DRB2 Modulates Leaf Rolling by Regulating Accumulation of MicroRNAs Related to Leaf Development in Rice
Abstract
:1. Introduction
2. Results
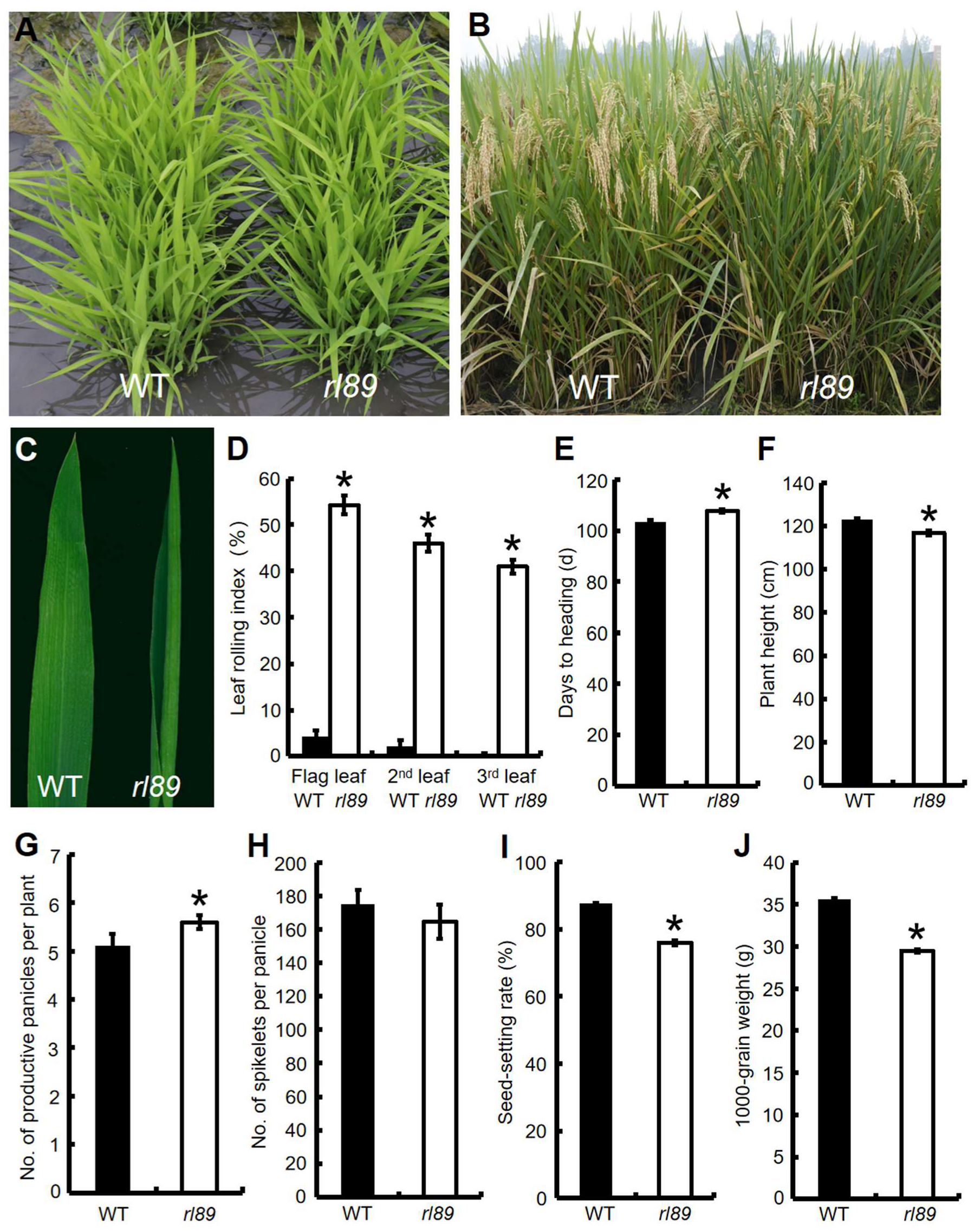
2.1. Phenotypes of rl89 Mutant
2.2. The rl89 Locus Was Mapped to a Putative Gene Encoding DRB2
2.3. Functional Confirmation of OsDRB2 through Complementation and Knockout Assays
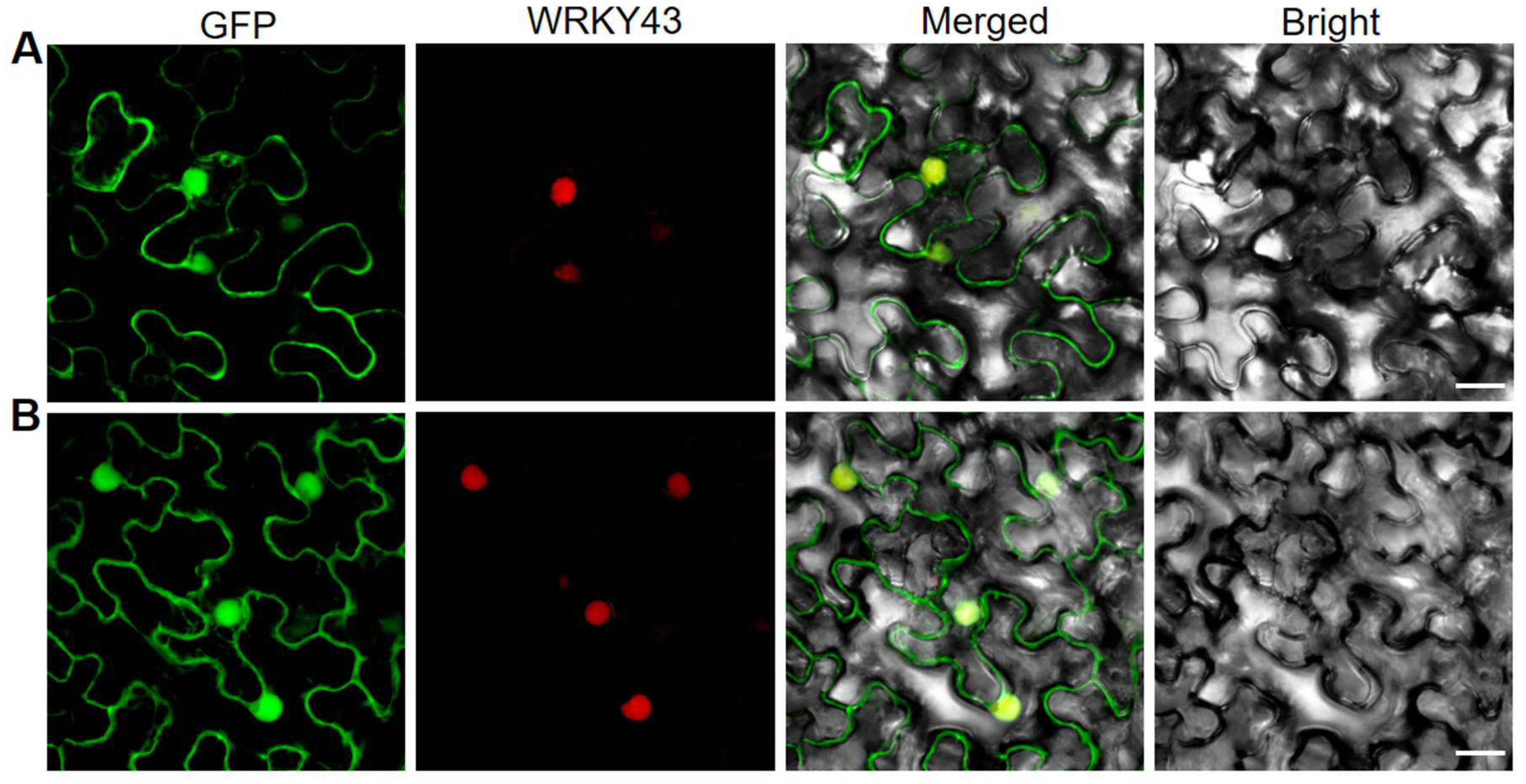
2.4. Subcellular Localization of OsDRB2 Protein
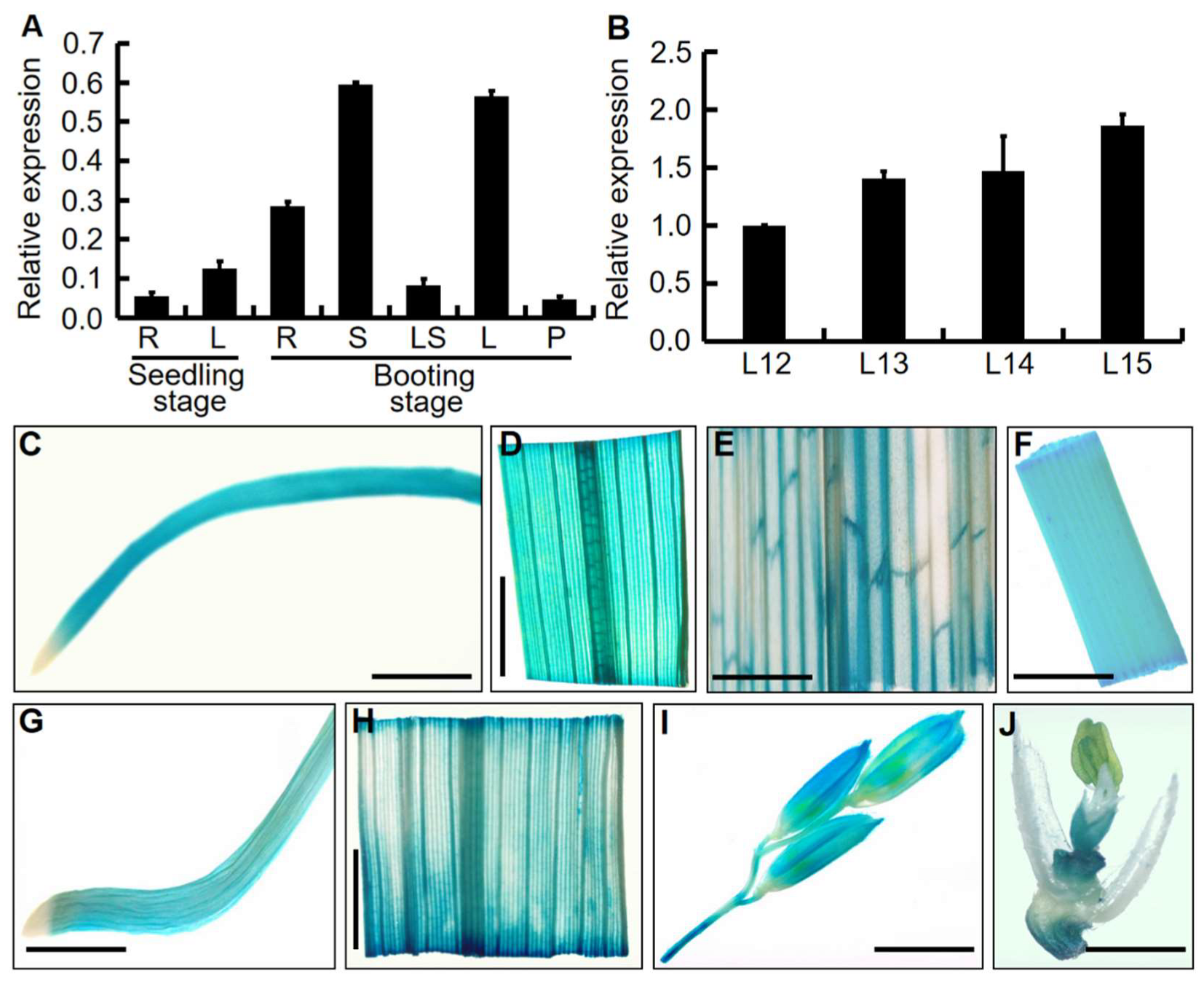
2.5. Expression Analyses of the OsDRB2 Gene
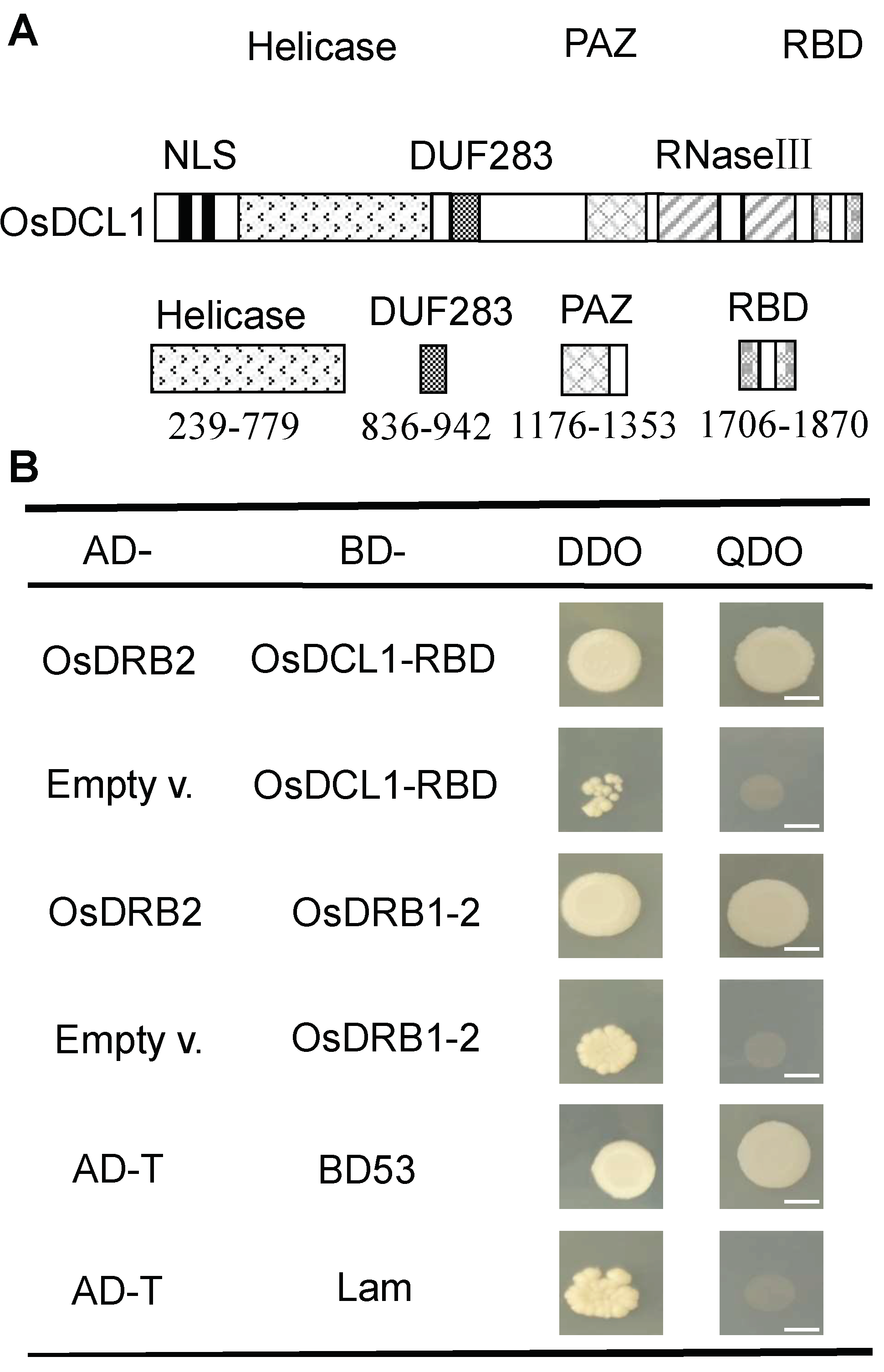
2.6. Protein Interaction of OsDRB2 with OsDCL1 and OsDRB1-2
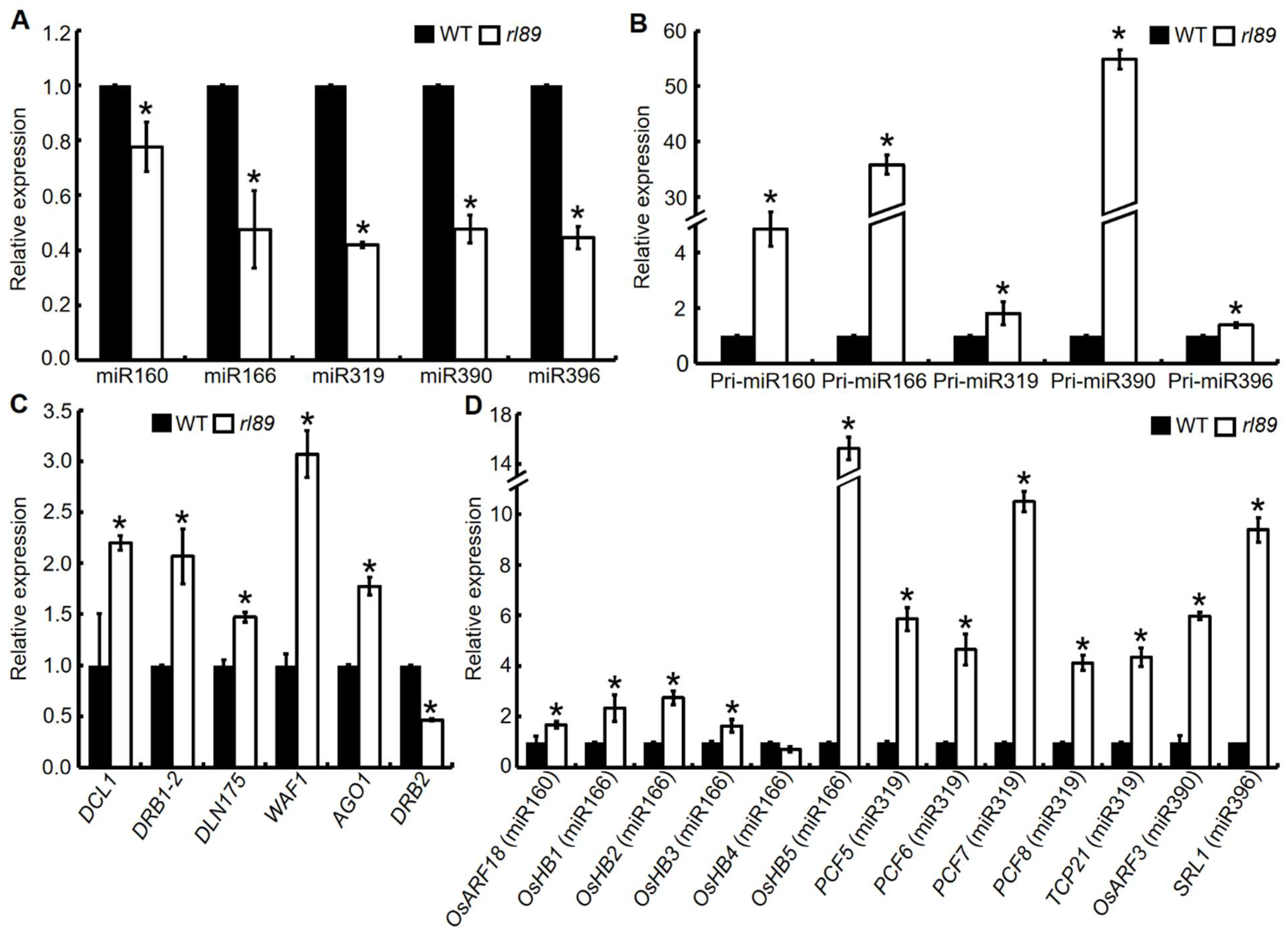
2.7. Expression Analyses of miRNAs and Their Target Genes Associated with Leaf Development
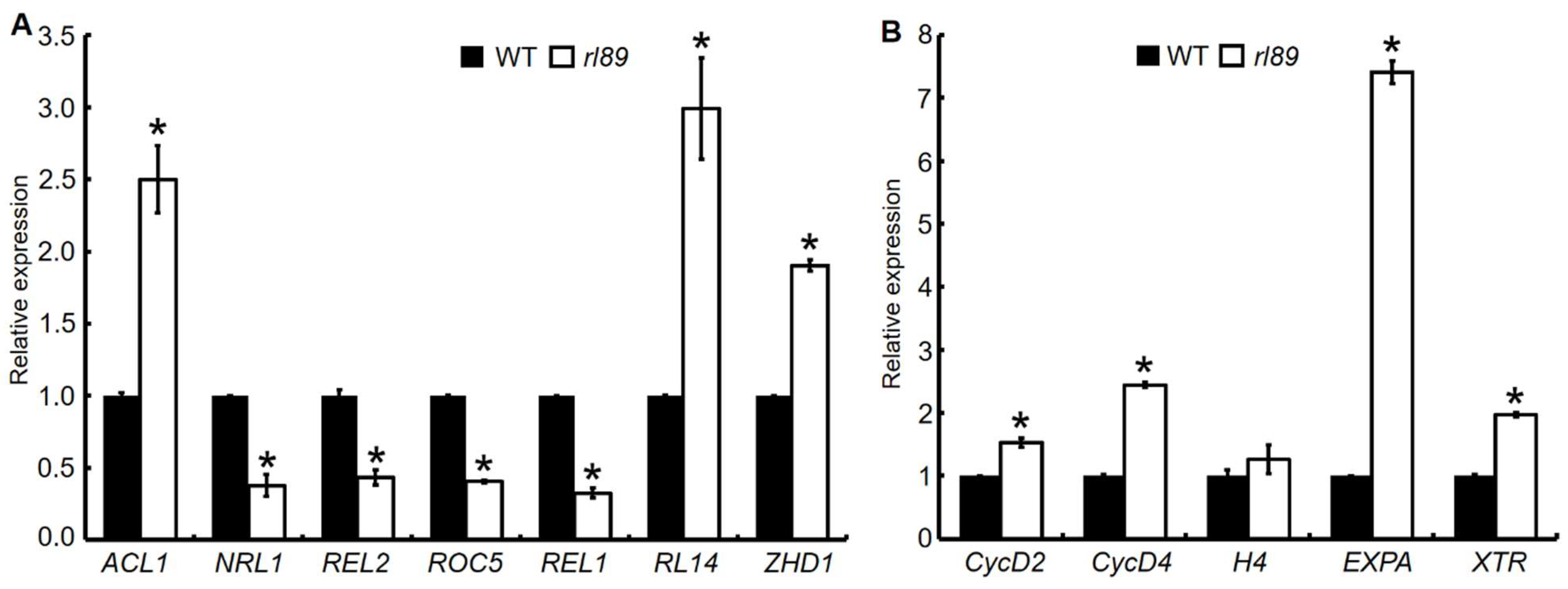
2.8. Expression Analyses of Some Genes Regulating Leaf Shape and Cell Growth
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Photosynthetic Parameter and Leaf-rolling Index (LRI)
4.3. Measurement of Photosynthetic Pigments
4.4. Histology Analysis
4.5. MutMap Analysis and Marker Development
4.6. Sequence Analysis
4.7. Vector Construction and Rice Transformation
4.8. Histochemical GUS Assay
4.9. Subcellular Localization
4.10. Yeast-Two-Hybrid (Y2H) Assay
4.11. qRT-PCR Analyses
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lang, Y.Z.; Zhang, Z.J.; Gu, X.Y.; Yang, J.C.; Zhu, Q.S. Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) I. Leaf orientation, canopy structure and light distribution. Acta Agron. Sin. 2004, 30, 806–810. [Google Scholar]
