Role of RNAIII in Resistance to Antibiotics and Antimicrobial Agents in Staphylococcus epidermidis Biofilms
Abstract
1. Introduction
2. Results
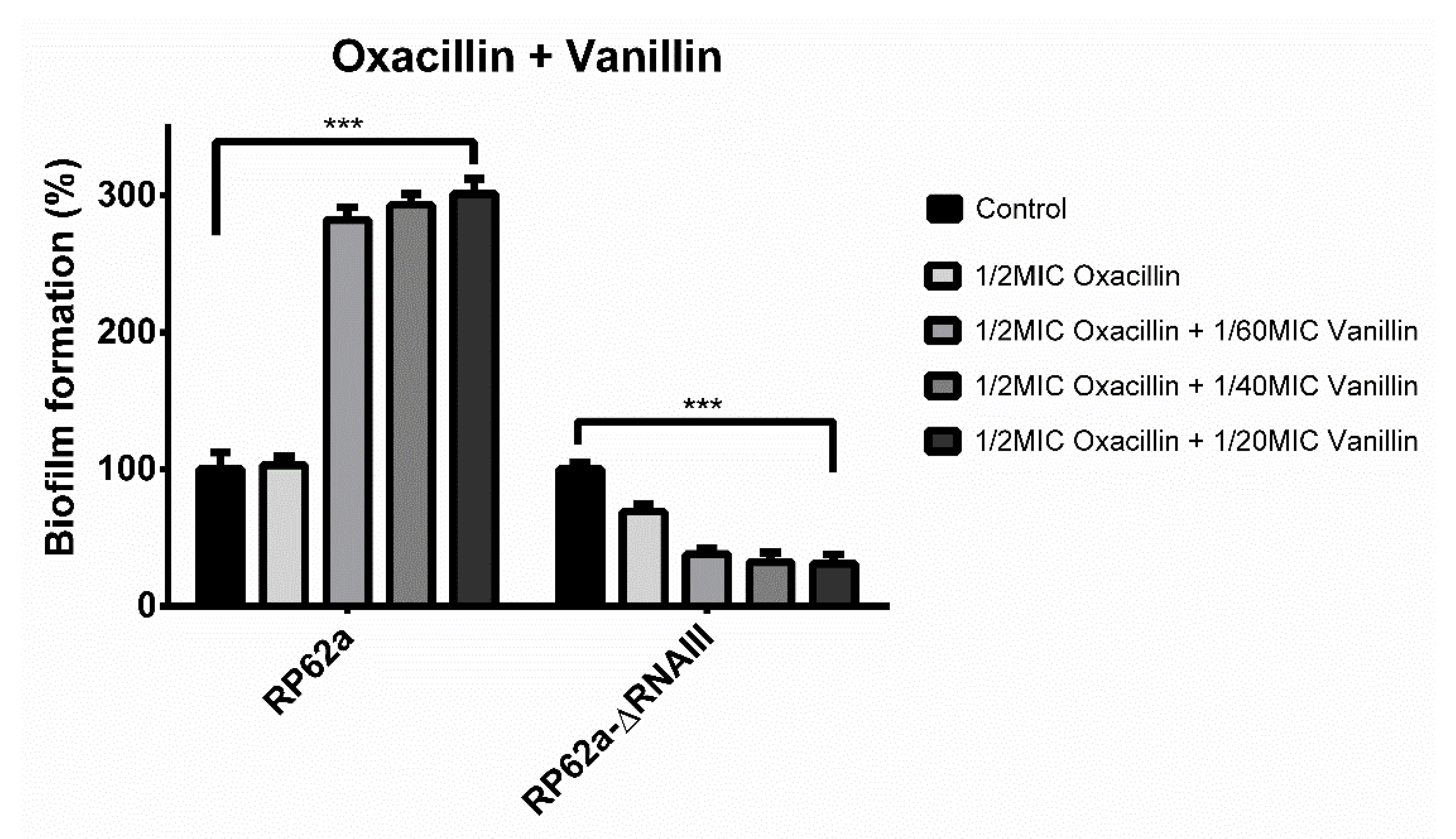
2.1. Biofilm Formation in Presence of Oxacillin and Phenolic Compounds
2.2. Construction of S. epidermidis RP62a-ΔRNAIII
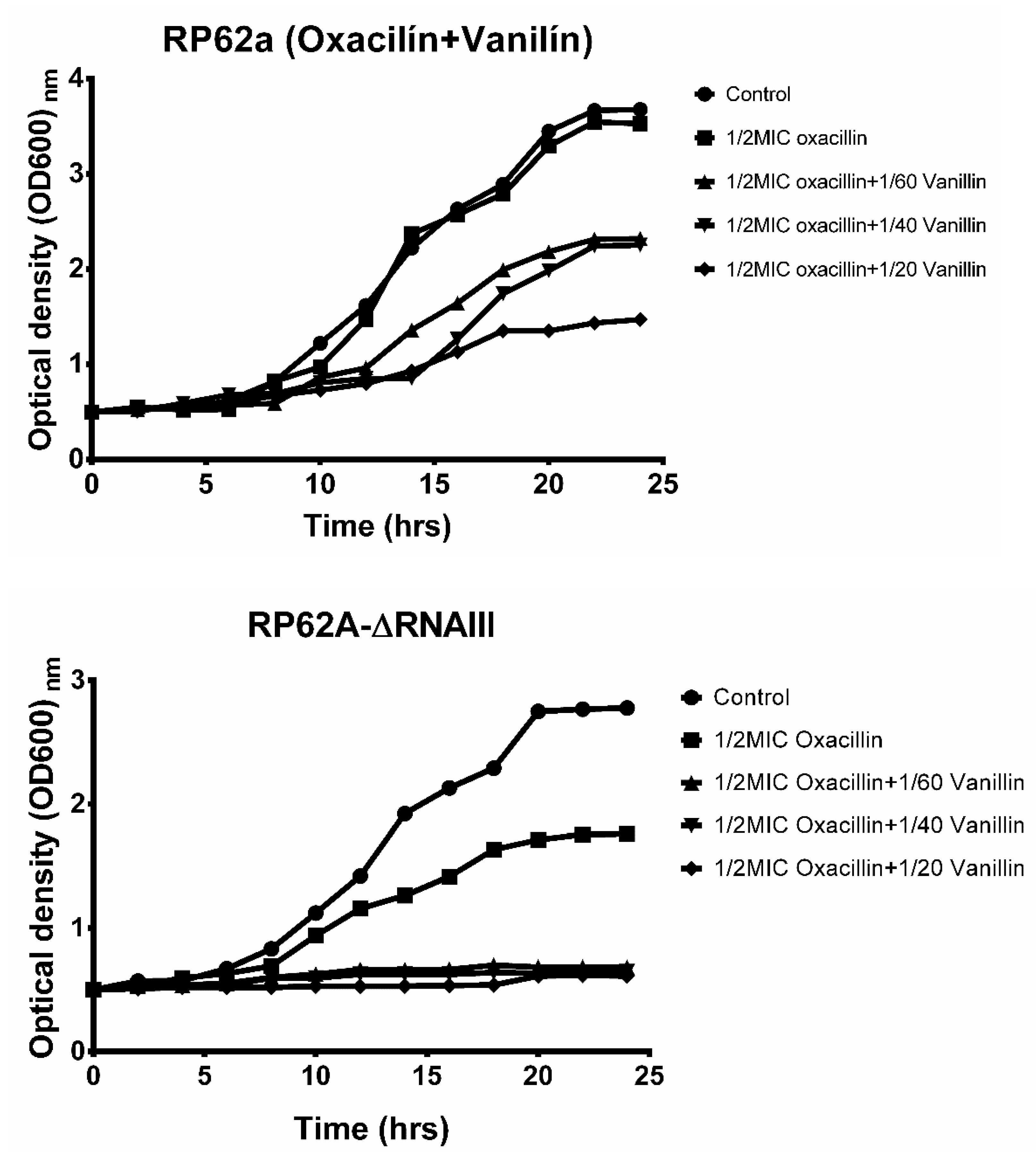
2.3. Effect of Vanillin and Oxacillin on S. epidermidis Biofilm
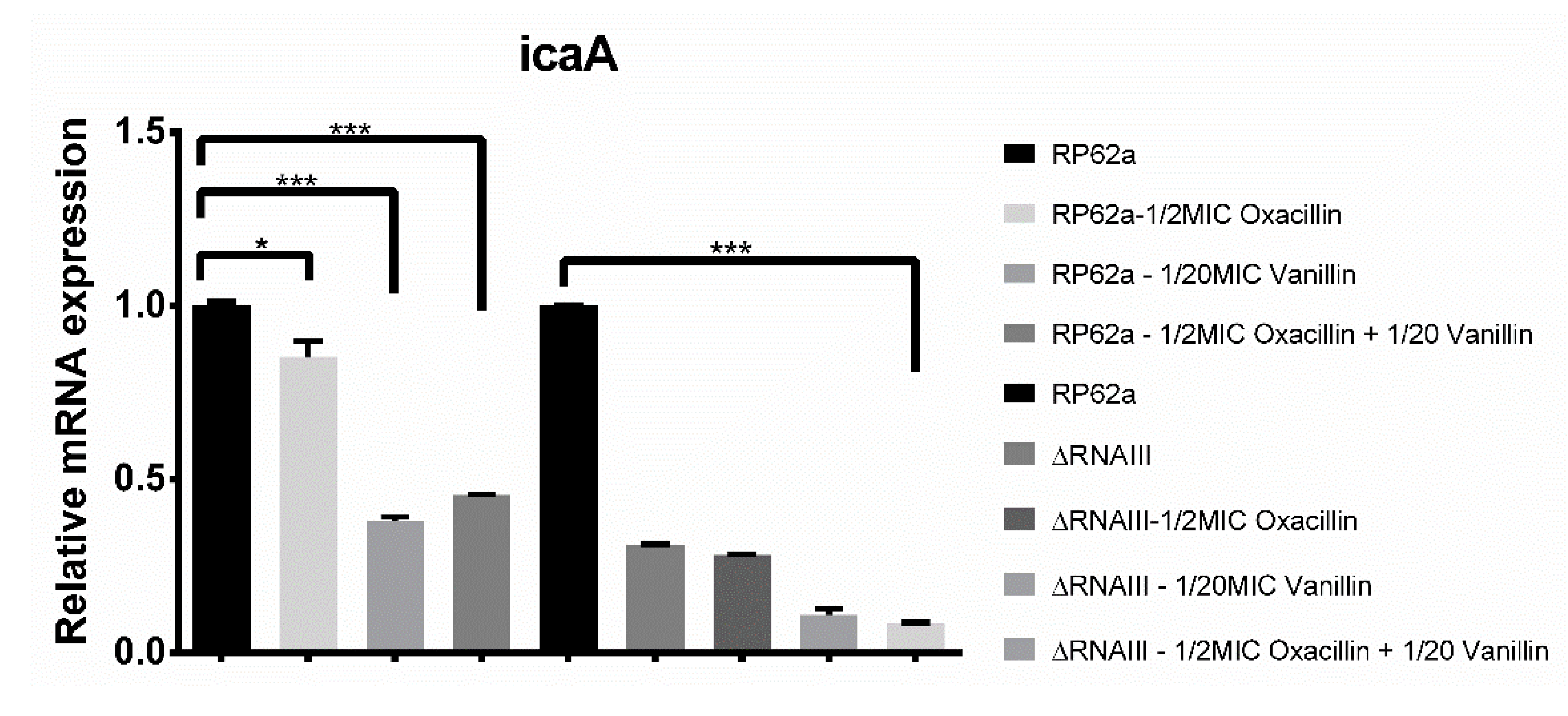
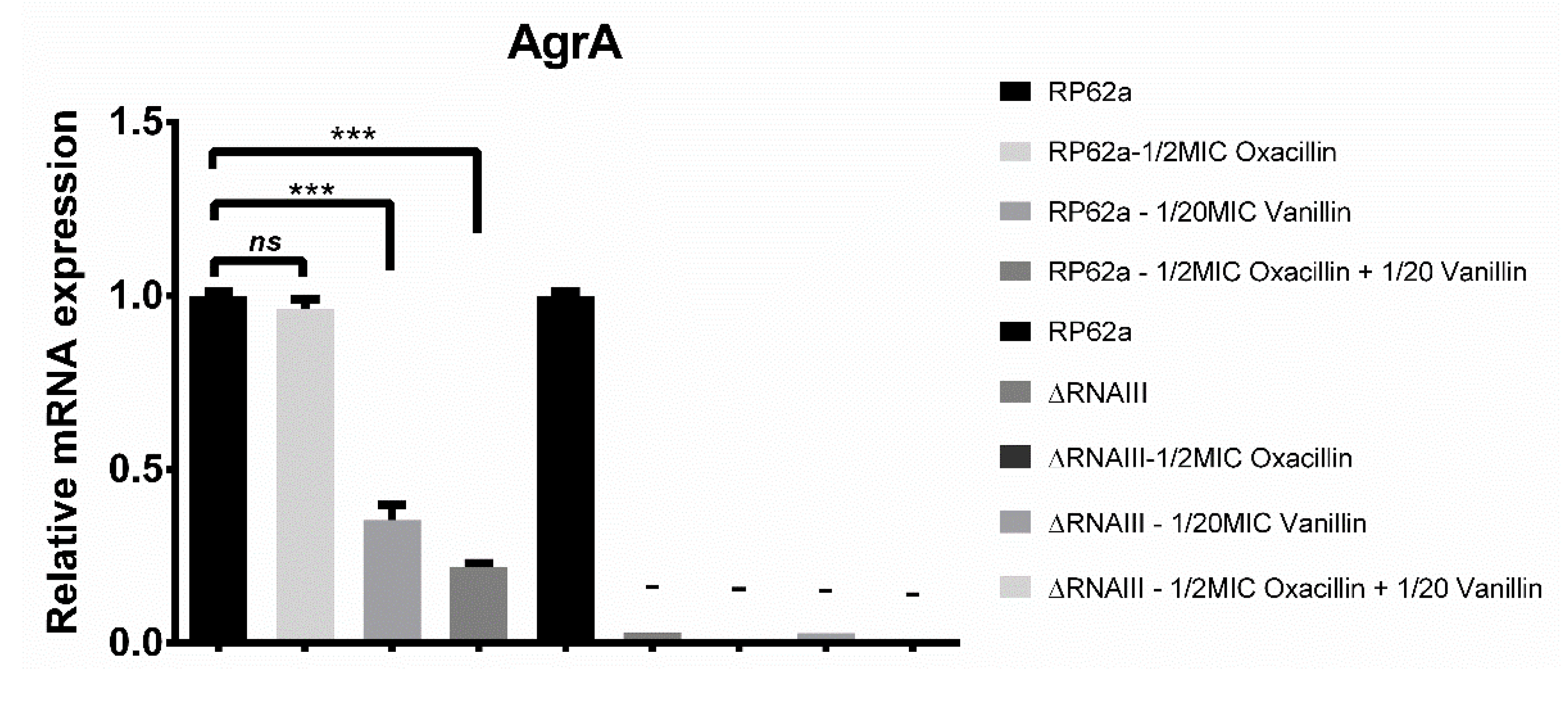
2.4. Real-Time qPCR Analysis of Biofilm-Associated Genes
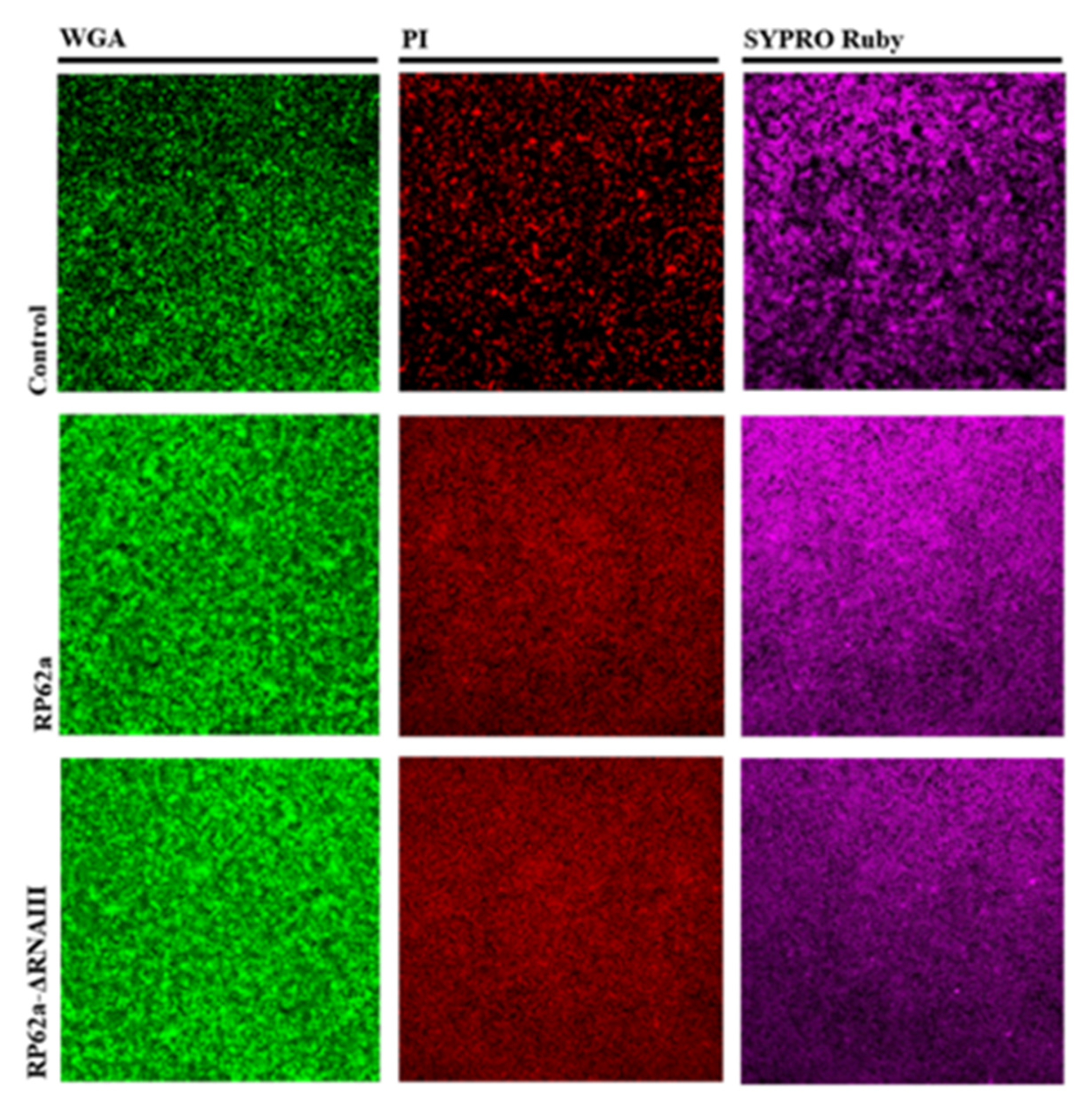
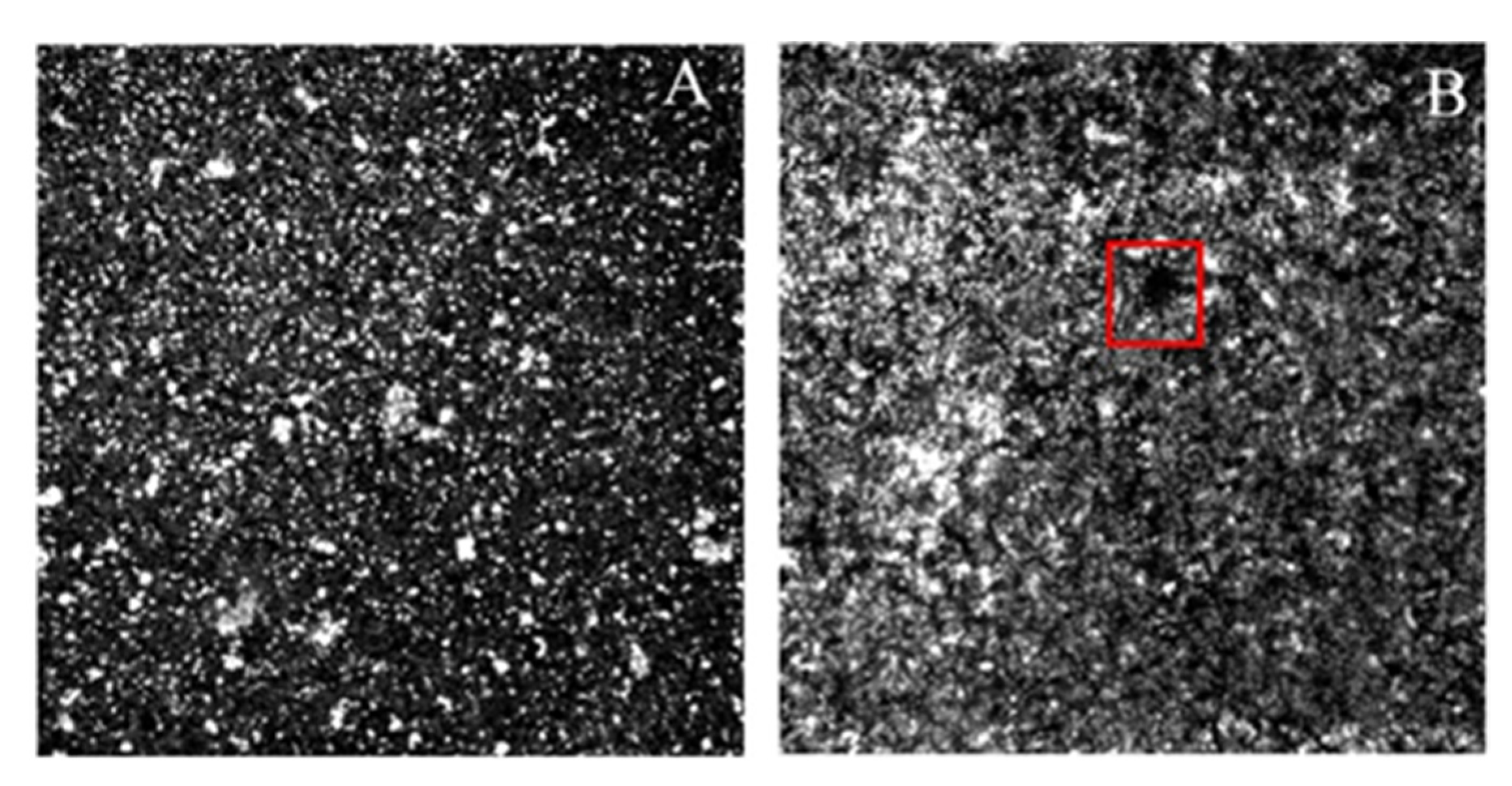
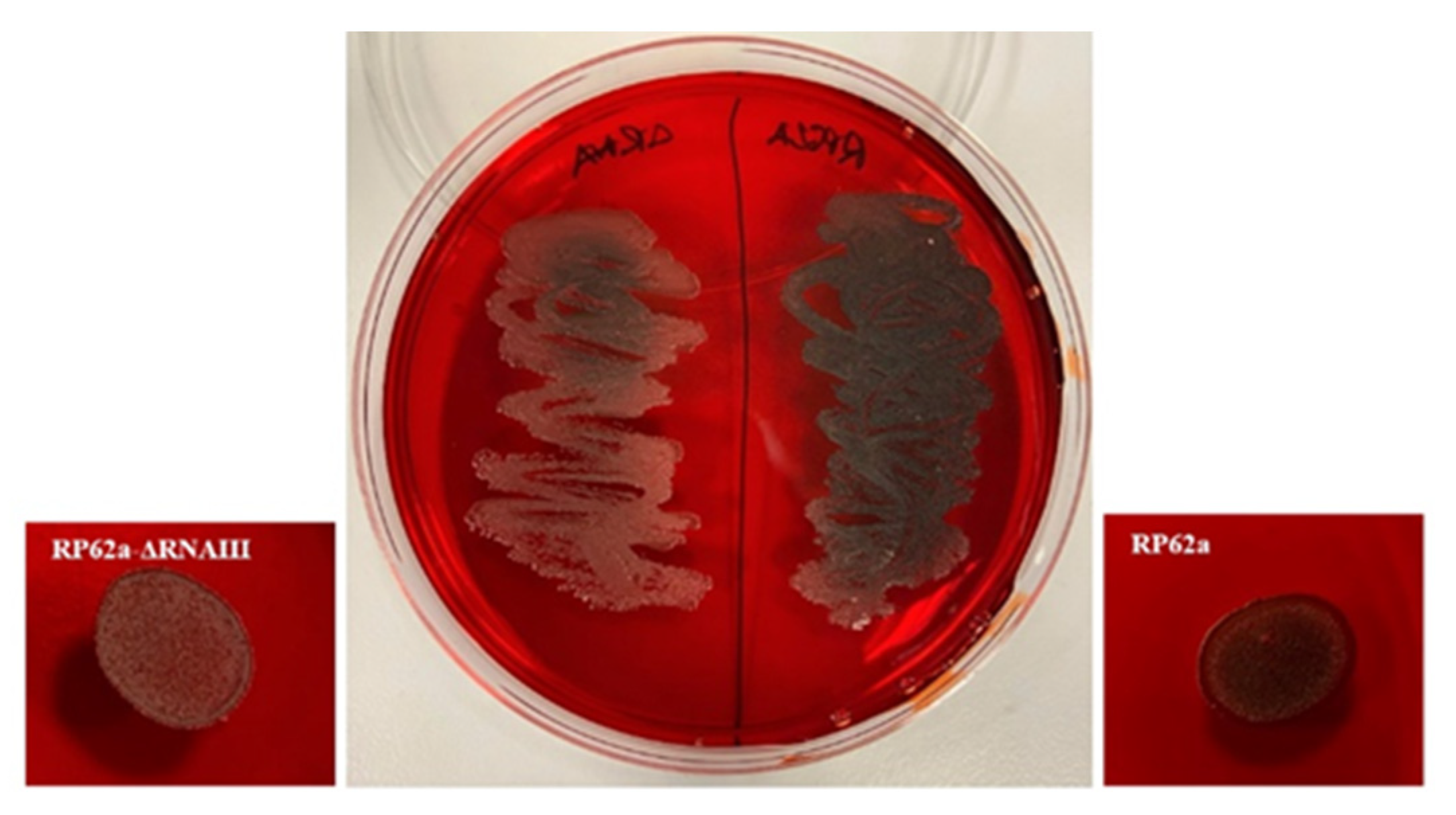
2.5. Analysis of Change in the Phenotype of the RNAIII-Deficient Strain
3. Discussion
4. Materials and Methods
4.1. Construction of S. epidermidis RP62a-ΔRNAIII
4.2. Bacterial Strains and Culture Conditions
4.3. Antimicrobial Agents and Antibiotics
4.4. Assessment of Biofilm Formation by Crystal Violet Staining
4.5. Determination of the Growth Curve of Treated and Untreated Strains
4.6. Confocal Laser Scanning Microscopy (CLSM)
4.7. Congo Red Agar Analysis
4.8. Phenol-Chloroform RNA Extraction
4.9. Real-Time qPCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hall-Stoodley, L.; Costerton, J.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Heilmann, C.; Ziebuhr, W.; Becker, K. Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infect. 2018, 29, 1198–1743. [Google Scholar] [CrossRef]
