hartman–Hahn transfer by a MILEV17 mixing sequence with a mixing time (tm) of 70 ms and 10 KHz spin-lock field strength; The 2D [1H-1H] NOESY spectra were recorded using a mixing time of 250 ms and 2D [1H-1H] ROESY were carried out with a cw spin-lock field strength of 4 kHz, a mixing time of 250 ms, and a relaxation delay of 5.0 s. All homonuclear 2D experiments were apodized with a square cosine window function and zero fill to a matrix of size 4096 × 1024 before Fourier transform and baseline correction.
- (ii)
- Heteronuclear natural-abundance 2D [1H–15N] HSQC (heteronuclear single quantum coherence) experiments were performed with 880 scans per t1 increment, SW of 1581.32 and 6001.30 Hz along t1 and t2, respectively, 2048 × 128 complex points in t2 and t1, respectively, and a 1.0 s relaxation delay. In particular, the 2D [1H–15N] HSQC pulse sequence was optimized as previously reported [16] using a Bruker standard gradient echo–antiecho pulse program [58]. Two-dimensional [1H–15N] HSQC spectra were apodized with a square cosine window function and a zero filling to a matrix of size a 2048 × 128 before Fourier transform and baseline correction. Natural-abundance 2D [1H–13C] HSQC spectra were acquired with 880 scans per t1 increment, SW of 5130.94 Hz along t1 and 6001.30 Hz along t2, 2048 × 200 complex points in t2 and t1, respectively, and a relaxation delay 1.0 s. The 2D constant time (CT) [1H–13C] HSQC pulse sequence was optimized as reported is a previous publication [16]. Two-dimensional CT [1H–13C] HSQC were acquired using a heteronuclear coupling constant (JXH) of 145 Hz, CT period of 26.6 ms, and shaped pulses for all 180° pulses on f2 channel with decoupling during f1 acquisition. Two-dimensional CT [1H–13C] HSQC spectra were apodized with a square cosine windows function and zero fill to a final matrix of 4096 × 4096 before Fourier transform and baseline correction.
- (i)
- Two-dimensional T2-filter [1H–15N] HSQC experiments were carried out using 880 scans per t1 increment, SW of 1581.32 Hz along t1 and 6001.30 Hz along t2, 2048 × 128 complex points in t2 and t1, respectively, and 1.0 s relaxation delay. In particular, for each peptide, the dynamics characterization was performed by two 2D T2-filter [1H–15N] HSQC spectra acquired using different relaxation compensated CPMG (Carr–Purcell–Meiboom–Gill) sequence periods (125 and 250 ms). Notably, in this [1H–15N] HSQC-based experiment only the 1H-15N cross-peaks of the residues for which the 15N transversal relaxation time (T2) was longer than the applied filter were observable. The pulse sequence was optimized as previously reported [16]. Two-dimensional T2-filter [1H–15N] HSQC spectra were apodized with a square cosine window function and zero filling to a matrix of size 4096 × 1024 before Fourier transform and baseline correction.
3.3. Chemical Shifts Analysis
3.4. Amide Temperature Coefficients and 3JHNHα Scalar Couplings Analysis
3.5. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petsalaki, E.; Russell, R.B. Peptide-mediated interactions in biological systems: New discoveries and applications. Curr. Opin. Biotechnol. 2008, 19, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, R.B.; Esteban-Martín, S.; Salvatella, X. Understanding biomolecular motion, recognition, and allostery by use of conformational ensembles. Eur. Biophys. J. 2011, 40, 1339–1355. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz, L.; Bajaj, R.; Becker, S.; Zweckstetter, M. The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein. Sci. 2010, 19, 2045–2054. [Google Scholar] [CrossRef] [PubMed]
- Choy, W.Y.; Forman-Kay, J.D. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J. Mol. Biol. 2001, 308, 1011–1032. [Google Scholar] [CrossRef]
- Russo, L.; Raiola, L.; Campitiello, M.A.; Magrì, A.; Fattorusso, R.; Malgieri, G.; Pappalardo, G.; La Mendola, D.; Isernia, C. Probing the residual structure in avian prion hexarepeats by CD, NMR and MD techniques. Molecules 2013, 18, 11467–11484. [Google Scholar] [CrossRef]
- Russo, L.; Maestre-Martinez, M.; Wolff, S.; Becker, S.; Griesinger, C. Interdomain dynamics explored by paramagnetic NMR. J. Am. Chem. Soc. 2013, 135, 17111–17120. [Google Scholar] [CrossRef]