- Xiang, J.J.; Zhang, G.H.; Qian, Q.; Xue, H.W. SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012, 159, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Q.; Li, W.Q.; Miao, H.; Gan, P.F.; Qiao, L.; Chang, Y.L.; Shi, C.H.; Chen, K.M. REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice 2016, 9, 37. [Google Scholar] [CrossRef]
- Yang, C.H.; Li, D.Y.; Liu, X.; Ji, C.J.; Hao, L.L.; Zhao, X.F.; Li, X.B.; Chen, C.Y.; Cheng, Z.K.; Zhu, L.H. OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol. 2014, 14, 158. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Li, M.; Liu, K.; Tang, D.; Sun, M.F.; Li, Y.F.; Shen, Y.; Du, G.J.; Cheng, Z.K. Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 2016, 67, 2139–2150. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Q.; Zhang, M.J.; Gan, P.F.; Qiao, L.; Yang, S.Q.; Miao, H.; Wang, G.F.; Zhang, M.M.; Liu, W.T.; Li, H.F.; et al. CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 2017, 92, 904–923. [Google Scholar] [CrossRef]
- Fang, J.J.; Guo, T.T.; Xie, Z.W.; Chun, Y.; Zhao, J.F.; Peng, L.X.; Zafar, S.A.; Yuan, S.J.; Xiao, L.T.; Li, X.Y. The URL1-ROC5-TPL2 transcriptional repressor complex represses the ACL1 gene to modulate leaf rolling in rice. Plant Physiol. 2021, 185, 1722–1744. [Google Scholar] [CrossRef]
