An Amidase Contributes to Full Virulence of Sclerotinia sclerotiorum
Abstract
1. Introduction
2. Results
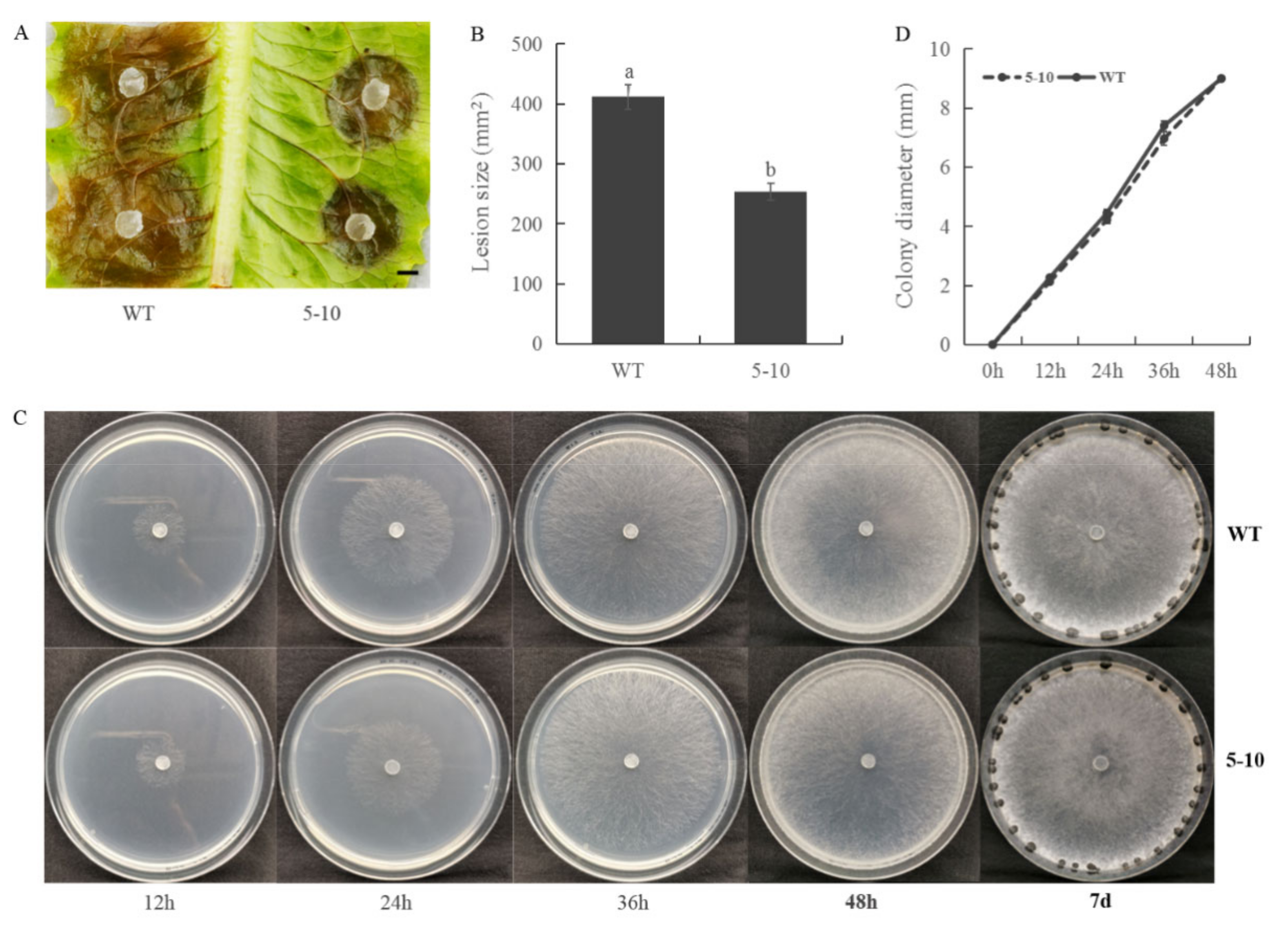
2.1. Screening for S. sclerotiorum Mutants with Reduced Virulence
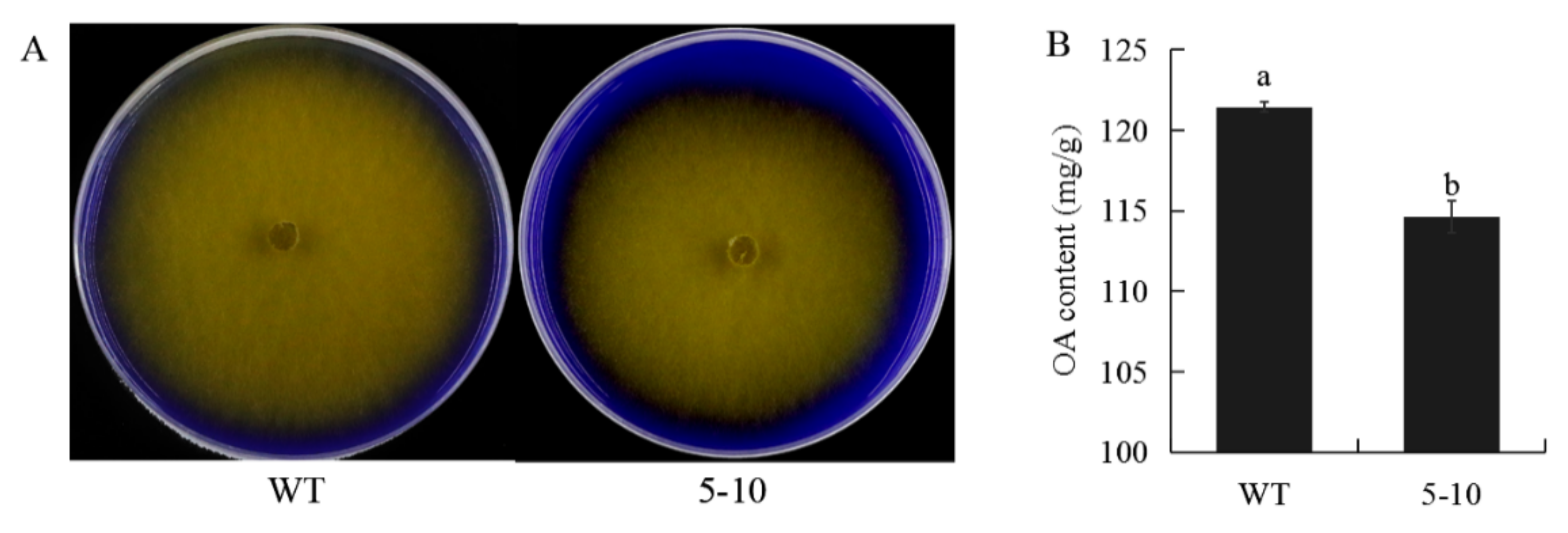
2.2. Less Oxalic Acid Is Observed in 5–10 Mutant
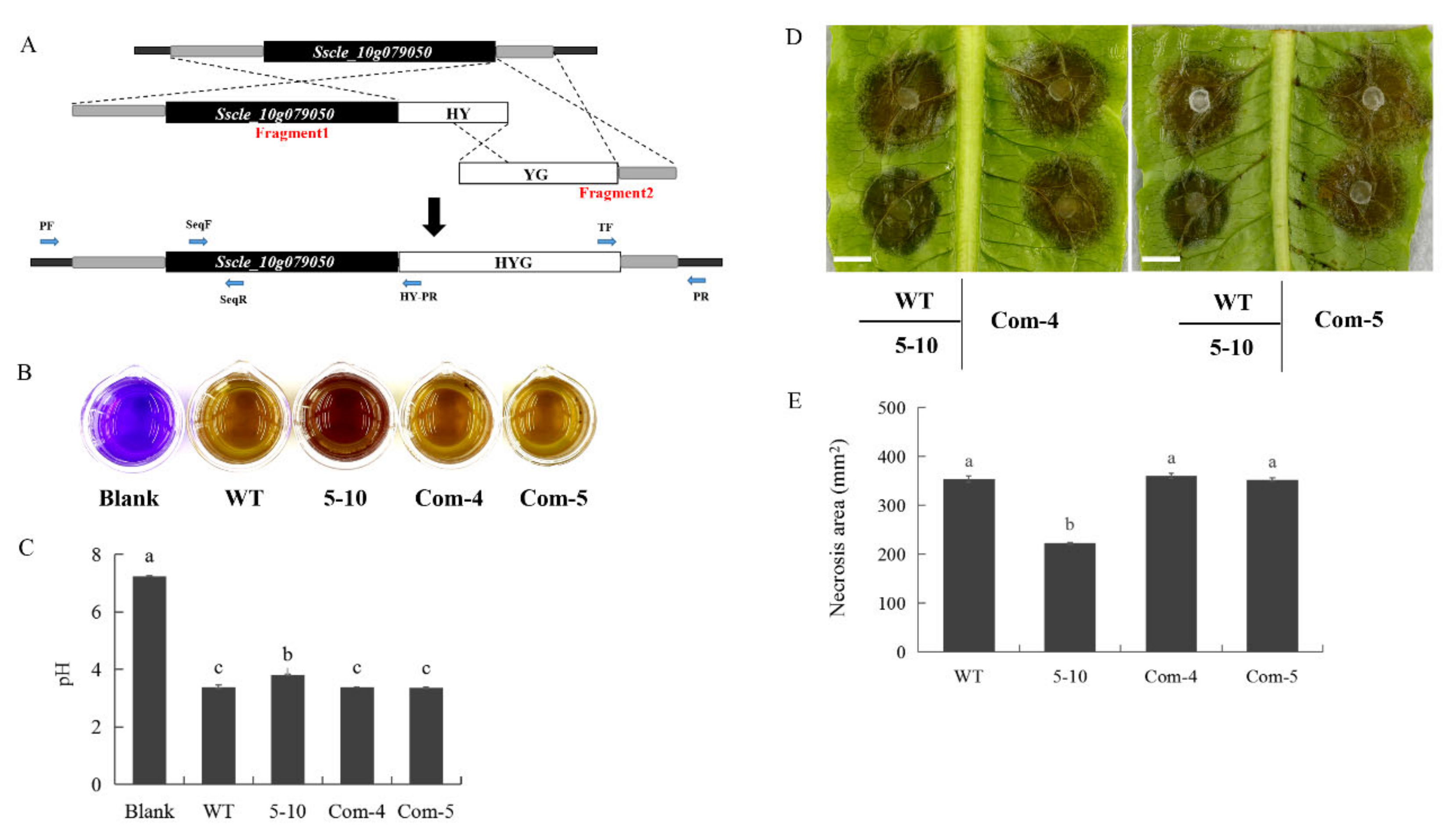
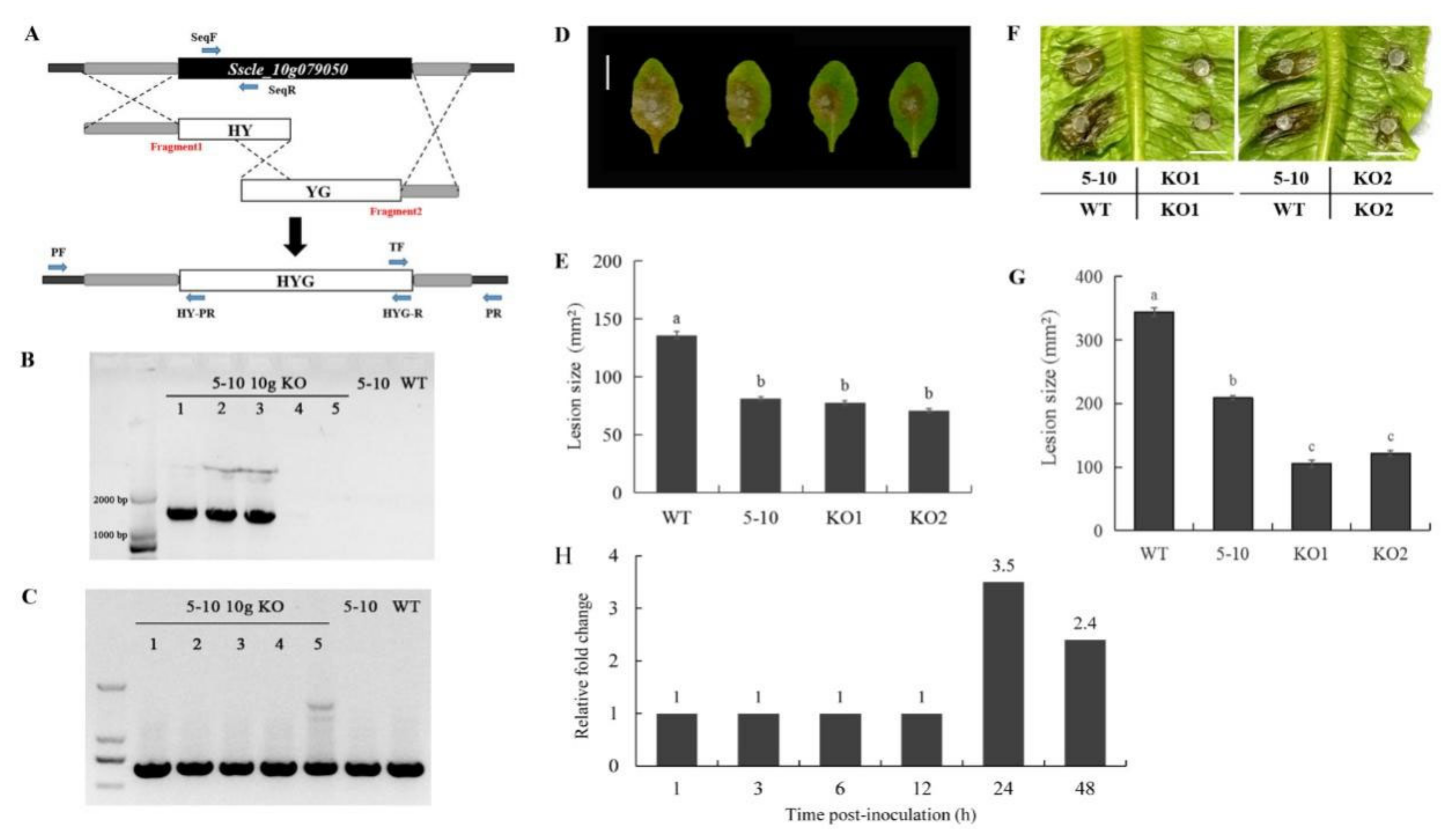
2.3. Mutation in Sscle_10g079050 Is Responsible for the Mutant Phenotypes of 5–10
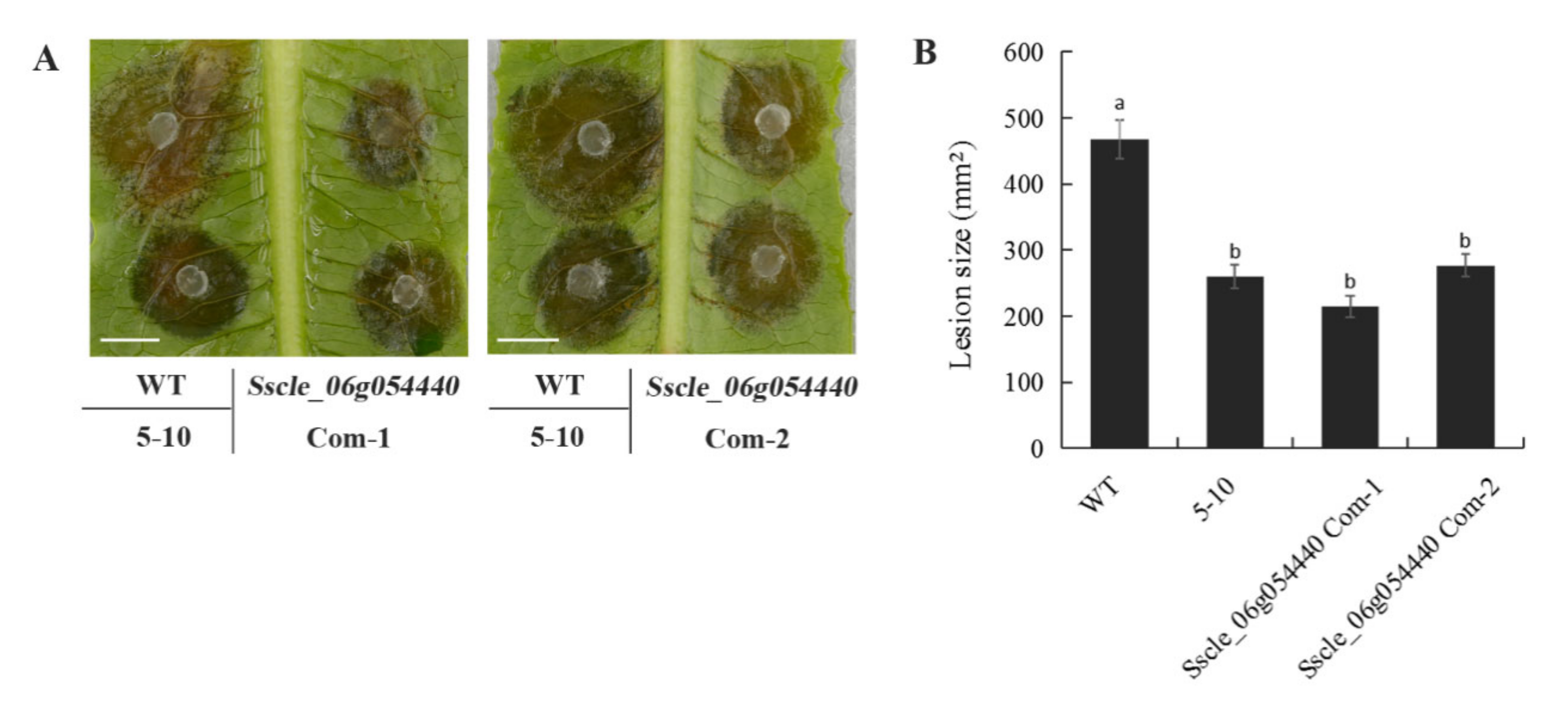
2.4. 5–10 and Sscle_10g079050 Knockout Mutants Exhibit Similar Hypovirulence
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Culture Condition
4.2. Plant Infection Assay
4.3. Screening for Mutants with Deficiency in Virulence
4.4. Genomic DNA Extraction and NGS
4.5. Candidate Genes Identification
4.6. Sanger Sequencing of Sscle_10g079050 in 5–10
4.7. Target Gene Knockout and Transgene Complementation
4.8. OA Analysis
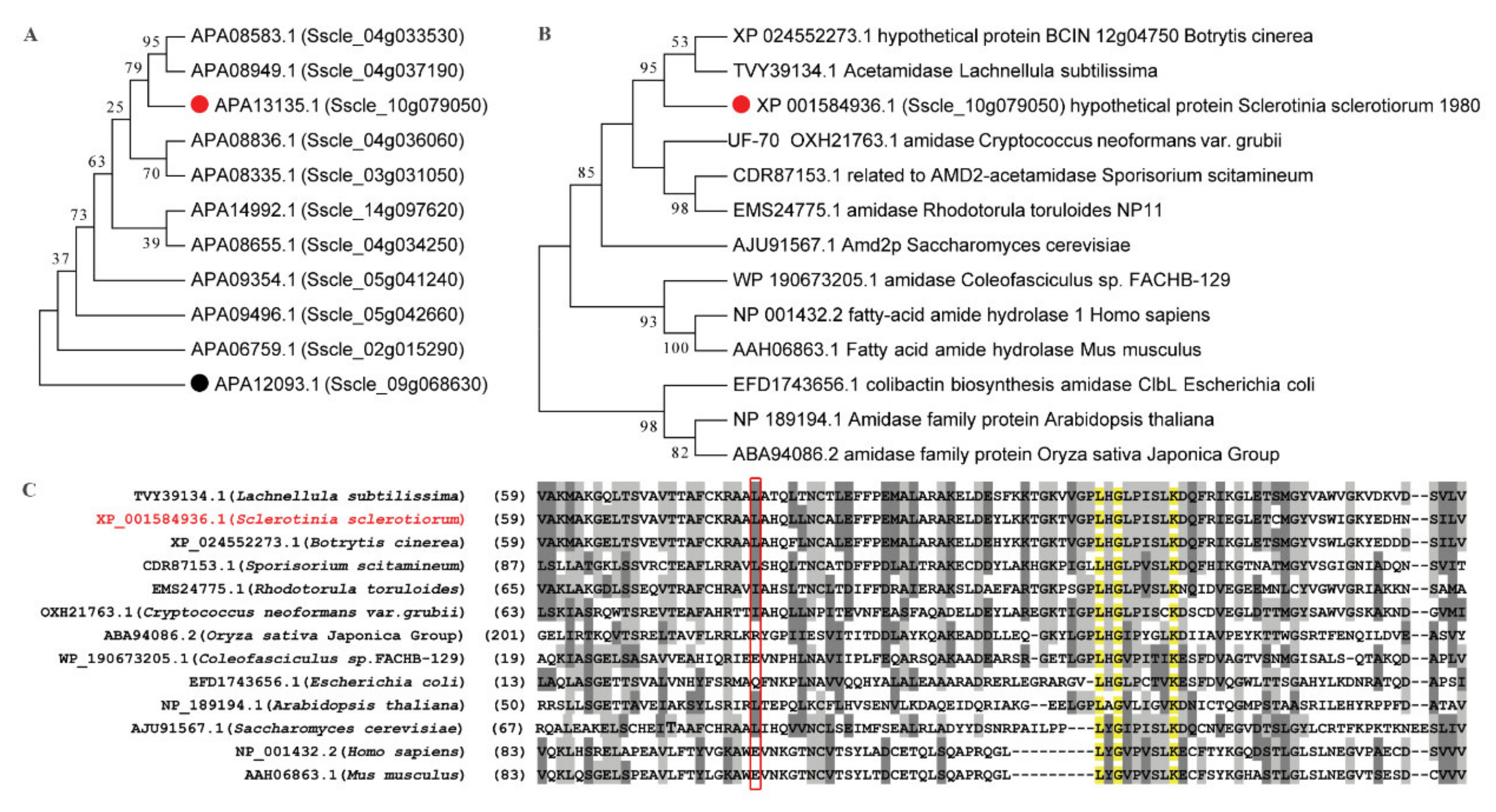
4.9. Phylogenetic Analyses
4.10. Protein Subcellular Localization and 3D Structure Prediction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, X.; Rollins, J.A. Mechanisms of Broad Host Range Necrotrophic Pathogenesis in Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1128–1140. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mu, W.; Veluchamy, S.; Liu, Y.; Zhang, Y.; Pan, H.; Rollins, J.A. The GATA-Type IVb Zinc-Finger Transcription Factor SsNsd1 Regulates Asexual-Sexual Development and Appressoria Formation in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2018, 19, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xiao, J.; Du, J.; Yang, Y.; Bi, C.; Qing, L. Disruption of the Gene Encoding Endo-β-1, 4-Xylanase Affects the Growth and Virulence of Sclerotinia sclerotiorum. Front. Microbiol. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Doughan, B.; Rollins, J.A. Characterization of MAT Gene Functions in the Life Cycle of Sclerotinia sclerotiorum Reveals a Lineage-Specific MAT Gene Functioning in Apothecium Morphogenesis. Fungal Biol. 2016, 120, 1105–1117. [Google Scholar] [CrossRef]