- N’Diaye, A.; Mijouin, L.; Hillion, M.; Diaz, S.; Konto-Ghiorghi, Y.; Percoco, G.; Chevalier, S.; Lefeuvre, L.; Harmer, N.J.; Lesouhaitier, O.; et al. Effect of substance P in StaphylocStaphylococcus epideroccus aureus and midis virulence: Implication for skin homeostasis. Front. Microbiol. 2016, 7, 506. [Google Scholar] [CrossRef]
- Cheung, A.L.; Projan, S.J. Clooning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 1994, 176, 4168–4172. [Google Scholar] [CrossRef]
- Beenken, K.E.; Dunman, P.M.; McAleese, F.; Macapagal, D.; Murphy, E.; Projan, S.J.; Blevins, J.S.; Smeltzer, M.S. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 2004, 86, 4665–4684. [Google Scholar] [CrossRef]
- Kornblum, J.; Kreiswirth, B.N.; Projan, S.J.; Ross, H.F.; Novick, R.P. Agr: A polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In Molecular Biology of the Staphylococci; Novick, R.P., Ed.; VCH Publishers: New York, NY, USA, 1990; pp. 373–402. [Google Scholar]
- Janzon, L.; Löfdahl, S.; Arvidson, S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Genet. Genom. 1989, 219, 480–485. [Google Scholar] [CrossRef]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007, 11, 1353–1366. [Google Scholar] [CrossRef]
- Saïd-Salim, B.; Dunman, P.M.; McAleese, F.M.; Macapagal, D.; Murphy, E.; McNamara, P.J.; Arvidson, S.; Foster, T.J.; Projan, S.J.; Kreiswirth, B.N. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 2003, 185, 610–619. [Google Scholar] [CrossRef]