- Zou, L.P.; Sun, X.H.; Zhang, Z.G.; Liu, P.; Wu, J.X.; Tian, C.J.; Qiu, J.L.; Lu, T.G. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 2011, 156, 1589–1602. [Google Scholar] [CrossRef]
- Li, L.; Shi, Z.Y.; Li, L.; Shen, G.Z.; Wang, X.Q.; An, L.S.; Zhang, J.L. Overexpression of ACL1 (abaxially curled leaf1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant 2010, 3, 807–817. [Google Scholar] [CrossRef]
- Cho, S.H.; Yoo, S.C.; Zhang, H.T.; Pandeya, D.; Koh, H.J.; Hwang, J.Y.; Kim, G.T.; Paek, N.C. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 2013, 198, 1071–1084. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.H.; Long, Q.Z.; Huang, J.X.; Wang, Y.L.; Zhou, K.N.; Zheng, M.; Sun, J.; Chen, H.; Chen, S.H.; et al. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 2014, 239, 803–816. [Google Scholar] [CrossRef]
- Chen, Q.L.; Xie, Q.J.; Gao, J.; Wang, W.Y.; Sun, B.; Liu, B.H.; Zhu, H.T.; Peng, H.F.; Zhao, H.B.; Liu, C.H.; et al. Characterization of Rolled and Erect Leaf1 in regulating leave morphology in rice. J. Exp. Bot. 2015, 66, 6047–6058. [Google Scholar] [CrossRef]
- Fang, L.K.; Zhao, F.M.; Cong, Y.F.; Sang, X.C.; Du, Q.; Wang, D.Z.; Li, Y.F.; Ling, Y.H.; Yang, Z.L.; He, G.H. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 2012, 10, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.H.; Hou, X.; Wang, L.; Xu, J.; Chen, J.; Fu, X.; Shen, N.W.; Nian, J.Q.; Jiang, Z.Z.; Hu, J.; et al. PHOTO-SENSITIVE LEAF ROLLING 1 encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice. New Phytol. 