- Liang, Y.; Xiong, W.; Steinkellner, S.; Feng, J. Deficiency of the Melanin Biosynthesis Genes SCD1 and THR1 Affects Sclerotial Development and Vegetative Growth, but Not Pathogenicity, in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2018, 19, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xiao, J.; Yang, Y.; Bi, C.; Qing, L.; Tan, W. Ss-Bi1 Encodes a Putative BAX Inhibitor-1 Protein That Is Required for Full Virulence of Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 2015, 90, 115–122. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Zhang, Y.; Yu, P.-L.; Pan, H.; Rollins, J.A. Introduction of Large Sequence Inserts by CRISPR-Cas9 to Create Pathogenicity Mutants in the Multinucleate Filamentous Pathogen Sclerotinia sclerotiorum. mBio 2018, 9, e00567-18. [Google Scholar] [CrossRef]
- Liang, X.; Moomaw, E.W.; Rollins, J.A. Fungal Oxalate Decarboxylase Activity Contributes to Sclerotinia sclerotiorum Early Infection by Affecting Both Compound Appressoria Development and Function. Mol. Plant Pathol. 2015, 16, 825–836. [Google Scholar] [CrossRef]
- Bashi, Z.D.; Gyawali, S.; Bekkaoui, D.; Coutu, C.; Lee, L.; Poon, J.; Rimmer, S.R.; Khachatourians, G.G.; Hegedus, D.D. The Sclerotinia sclerotiorum Slt2 Mitogen-Activated Protein Kinase Ortholog, SMK3, Is Required for Infection Initiation but Not Lesion Expansion. Can. J. Microbiol. 2016, 62, 836–850. [Google Scholar] [CrossRef]
- Kim, H.; Chen, C.; Kabbage, M.; Dickman, M.B. Identification and Characterization of Sclerotinia sclerotiorum NADPH Oxidases. Appl. Environ. Microbiol. 2011, 77, 7721–7729. [Google Scholar] [CrossRef]