- Russo, L.; Giller, K.; Pfitzner, E.; Griesinger, C.; Becker, S. Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6. Sci. Rep. 2017, 7, 16845. [Google Scholar] [CrossRef]
- Russo, L.; Mascanzoni, F.; Farina, B.; Dolga, A.M.; Monti, A.; Caporale, A.; Culmsee, C.; Fattorusso, R.; Ruvo, M.; Doti, N. Design, Optimization, and Structural Characterization of an Apoptosis-Inducing Factor Peptide Targeting Human Cyclophilin A to Inhibit Apoptosis Inducing Factor-Mediated Cell Death. J. Med. Chem. 2021, 64, 11445–11459. [Google Scholar] [CrossRef]
- Eisenmesser, E.Z.; Bosco, D.A.; Akke, M.; Kern, D. Enzyme dynamics during catalysis. Science 2002, 295, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Ryabov, Y.E.; Fushman, D. A model of interdomain mobility in a multidomain protein. J. Am. Chem. Soc. 2007, 129, 3315–3327. [Google Scholar] [CrossRef] [PubMed]
- Mulder, F.A.; Mittermaier, A.; Hon, B.; Dahlquist, F.W.; Kay, L.E. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 2001, 8, 932–935. [Google Scholar] [CrossRef] [PubMed]
- Grzesiek, S.; Döbeli, H.; Gentz, R.; Garotta, G.; Labhardt, A.M.; Bax, A. 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-gamma. Biochemistry 1992, 31, 8180–8190. [Google Scholar] [CrossRef]
- Kosol, S.; Contreras-Martos, S.; Cedeño, C.; Tompa, P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules 2013, 18, 10802–10828. [Google Scholar] [CrossRef]
- Farina, B.; Del Gatto, A.; Comegna, D.; Di Gaetano, S.; Capasso, D.; Isernia, C.; Saviano, M.; Fattorusso, R.; Zaccaro, L.; Russo, L. Conformational studies of RGDechi peptide by natural-abundance NMR spectroscopy. J. Pept. Sci. 2019, 25, e3166. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef]
- Del Gatto, A.; Zaccaro, L.; Grieco, P.; Novellino, E.; Zannetti, A.; Del Vecchio, S.; Iommelli, F.; Salvatore, M.; Pedone, C.; Saviano, M. Novel and selective alpha(v)beta3 receptor peptide antagonist: Design, synthesis, and biological behavior. J. Med. Chem. 2006, 49, 3416–3420. [Google Scholar] [CrossRef]
- Takada, Y.; Ye, X.; Simon, S. The integrins. Genome. Biol. 2007, 8, 215. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Brooks, P.C.; Clark, R.A.; Cheresh, D.A. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science 1994, 264, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Giancotti, F.G.; Ruoslahti, E. Integrin signaling. Science 1999, 285, 1028–1032. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Zannetti, A.; Del Vecchio, S.; Iommelli, F.; Del Gatto, A.; De Luca, S.; Zaccaro, L.; Papaccioli, A.; Sommella, J.; Panico, M.; Speranza, A.; et al. Imaging of alpha(v)beta(3) expression by a bifunctional chimeric RGD peptide not cross-reacting with alpha(v)beta(5). Clin. Cancer Res. 2009, 15, 5224–5233. [Google Scholar] [CrossRef]
- Santulli, G.; Basilicata, M.F.; De Simone, M.; Del Giudice, C.; Anastasio, A.; Sorriento, D.; Saviano, M.; Del Gatto, A.; Trimarco, B.; Pedone, C.; et al. Evaluation of the anti-angiogenic properties of the new selective αVβ3 integrin antagonist RGDechiHCit. J. Transl. Med. 2011, 9, 7. [Google Scholar] [CrossRef]
- Pisano, M.; DE Paola, I.; Nieddu, V.; Sassu, I.; Cossu, S.; Galleri, G.; Del Gatto, A.; Budroni, M.; Cossu, A.; Saviano, M.; et al. In vitro activity of the αvβ3 integrin antagonist RGDechi-hCit on malignant melanoma cells. Anticancer Res. 2013, 33, 871–879. [Google Scholar]
- Capasso, D.; de Paola, I.; Liguoro, A.; Del Gatto, A.; Di Gaetano, S.; Guarnieri, D.; Saviano, M.; Zaccaro, L. RGDechi-hCit: αvβ3 selective pro-apoptotic peptide as potential carrier for drug delivery into melanoma metastatic cells. PLoS ONE 2014, 9, e106441. [Google Scholar] [CrossRef] [PubMed]
- Farina, B.; de Paola, I.; Russo, L.; Capasso, D.; Liguoro, A.; Gatto, A.D.; Saviano, M.; Pedone, P.V.; Di Gaetano, S.; Malgieri, G.; et al. A Combined NMR and Computational Approach to Determine the RGDechi-hCit-αv β3 Integrin Recognition Mode in Isolated Cell Membranes. Chemistry 2016, 22, 681–693. [Google Scholar] [CrossRef]