2021, 229, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiong, G.Y.; Li, R.; Cui, J.J.; Tang, D.; Zhang, B.C.; Pauly, M.; Cheng, Z.K.; Zhou, Y.H. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009, 60, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, L.; Zeng, D.L.; Gao, Z.Y.; Guo, L.B.; Fang, Y.X.; Zhang, G.H.; Dong, G.J.; Yan, M.X.; Liu, J.; et al. Identification and characterization of NARROW AND ROLLED LEAF1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010, 73, 283–292. [Google Scholar] [CrossRef]
- Zhang, G.H.; Xu, Q.; Zhu, X.D.; Qian, Q.; Xue, H.W. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 2009, 21, 719–735. [Google Scholar] [CrossRef]
- Wu, R.H.; Li, S.B.; He, S.; Wassmann, F.; Yu, C.H.; Qin, G.J.; Schreiber, L.; Qu, L.J.; Gu, H.Y. CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 2011, 23, 3392–3411. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Reis, R.S.; Eamens, A.L.; Waterhouse, P.M. Missing pieces in the puzzle of plant microRNAs. Trends Plant Sci. 2015, 20, 721–728. [Google Scholar] [CrossRef]
- Reis, R.S.; Hart-Smith, G.; Eamens, A.L.; Wilkins, M.R.; Waterhouse, P.M. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat. Plants 2015, 1, 14027. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dong, W.G.; Ren, W.Q.; Zhao, Q.X.; Wu, F.J.; He, Y.K. Cytoplasmic HYL1 modulates miRNA-mediated translational repression. Plant Cell 2021, 33, 1980–1996. [Google Scholar] [CrossRef] [PubMed]