- Fan, H.; Yu, G.; Liu, Y.; Zhang, X.; Liu, J.; Zhang, Y.; Rollins, J.A.; Sun, F.; Pan, H. An Atypical Forkhead-Containing Transcription Factor SsFKH1 Is Involved in Sclerotial Formation and Is Essential for Pathogenicity in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2017, 18, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Liberti, D.; Li, M.; Kim, Y.-T.; Hutchens, A.; Wilson, R.; Rollins, J.A. Oxaloacetate Acetylhydrolase Gene Mutants of Sclerotinia sclerotiorum Do Not Accumulate Oxalic Acid, but Do Produce Limited Lesions on Host Plants. Mol. Plant Pathol. 2015, 16, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xiang, M.; White, D.; Chen, W. PH Dependency of Sclerotial Development and Pathogenicity Revealed by Using Genetically Defined Oxalate-Minus Mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 2015, 17, 2896–2909. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D.; Thomma, B.P.H.J.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and Molecular Traits of a Cosmopolitan Pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef]
- Kabbage, M.; Yarden, O.; Dickman, M.B. Pathogenic Attributes of Sclerotinia sclerotiorum: Switching from a Biotrophic to Necrotrophic Lifestyle. Plant Sci. 2015, 233, 53–60. [Google Scholar] [CrossRef]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. A Small Secreted Virulence-Related Protein Is Essential for the Necrotrophic Interactions of Sclerotinia sclerotiorum with Its Host Plants. PLoS Pathog. 2016, 12, e1005435. [Google Scholar] [CrossRef]
- Favaron, F.; Sella, L.; D’Ovidio, R. Relationships among Endo-Polygalacturonase, Oxalate, PH, and Plant Polygalacturonase-Inhibiting Protein (PGIP) in the Interaction between Sclerotinia sclerotiorum and Soybean. Mol. Plant Microbe Interact. 2004, 17, 1402–1409. [Google Scholar] [CrossRef]