- Capasso, D.; Di Gaetano, S.; Celentano, V.; Diana, D.; Festa, L.; Di Stasi, R.; De Rosa, L.; Fattorusso, R.; D’Andrea, L.D. Unveiling a VEGF-mimetic peptide sequence in the IQGAP1 protein. Mol. Biosyst. 2017, 13, 1619–1629. [Google Scholar] [CrossRef]
- Russo, L.; Farina, B.; Del Gatto, A.; Comegna, D.; Di Gaetano, S.; Capasso, D.; Liguoro, A.; Malgieri, G.; Saviano, M.; Fattorusso, R.; et al. Deciphering RGDechi peptide-α5β1 integrin interaction mode in isolated cell membranes. Pept. Sci. 2018, 110, e24065. [Google Scholar] [CrossRef]
- Hill, B.S.; Sarnella, A.; Capasso, D.; Comegna, D.; Del Gatto, A.; Gramanzini, M.; Albanese, S.; Saviano, M.; Zaccaro, L.; Zannetti, A. Therapeutic Potential of a Novel αvβ3 Antagonist to Hamper the Aggressiveness of Mesenchymal Triple Negative Breast Cancer Sub-Type. Cancer 2019, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Comegna, D.; Zannetti, A.; Del Gatto, A.; de Paola, I.; Russo, L.; Di Gaetano, S.; Liguoro, A.; Capasso, D.; Saviano, M.; Zaccaro, L. Chemical Modification for Proteolytic Stabilization of the Selective α. J. Med. Chem. 2017, 60, 9874–9884. [Google Scholar] [CrossRef] [PubMed]
- Kjaergaard, M.; Brander, S.; Poulsen, F.M. Random coil chemical shift for intrinsically disordered proteins: Effects of temperature and pH. J. Biomol. NMR 2011, 49, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Kjaergaard, M.; Poulsen, F.M. Sequence correction of random coil chemical shifts: Correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 2011, 50, 157–165. [Google Scholar] [CrossRef]
- Marsh, J.A.; Singh, V.K.; Jia, Z.; Forman-Kay, J.D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein. Sci. 2006, 15, 2795–2804. [Google Scholar] [CrossRef]
- Xu, X.P.; Case, D.A. Automated prediction of 15N, 13Calpha, 13Cbeta and 13C’ chemical shifts in proteins using a density functional database. J. Biomol. NMR 2001, 21, 321–333. [Google Scholar] [CrossRef]
- Neal, S.; Nip, A.M.; Zhang, H.; Wishart, D.S. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR 2003, 26, 215–240. [Google Scholar] [CrossRef]
- Meiler, J. PROSHIFT: Protein chemical shift prediction using artificial neural networks. J. Biomol. NMR 2003, 26, 25–37. [Google Scholar] [CrossRef]
- Dorman, D.E.; Torchia, D.A.; Bovey, F.A. Carbon-13 and proton nuclear magnetic resonance observations of the conformation of poly(L-proline) in aqueous salt solutions. Macromolecules 1973, 6, 80–82. [Google Scholar] [CrossRef]
- Schubert, M.; Labudde, D.; Oschkinat, H.; Schmieder, P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J. Biomol. NMR 2002, 24, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Siemion, I.Z.; Wieland, T.; Pook, K.H. Influence of the distance of the proline carbonyl from the beta and gamma carbon on the 13C chemical shifts. Angew. Chem. Int. Ed. Engl. 1975, 14, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bax, A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J. Biomol. NMR 2010, 46, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.N.; Ramadrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Pardi, A.; Billeter, M.; Wüthrich, K. Calibration of the angular dependence of the amide proton-C alpha proton coupling constants, 3JHN alpha, in a globular protein. Use of 3JHN alpha for identification of helical secondary structure. J. Mol. Biol. 1984, 180, 741–751. [Google Scholar] [CrossRef]
- Karplus, M. Contact Electron-Spin Coupling of Nuclear Magnetic Moments. J. Chem. Phys. 1959, 30, 11–15. [Google Scholar] [CrossRef]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; Wiley: New York, NY, USA, 1986. [Google Scholar]
- Smith, L.J.; Fiebig, K.M.; Schwalbe, H.; Dobson, C.M. The concept of a random coil. Residual structure in peptides and denatured proteins. Fold. Des. 1996, 1, R95–R106. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Defining solution conformations of small linear peptides. Annu. Rev. Biophys. Biophys. Chem. 1991, 20, 519–538. [Google Scholar] [CrossRef]
- Andersen, N.H.; Neidigh, J.W.; Harris, S.M.; Lee, G.M.; Liu, Z.; Tong, H. Extracting information from the temperature gradients of polypeptide NH chemical shifts. 1. The importance of conformational averaging. J. Am. Chem. Soc. 1997, 119, 8547–8561. [Google Scholar] [CrossRef]