- Lobbes, D.; Rallapalli, G.; Schmidt, D.D.; Martin, C.; Clarke, J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006, 7, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Wagner, D.R. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 2001, 13, 1263–1279. [Google Scholar] [CrossRef]
- Schauer, S.E.; Jacobsen, S.E.; Meinke, D.W.; Ray, A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002, 7, 487–491. [Google Scholar] [CrossRef]
- Vazquez, F.; Gasciolli, V.; Crété, P.; Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004, 14, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Grigg, S.P.; Canales, C.; Hay, A.; Tsiantis, M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 2005, 437, 1022–1026. [Google Scholar] [CrossRef] [PubMed]
- Eamens, A.L.; Kim, K.W.; Curtin, S.J.; Waterhouse, P.M. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS ONE 2012, 7, e35933. [Google Scholar] [CrossRef]
- Curtin, S.J.; Xiong, Y.; Michno, J.M.; Campbell, B.W.; Stec, A.O.; Čermák, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef]
- Ma, X.L.; Zhang, Q.Y.; Zhu, Q.L.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.F.; Li, H.Y.; Lin, Y.R.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Clavel, M.; Pélissier, T.; Descombin, J.; Jean, V.; Picart, C.; Charbonel, C.; Saez-Vásquez, J.; Bousquet-Antonelli, C.; Deragon, J.M. Parallel action of AtDRB2 and RdDM in the control of transposable element expression. BMC Plant Biol. 2015, 15, 70. [Google Scholar] [CrossRef] [Green Version]