- Rollins, J.A. The Sclerotinia sclerotiorum Pac1 Gene Is Required for Sclerotial Development and Virulence. Mol. Plant Microbe Interact. 2003, 16, 785–795. [Google Scholar] [CrossRef]
- Cotton, P.; Kasza, Z.; Bruel, C.; Rascle, C.; Fèvre, M. Ambient PH Controls the Expression of Endopolygalacturonase Genes in the Necrotrophic Fungus Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 2003, 227, 163–169. [Google Scholar] [CrossRef]
- Dutton, M.V.; Evans, C.S. Oxalate Production by Fungi: Its Role in Pathogenicity and Ecology in the Soil Environment. Can. J. Microbiol. 1996, 42, 881–895. [Google Scholar] [CrossRef]
- Heller, A.; Witt-Geiges, T. Oxalic Acid Has an Additional, Detoxifying Function in Sclerotinia sclerotiorum Pathogenesis. PLoS ONE 2013, 8, e72292. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Min, J.-Y.; Dickman, M.B. Oxalic Acid Is an Elicitor of Plant Programmed Cell Death during Sclerotinia sclerotiorum Disease Development. Mol. Plant Microbe Interact. 2008, 21, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Noyes, R.D.; Hancock, J.G. Role of Oxalic Acid in the Sclerotinia Wilt of Sunflower. Physiol. Plant Pathol. 1981, 18, 123–132. [Google Scholar] [CrossRef]
- Williams, B.; Kabbage, M.; Kim, H.-J.; Britt, R.; Dickman, M.B. Tipping the Balance: Sclerotinia sclerotiorum Secreted Oxalic Acid Suppresses Host Defenses by Manipulating the Host Redox Environment. PLoS Pathog. 2011, 7, e1002107. [Google Scholar] [CrossRef]
- Kabbage, M.; Williams, B.; Dickman, M.B. Cell Death Control: The Interplay of Apoptosis and Autophagy in the Pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 2013, 9, e1003287. [Google Scholar] [CrossRef]
- Xu, Y.; Ao, K.; Tian, L.; Qiu, Y.; Huang, X.; Liu, X.; Hoy, R.; Zhang, Y.; Rashid, K.Y.; Xia, S.; et al. A Forward Genetic Screen in Sclerotinia sclerotiorum Revealed the Transcriptional Regulation of Its Sclerotial Melanization Pathway. Mol. Plant Microbe Interact. 2022, 35, 244–256. [Google Scholar] [CrossRef]