- Zielinska, A.K.; Beenken, K.E.; Mrak, L.N.; Spencer, H.J.; Post, G.R.; Skinner, R.A.; Tackett, A.J.; Horswill, A.R.; Smeltzer, M.S. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol. Microbiol. 2012, 86, 1183–1196. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef]
- Surewaard, B.G.; Nijland, R.; Spaan, A.N.; Kruijtzer, J.A.; de Haas, C.J.; van Strijp, J.A. Inactivation of staphylococcal phenol soluble modulins by serum lipoprotein particles. PLoS Pathog. 2012, 8, e1002606. [Google Scholar] [CrossRef]
- Wang, R.; Khan, B.A.; Cheung, G.Y.; Bach, T.H.; Jameson-Lee, M.; Kong, K.F.; Queck, S.Y.; Otto, M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Investig. 2011, 121, 238–248. [Google Scholar] [CrossRef]
- Lázaro-Díez, M.; Remuzgo-Martínez, S.; Rodríguez-Mirones, C.; Acosta, F.; Icardo, J.M.; Martínez-Martínez, L.; Ramos-Vivas, J. Effects of Subinhibitory Concentrations of Ceftaroline on Methicillin-Resistant Staphylococcus aureus (MRSA) Biofilms. PLoS ONE 2016, 11, e0147569. [Google Scholar] [CrossRef]
- Akinbobola, A.B.; Sherry, L.; Mckay, W.G.; Ramage, G.; Williams, C. Tolerance of Pseudomonas aeruginosa in in-vitro biofilms to high-level peracetic acid disinfection. J. Hosp. Infect. 2017, 97, 162–168. [Google Scholar] [CrossRef]
- Gov, Y.; Bitler, A.; Dell’Acqua, G.; Torres, J.V.; Balaban, N. RNAIII inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: Structure and function analysis. Peptides 2001, 22, 1609–1620. [Google Scholar] [CrossRef]