- Hiraguri, A.; Itoh, R.; Kondo, N.; Nomura, Y.; Aizawa, D.; Murai, Y.; Koiwa, H.; Seki, M.; Shinozaki, K.; Fukuhara, T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 173–188. [Google Scholar] [CrossRef]
- Raghuram, B.; Sheikh, A.H.; Rustagi, Y.; Sinha, A.K. MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 2015, 282, 521–536. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, Z.Y.; Zhao, D.Z. Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 2016, 6, 29938. [Google Scholar] [CrossRef]
- Li, Y.H.; Yang, Y.Q.; Liu, Y.; Li, D.X.; Zhao, Y.H.; Li, Z.J.; Liu, Y.; Jiang, D.G.; Li, J.; Zhou, H.; et al. Overexpression of OsAGO1b induces adaxially rolled leaves by affecting leaf abaxial sclerenchymatous cell development in rice. Rice 2019, 12, 60. [Google Scholar] [CrossRef]
- Han, M.H.; Goud, S.; Song, L.; Fedoroff, N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 2004, 101, 1093–1098. [Google Scholar] [CrossRef]
- Itoh, J.I.; Hibara, K.I.; Sato, Y.; Nagato, Y. Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.S.; Zhang, H.; Srivastava, A.K.; Pan, Y.J.; Bai, J.J.; Fang, J.J.; Shi, H.Z.; Zhu, J.K. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.L.; Tang, D.Y.; Yang, Y.Z.; Li, Y.X.; Wang, W.W.; Lu, H.; Liu, X.M.; Lin, J.Z. Preliminary study on the rice OsYABBY6 gene involving in the regulation of leaf development. Life Sci. Res. 2017, 21, 23–31. [Google Scholar]