- Seifbarghi, S.; Borhan, M.H.; Wei, Y.; Coutu, C.; Robinson, S.J.; Hegedus, D.D. Changes in the Sclerotinia sclerotiorum Transcriptome during Infection of Brassica napus. BMC Genom. 2017, 18, 266. [Google Scholar] [CrossRef]
- Shaw, N.M.; Naughton, A.; Robins, K.; Tinschert, A.; Schmid, E.; Hischier, M.-L.; Venetz, V.; Werlen, J.; Zimmermann, T.; Brieden, W.; et al. Selection, Purification, Characterisation, and Cloning of a Novel Heat-Stable Stereo-Specific Amidase from Klebsiella oxytoca, and Its Application in the Synthesis of Enantiomerically Pure (R)- and (S)-3,3,3-Trifluoro-2-Hydroxy-2-Methylpropionic Acids and (S)-3,3,3-Trifluoro-2-Hydroxy-2-Methylpropionamide. Org. Process Res. Dev. 2002, 6, 497–504. [Google Scholar] [CrossRef]
- Kang, X.-M.; Cai, X.; Liu, Z.-Q.; Zheng, Y.-G. Identification and Characterization of an Amidase from Leclercia Adecarboxylata for Efficient Biosynthesis of L-Phosphinothricin. Bioresour. Technol. 2019, 289, 121658. [Google Scholar] [CrossRef]
- Pace, H.C.; Brenner, C. The Nitrilase Superfamily: Classification, Structure and Function. Genome Biol. 2001, 2, REVIEWS0001. [Google Scholar] [CrossRef]
- Wu, Z.-M.; Zheng, R.-C.; Tang, X.-L.; Zheng, Y.-G. Identification and Characterization of a Thermostable and Cobalt-Dependent Amidase from Burkholderia phytofirmans ZJB-15079 for Efficient Synthesis of (R)-3,3,3-Trifluoro-2-Hydroxy-2-Methylpropionic Acid. Appl. Microbiol. Biotechnol. 2017, 101, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-M.; Zheng, R.-C.; Zheng, Y.-G. Identification and Characterization of a Novel Amidase Signature Family Amidase from Parvibaculum Lavamentivorans ZJB14001. Protein Expr. Purif. 2017, 129, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Labahn, J.; Neumann, S.; Büldt, G.; Kula, M.-R.; Granzin, J. An Alternative Mechanism for Amidase Signature Enzymes. J. Mol. Biol. 2002, 322, 1053–1064. [Google Scholar] [CrossRef]