- Walsh, D.J.; Livinghouse, T.; Durling, G.M.; Arnold, A.D.; Brasier, W.; Berry, L.; Goeres, D.M.; Stewart, P.S. Novel phenolic antimicrobials enhanced activity of iminodiacetate prodrugs against biofilm and planktonic bacteria. Chem. Biol. Drug Des. 2021, 97, 134–147. [Google Scholar] [CrossRef]
- Diniz-Silva, H.T.; Magnani, M.; de Siqueira, S.; de Souza, E.L.; de Siqueira Júnior, J.P. Fruit flavonoids as modulators of norfloxacin resistance in Staphylococcus aureus that overexpresses norA. LWT—Food Sci. Technol. 2017, 85, 324–326. [Google Scholar] [CrossRef]
- Minich, A.; Levarski, Z.; Mikulášová, M.; Straka, M.; Liptáková, A.; Stuchlík, S. Complex Analysis of Vanillin and Syringic Acid as Natural Antimicrobial Agents against Staphylococcus epidermidis Biofilms. Int. J. Mol. Sci. 2022, 23, 1816. [Google Scholar] [CrossRef]
- Monk, I.R.; Shah, I.M.; Xu, M.; Tan, M.W.; Foster, T.J. Transforming the untransformable: Application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 2012, 3, e00277-11. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal infections: Mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 2013, 64, 175–188. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Ehrlich, G.D.; Montanaro, L. Biofilm based implant infections in orthopaedics. Adv. Exp. Med. Biol. 2015, 830, 29–46. [Google Scholar]
- Sandasi, M.; Leonard, C.M.; Viljoen, A.M. The effect of five common essential oil components on Listeria monocytogenes biofilms. Food Control 2008, 19, 1070–1075. [Google Scholar] [CrossRef]