- Liu, B.; Li, P.C.; Li, X.; Liu, C.Y.; Cao, S.Y.; Chu, C.C.; Cao, X.F. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005, 139, 296–305. [Google Scholar] [CrossRef]
- Abe, M.; Yoshikawa, T.; Nosaka, M.; Sakakibara, H.; Sato, Y.; Nagato, Y.; Itoh, J.I. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining microRNA and transacting small interfering RNA accumulation in rice. Plant Physiol. 2010, 154, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juarez, M.T.; Kui, J.S.; Thomas, J.; Heller, B.A.; Timmermans, M.C.P. MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004, 428, 84–88. [Google Scholar] [CrossRef]
- Zhang, T.; You, J.; Zhang, Y.; Yao, W.Y.; Chen, W.B.; Duan, Q.N.; Xiao, W.W.; Ye, L.; Zhou, Y.; Sang, X.C.; et al. LF1 regulates the lateral organs polarity development in rice. New Phytol. 2021, 231, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Hart-Smith, G.; Eamens, A.L.; Wilkins, M.R.; Waterhouse, P.M. MicroRNA regulatory mechanisms play different roles in Arabidopsis. J. Proteome Res. 2015, 14, 4743–4751. [Google Scholar] [CrossRef]
- Sun, Y.J.; Ma, J.; Sun, Y.Y.; Xu, H.; Yang, Z.Y.; Liu, S.J.; Jia, X.W.; Zheng, H.Z. The effects of different water and nitrogen managements on yield and nitrogen use efficiency in hybrid rice of China. Field Crop Res. 2012, 127, 85–98. [Google Scholar] [CrossRef]