- Lee, S.; Park, E.-H.; Ko, H.-J.; Bang, W.G.; Kim, H.-Y.; Kim, K.H.; Choi, I.-G. Crystal Structure Analysis of a Bacterial Aryl Acylamidase Belonging to the Amidase Signature Enzyme Family. Biochem. Biophys. Res. Commun. 2015, 467, 268–274. [Google Scholar] [CrossRef]
- Bracey, M.H.; Hanson, M.A.; Masuda, K.R.; Stevens, R.C.; Cravatt, B.F. Structural Adaptations in a Membrane Enzyme That Terminates Endocannabinoid Signaling. Science 2002, 298, 1793–1796. [Google Scholar] [CrossRef]
- Amselem, J.; Cuomo, C.A.; van Kan, J.A.L.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S.; et al. Genomic Analysis of the Necrotrophic Fungal Pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef]
- Munir, E.; Yoon, J.-J.; Tokimatsu, T.; Hattori, T.; Shimada, M. New Role for Glyoxylate Cycle Enzymes in Wood-Rotting Basidiomycetes in Relation to Biosynthesis of Oxalic Acid. J. Wood Sci. 2001, 47, 368–373. [Google Scholar] [CrossRef]
- Sakai, S.; Nishide, T.; Munir, E.; Baba, K.; Inui, H.; Nakano, Y.; Hattori, T.; Shimada, M. Subcellular Localization of Glyoxylate Cycle Key Enzymes Involved in Oxalate Biosynthesis of Wood-Destroying Basidiomycete Fomitopsis Palustris Grown on Glucose. Microbiology 2006, 152, 1857–1866. [Google Scholar] [CrossRef][Green Version]
- Liberti, D.; Rollins, J.A.; Dobinson, K.F. Peroxysomal Carnitine Acetyl Transferase Influences Host Colonization Capacity in Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 2013, 26, 768–780. [Google Scholar] [CrossRef]
- Asakura, M.; Okuno, T.; Takano, Y. Multiple Contributions of Peroxisomal Metabolic Function to Fungal Pathogenicity in Colletotrichum lagenarium. Appl. Environ. Microbiol. 2006, 72, 6345–6354. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Thornton, C.R.; Kershaw, M.J.; Debao, L.; Talbot, N.J. The Glyoxylate Cycle Is Required for Temporal Regulation of Virulence by the Plant Pathogenic Fungus Magnaporthe grisea. Mol. Microbiol. 2003, 47, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Solomon, P.S.; Lee, R.C.; Wilson, T.J.G.; Oliver, R.P. Pathogenicity of Stagonospora nodorum Requires Malate Synthase. Mol. Microbiol. 2004, 53, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Liberti, D.; Grant, S.J.; Benny, U.; Rollins, J.A.; Dobinson, K.F. Development of an Agrobacterium Tumefaciens-Mediated Gene Disruption Method for Sclerotinia sclerotiorum. Can. J. Plant Pathol. 2007, 29, 394–400. [Google Scholar] [CrossRef]
- Ruan, L.-T.; Zheng, R.-C.; Zheng, Y.-G.; Shen, Y.-C. Purification and Characterization of R-Stereospecific Amidase from Brevibacterium epidermidis ZJB-07021. Int. J. Biol. Macromol. 2016, 86, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.; Liang, B.; Qiu, J.; Zhang, L.; Zhao, Y.; Jiang, J.; Wang, A. Functional Characterization of a Novel Amidase Involved in Biotransformation of Triclocarban and Its Dehalogenated Congeners in Ochrobactrum sp. TCC-2. Environ. Sci. Technol. 2017, 51, 291–300. [Google Scholar] [CrossRef]
- Trott, S.; Bürger, S.; Calaminus, C.; Stolz, A. Cloning and Heterologous Expression of an Enantioselective Amidase from Rhodococcus erythropolis Strain MP50. Appl. Environ. Microbiol. 2002, 68, 3279–3286. [Google Scholar] [CrossRef]
- Kunau, W.H.; Dommes, V.; Schulz, H. Beta-Oxidation of Fatty Acids in Mitochondria, Peroxisomes, and Bacteria: A Century of Continued Progress. Prog. Lipid Res. 1995, 34, 267–342. [Google Scholar] [CrossRef]
- Bhambra, G.K.; Wang, Z.-Y.; Soanes, D.M.; Wakley, G.E.; Talbot, N.J. Peroxisomal Carnitine Acetyl Transferase Is Required for Elaboration of Penetration Hyphae during Plant Infection by Magnaporthe grisea. Mol. Microbiol. 2006, 61, 46–60. [Google Scholar] [CrossRef]