- Adonizio, A.; Kong, K.F.; Mathee, K. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrob. Agents Chemother. 2008, 52, 198–203. [Google Scholar] [CrossRef]
- Song, J.H.; Yang, T.C.; Chang, K.W.; Han, S.K.; Yi, H.K.; Jeon, J.G. In vitro effects of a fraction separated from Polygonum cuspidatum root on the viability, in suspension and biofilms, and biofilm formation of mutans streptococci. J. Ethnopharmacol. 2007, 112, 419–425. [Google Scholar] [CrossRef]
- Nostro, A.; Roccaro, A.S.; Bisignano, G.; Marino, A.; Cannatelli, M.A.; Pizzimenti, F.C.; Cioni, P.L.; Procopio, F.; Blanco, A.R. Effects of oregano, carvacrol and thymol on Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Med. Microbiol. 2007, 56 Pt 4, 519–523. [Google Scholar] [CrossRef]
- Miklasińska-Majdanik, M.; Kępa, M.; Wojtyczka, R.D.; Idzi, D.; Wąsik, T.J. Phenolic Compounds Diminish Antibiotic Resistance of Staphylococcus aureus Clinical Strains. Int. J. Environ. Res. Public Health 2018, 15, 2321. [Google Scholar] [CrossRef]
- Ndezo, B.B.; Kuaté, C.R.T.; Dzoyem, J.P. Synergistic Antibiofilm Efficacy of Thymol and Piperine in Combination with Three Aminoglycoside Antibiotics against Klebsiella pneumoniae Biofilms. Can. J. Infect. Dis. Med. Microbiol. 2021, 8, 7029944. [Google Scholar] [CrossRef]
