Identification and Characterization of Circular RNAs Involved in the Flower Development and Senescence of Rhododendron delavayi Franch
Abstract
:1. Introduction
2. Results
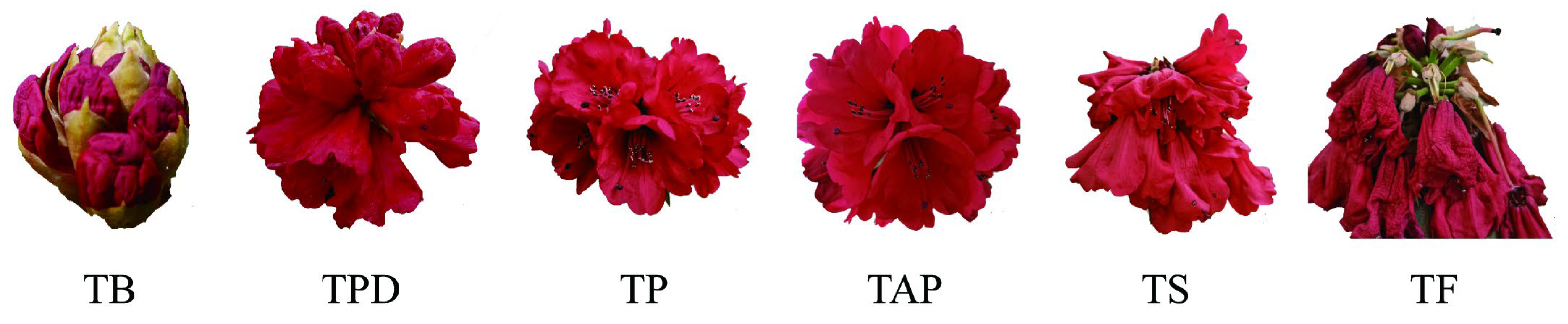
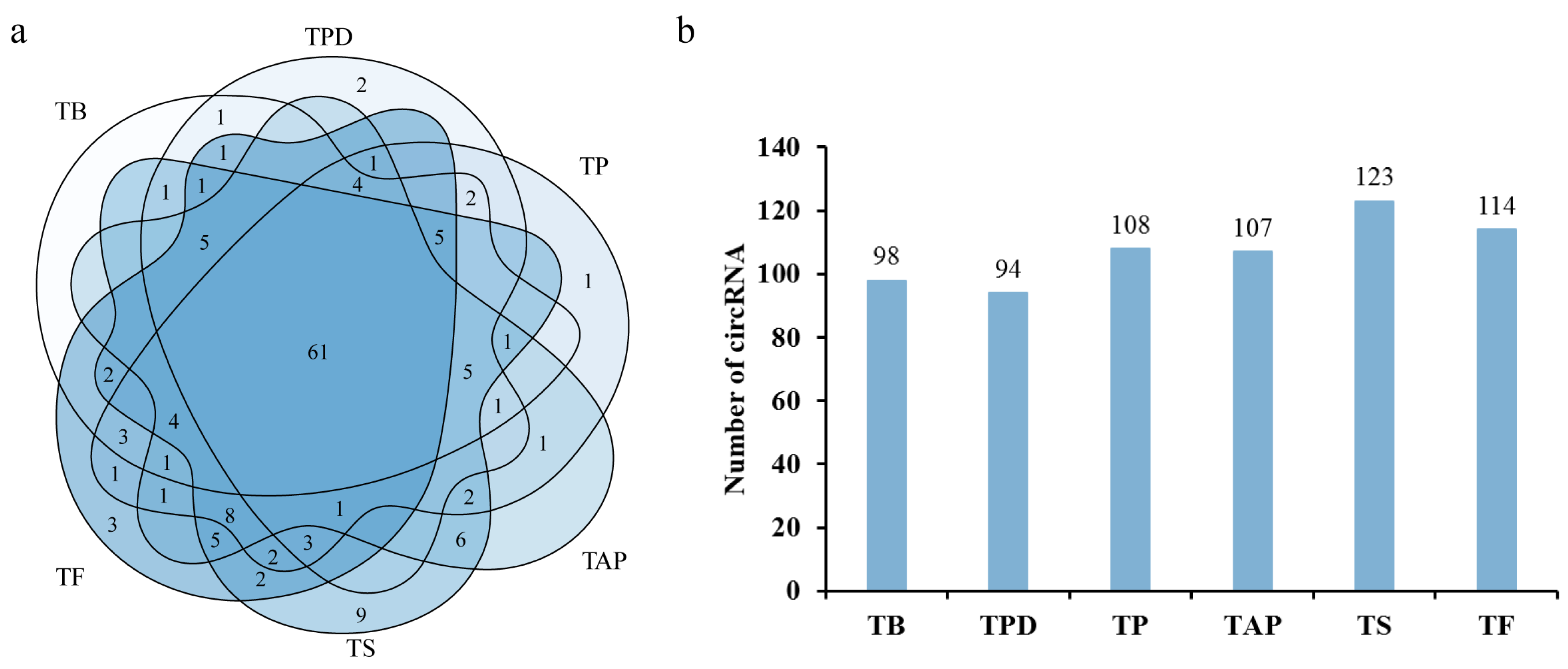