- Cubero, O.F.; Crespo, A.; Fatehi, J.; Bridge, P.D. DNA Extraction and PCR Amplification Method Suitable for Fresh, Herbarium-Stored, Lichenized, and Other Fungi. Plant Syst. Evol. 1999, 216, 243–249. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Huang, J.; Sun, Y.; Orduna, A.R.; Jetter, R.; Li, X. The Mediator Kinase Module Serves as a Positive Regulator of Salicylic Acid Accumulation and Systemic Acquired Resistance. Plant J. 2019, 98, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Catlett, N.; Lee, B.-N.; Yoder, O.; Turgeon, B. Split-Marker Recombination for Efficient Targeted Deletion of Fungal Genes. Fungal Genet. Rep. 2003, 50, 9–11. [Google Scholar] [CrossRef]
- Yu, J.-H.; Hamari, Z.; Han, K.-H.; Seo, J.-A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-Joint PCR: A PCR-Based Molecular Tool for Gene Manipulations in Filamentous Fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Lu, J.; Yang, C.; Arildsen, K.; Li, X.; Xia, S. An Amidase Contributes to Full Virulence of Sclerotinia sclerotiorum. Int. J. Mol. Sci. 2022, 23, 11207. https://doi.org/10.3390/ijms231911207
Li W, Lu J, Yang C, Arildsen K, Li X, Xia S. An Amidase Contributes to Full Virulence of Sclerotinia sclerotiorum. International Journal of Molecular Sciences. 2022; 23(19):11207. https://doi.org/10.3390/ijms231911207
Chicago/Turabian StyleLi, Wei, Junxing Lu, Chenghuizi Yang, Kate Arildsen, Xin Li, and Shitou Xia. 2022. "An Amidase Contributes to Full Virulence of Sclerotinia sclerotiorum" International Journal of Molecular Sciences 23, no. 19: 11207. https://doi.org/10.3390/ijms231911207
APA StyleLi, W., Lu, J., Yang, C., Arildsen, K., Li, X., & Xia, S. (2022). An Amidase Contributes to Full Virulence of Sclerotinia sclerotiorum. International Journal of Molecular Sciences, 23(19), 11207. https://doi.org/10.3390/ijms231911207