- Penesyan, A.; Paulsen, I.T.; Gillings, M.R.; Kjelleberg, S.; Manefield, M.J. Secondary Effects of Antibiotics on Microbial Biofilms. Front. Microbiol. 2020, 11, 2109. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Xiao, K.; Li, B.; Jiang, W.; Peng, W.; Zheng, J.; Zhou, H. The combination of catechin and epicatechin gallate from Fructus Crataegi potentiates beta-lactam antibiotics against methicillin-resistant Staphylococcus aureus (MRSA) in vitro and in vivo. Int. J. Mol. Sci. 2013, 14, 1802–1821. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.; Moser, E.; Kaatz, G.W. Catechin gallates inhibit multidrug resistance (MDR) in Staphylococcus aureus. Planta Med. 2004, 70, 1240–1242. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Skindersoe, M.E.; Bjarnsholt, T.; Phipps, R.K.; Christensen, K.B.; Jensen, P.O. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 2005, 151, 1325–1340. [Google Scholar] [CrossRef] [PubMed]
- Plyuta, V.; Zaitseva, J.; Lobakova, E.; Zagoskina, N.; Kuznetsov, A.; Khmel, I. Effect of plant phenolic compounds on biofilm formation by Pseudomonas aeruginosa. APMIS 2013, 121, 1073–1081. [Google Scholar] [CrossRef]
- Lim, Y.; Jana, M.; Luong, T.T.; Lee, C.Y. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 2004, 186, 722–729. [Google Scholar] [CrossRef]
- You, Y.; Xue, T.; Cao, L.; Zhao, L.; Sun, H.; Sun, B. Staphylococcus aureus glucose-induced biofilm accessory proteins, GbaAB, influence biofilm formation in a PIA-dependent manner. Int. J. Med. Microbiol. 2014, 304, 603–612. [Google Scholar] [CrossRef]