- Baxter, N.J.; Williamson, M.P. Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR 1997, 9, 359–369. [Google Scholar] [CrossRef]
- Baxter, N.J.; Hosszu, L.L.; Waltho, J.P.; Williamson, M.P. Characterisation of low free-energy excited states of folded proteins. J. Mol. Biol. 1998, 284, 1625–1639. [Google Scholar] [CrossRef] [PubMed]
- Li, D.W.; Brüschweiler, R. PPM: A side-chain and backbone chemical shift predictor for the assessment of protein conformational ensembles. J. Biomol. NMR 2012, 54, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Brüschweiler, R. PPM_One: A static protein structure based chemical shift predictor. J. Biomol. NMR 2015, 62, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Brüschweiler, R.; Ernst, R.R. Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J. Magn. Reason. 1983, 53, 521–528. [Google Scholar]
- Kumar, A.; Ernst, R.R.; Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 95, 1–6. [Google Scholar] [CrossRef]
- Griesinger, C.; Ernst, R.R. Frequency Offset Effects and Their Elimination in NMR Rotating-Frame Cross-Relaxation Spectroscopy. J. Magn. Reason. 1987, 75, 261–271. [Google Scholar] [CrossRef]
- Sattler, M.; Schleucher, J.R.; Griesinger, C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999, 34, 93–158. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- Keller, R.L.J. Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment Dissertation; Swiss Federal Institute of Technology: Zürich, Switzerland, 2004. [Google Scholar]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory. Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.A.; Pedersen, L.G. Molecular modeling: An experimental tool. Environ. Health Perspect. 1993, 101, 410–412. [Google Scholar] [CrossRef]
- Elber, R.; Ruymgaart, A.P.; Hess, B. SHAKE parallelization. Eur. Phys. J. Spec. Top. 2011, 200, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Gardner, S.P.; Sutcliffe, M.J. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein. Eng. 1996, 9, 1063–1065. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Vriend, G. WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 1990, 8, 52–56, 29. [Google Scholar] [CrossRef]
- Vuister, G.W.; Bax, A. Quantitative J correlation: A new approach for measuring homonuclear three-bond J(HNHa) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 1993, 115, 7772–7777. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acconcia, C.; Paladino, A.; Valle, M.d.; Farina, B.; Del Gatto, A.; Gaetano, S.D.; Capasso, D.; Gentile, M.T.; Malgieri, G.; Isernia, C.; et al. High-Resolution Conformational Analysis of RGDechi-Derived Peptides Based on a Combination of NMR Spectroscopy and MD Simulations. Int. J. Mol. Sci. 2022, 23, 11039. https://doi.org/10.3390/ijms231911039
Acconcia C, Paladino A, Valle Md, Farina B, Del Gatto A, Gaetano SD, Capasso D, Gentile MT, Malgieri G, Isernia C, et al. High-Resolution Conformational Analysis of RGDechi-Derived Peptides Based on a Combination of NMR Spectroscopy and MD Simulations. International Journal of Molecular Sciences. 2022; 23(19):11039. https://doi.org/10.3390/ijms231911039
Chicago/Turabian StyleAcconcia, Clementina, Antonella Paladino, Maria della Valle, Biancamaria Farina, Annarita Del Gatto, Sonia Di Gaetano, Domenica Capasso, Maria Teresa Gentile, Gaetano Malgieri, Carla Isernia, and et al. 2022. "High-Resolution Conformational Analysis of RGDechi-Derived Peptides Based on a Combination of NMR Spectroscopy and MD Simulations" International Journal of Molecular Sciences 23, no. 19: 11039. https://doi.org/10.3390/ijms231911039
APA StyleAcconcia, C., Paladino, A., Valle, M. d., Farina, B., Del Gatto, A., Gaetano, S. D., Capasso, D., Gentile, M. T., Malgieri, G., Isernia, C., Saviano, M., Fattorusso, R., Zaccaro, L., & Russo, L. (2022). High-Resolution Conformational Analysis of RGDechi-Derived Peptides Based on a Combination of NMR Spectroscopy and MD Simulations. International Journal of Molecular Sciences, 23(19), 11039. https://doi.org/10.3390/ijms231911039