2.1. Identification of circRNAs in the Rhododendron delavayi Franch Flowers
2.2. Analysis of Differentially Expressed circRNAs
2.3. Time Series Profile Analysis of Transcript Expression
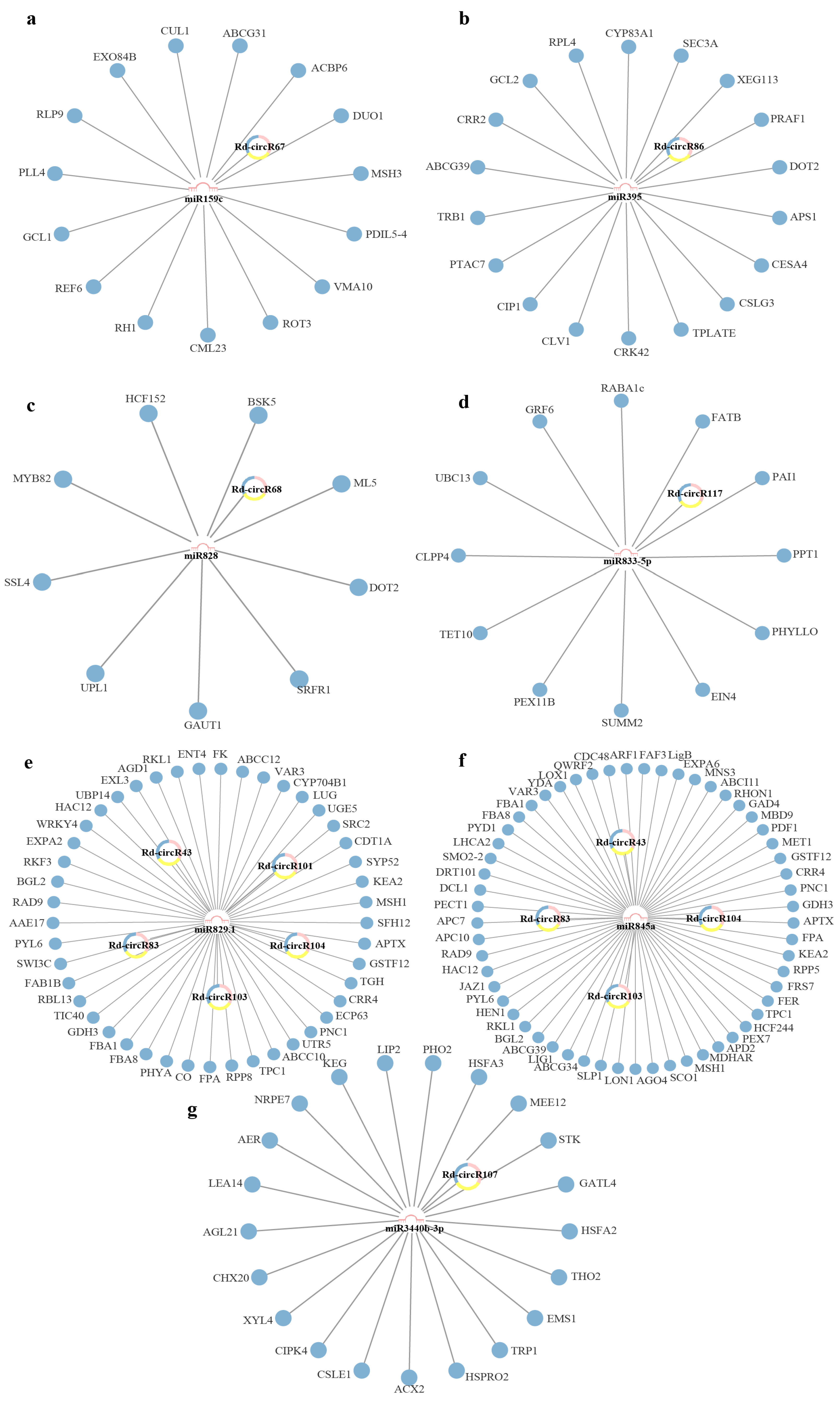