- Topaz, M.; Athamna, A.; Ashkenazi, I.; Shpitz, B.; Freimann, S. In-vitro model for bacterial growth inhibition of compartmentalized infection treated by an ultra-high concentration of antibiotics. PLoS ONE 2021, 16, e0252724. [Google Scholar] [CrossRef]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef]
- Kaplan, J.B. Antibiotic-induced biofilm formation. Int. J. Artif. Organs 2011, 34, 737–751. [Google Scholar] [CrossRef]
- Gupta, R.K.; Luong, T.T.; Lee, C.Y. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 14036–14041. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Horswill, A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef] [PubMed]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.H.; Carter, G.P.; Pidot, S.J.; Guérillot, R.; Seemann, T.; Gonçalves da Silva, A.; Foster, T.J.; Howden, B.P.; Stinear, T.P.; Monk, I.R. Mining the Methylome Reveals Extensive Diversity in Staphylococcus epidermidis Restriction Modification. MBio 2019, 10, e02451-19. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, S.; Joo, H.S.; Duong, A.C.; Bach, T.H.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Kaiser, T.D.; Pereira, E.M.; Dos Santos, K.R.; Maciel, E.L.; Schuenck, R.P.; Nunes, A.P. Modification of the Congo red agar method to detect biofilm production by Staphylococcus epidermidis. Diagn. Microbiol. Infect. Dis. 2013, 75, 235–239. [Google Scholar] [CrossRef]