- Zhu, D.F.; Lin, X.Q.; Cao, W.X. Comparison of leaf photosynthetic characteristics among rice hybrids with different leaf rolling index. Acta Agron. Sin. 2001, 27, 329–333. [Google Scholar]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef]
- Chen, X.S.; Li, T.T.; Zhou, S.L.; Zhao, Y. Transient expression of exogenous protein in tobacco leaves. Bio-101 2018, e1010127. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Z.; Pan, J.; Chen, C.; Tang, Y.; Zhang, H.; Guo, J.; Yang, X.; Chen, L.; Li, C.; Zhao, K.; et al. DRB2 Modulates Leaf Rolling by Regulating Accumulation of MicroRNAs Related to Leaf Development in Rice. Int. J. Mol. Sci. 2022, 23, 11147. https://doi.org/10.3390/ijms231911147
Yuan Z, Pan J, Chen C, Tang Y, Zhang H, Guo J, Yang X, Chen L, Li C, Zhao K, et al. DRB2 Modulates Leaf Rolling by Regulating Accumulation of MicroRNAs Related to Leaf Development in Rice. International Journal of Molecular Sciences. 2022; 23(19):11147. https://doi.org/10.3390/ijms231911147
Chicago/Turabian StyleYuan, Zhaodi, Jihong Pan, Congping Chen, Yulin Tang, Hongshan Zhang, Jia Guo, Xiaorong Yang, Longfei Chen, Chunyan Li, Ke Zhao, and et al. 2022. "DRB2 Modulates Leaf Rolling by Regulating Accumulation of MicroRNAs Related to Leaf Development in Rice" International Journal of Molecular Sciences 23, no. 19: 11147. https://doi.org/10.3390/ijms231911147