- Löfblom, J.; Kronqvist, N.; Uhlén, M.; Ståhl, S.; Wernérus, H. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J. Appl. Microbiol. 2007, 102, 736–747. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minich, A.; Lišková, V.; Kormanová, Ľ.; Krahulec, J.; Šarkanová, J.; Mikulášová, M.; Levarski, Z.; Stuchlík, S. Role of RNAIII in Resistance to Antibiotics and Antimicrobial Agents in Staphylococcus epidermidis Biofilms. Int. J. Mol. Sci. 2022, 23, 11094. https://doi.org/10.3390/ijms231911094
Minich A, Lišková V, Kormanová Ľ, Krahulec J, Šarkanová J, Mikulášová M, Levarski Z, Stuchlík S. Role of RNAIII in Resistance to Antibiotics and Antimicrobial Agents in Staphylococcus epidermidis Biofilms. International Journal of Molecular Sciences. 2022; 23(19):11094. https://doi.org/10.3390/ijms231911094
Chicago/Turabian StyleMinich, Andrej, Veronika Lišková, Ľubica Kormanová, Ján Krahulec, Júlia Šarkanová, Mária Mikulášová, Zdenko Levarski, and Stanislav Stuchlík. 2022. "Role of RNAIII in Resistance to Antibiotics and Antimicrobial Agents in Staphylococcus epidermidis Biofilms" International Journal of Molecular Sciences 23, no. 19: 11094. https://doi.org/10.3390/ijms231911094
APA StyleMinich, A., Lišková, V., Kormanová, Ľ., Krahulec, J., Šarkanová, J., Mikulášová, M., Levarski, Z., & Stuchlík, S. (2022). Role of RNAIII in Resistance to Antibiotics and Antimicrobial Agents in Staphylococcus epidermidis Biofilms. International Journal of Molecular Sciences, 23(19), 11094. https://doi.org/10.3390/ijms231911094