2.4. Construction of circRNA-miRNA-mRNA Networks
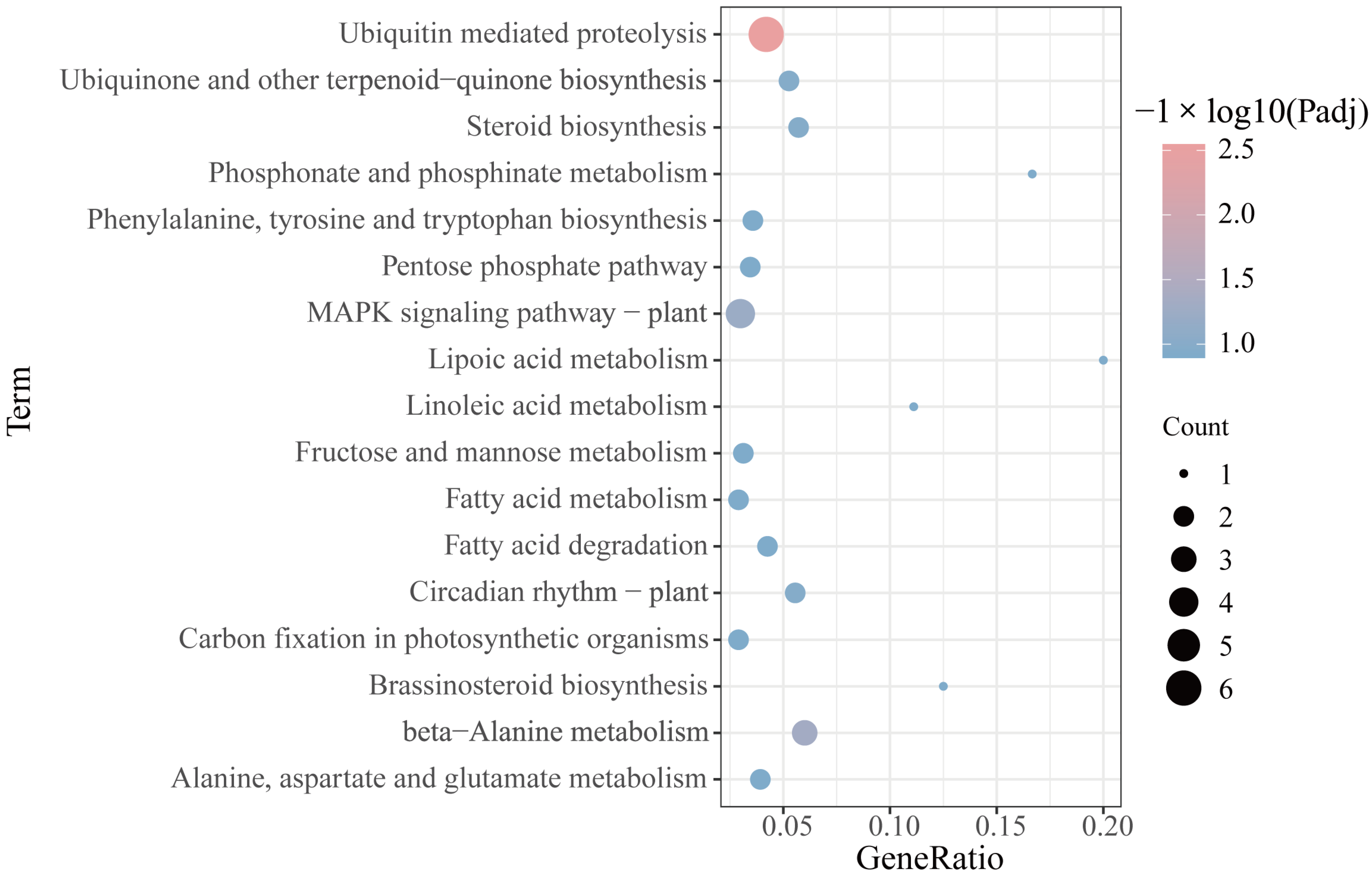
2.5. Functional Annotations of the mRNAs
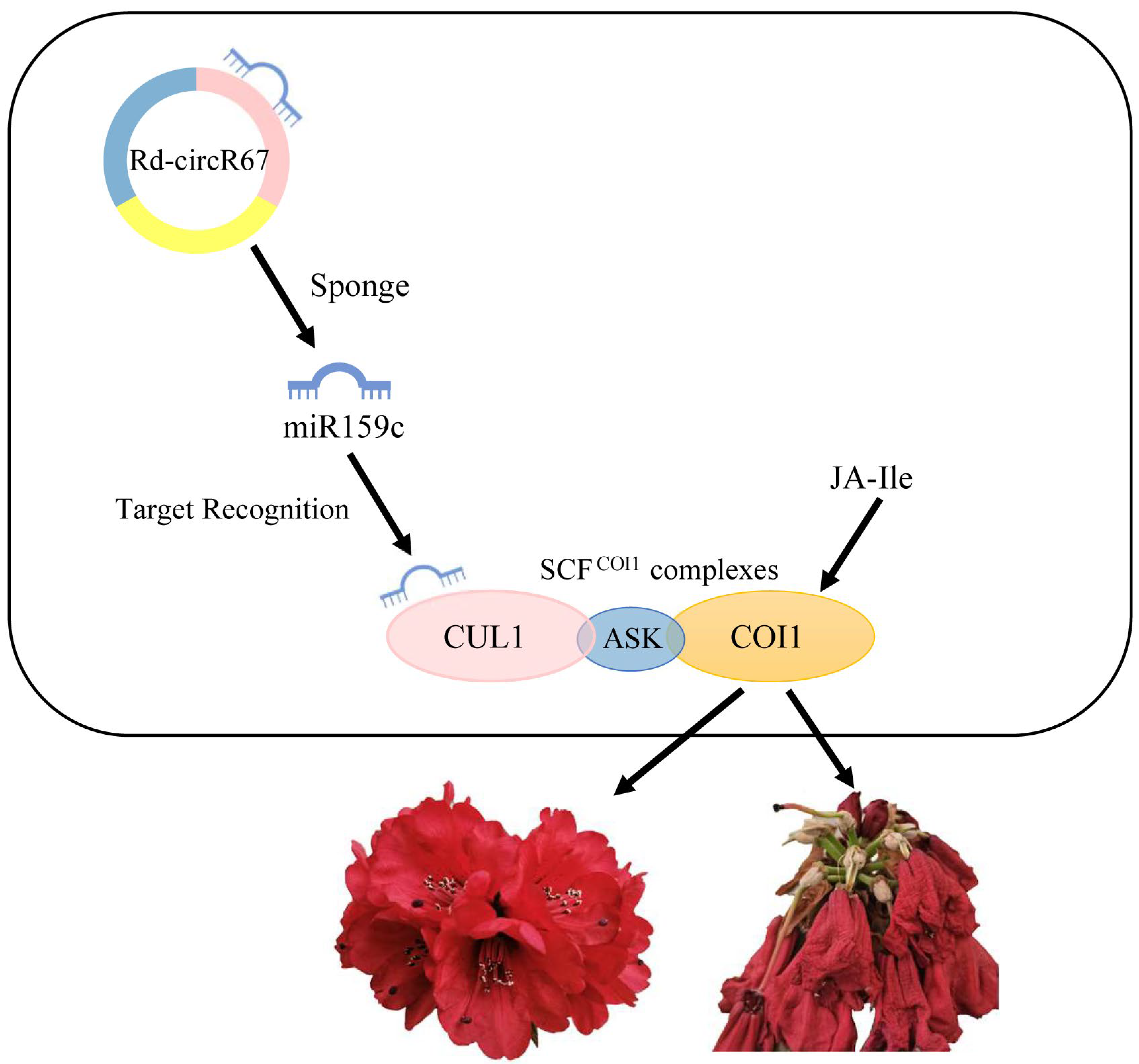
3. Discussion
4. Materials and Methods
4.1. Plant Sample Collection
4.2. High Throughput Sequencing and Identification of circRNAs
4.3. Identification of Differentially Expressed circRNAs (DEcircRNAs)
4.4. Time Series Profile Analysis of the Transcriptome
4.5. Construction of circRNA-miRNA-mRNA Networks
4.6. Functional Annotations
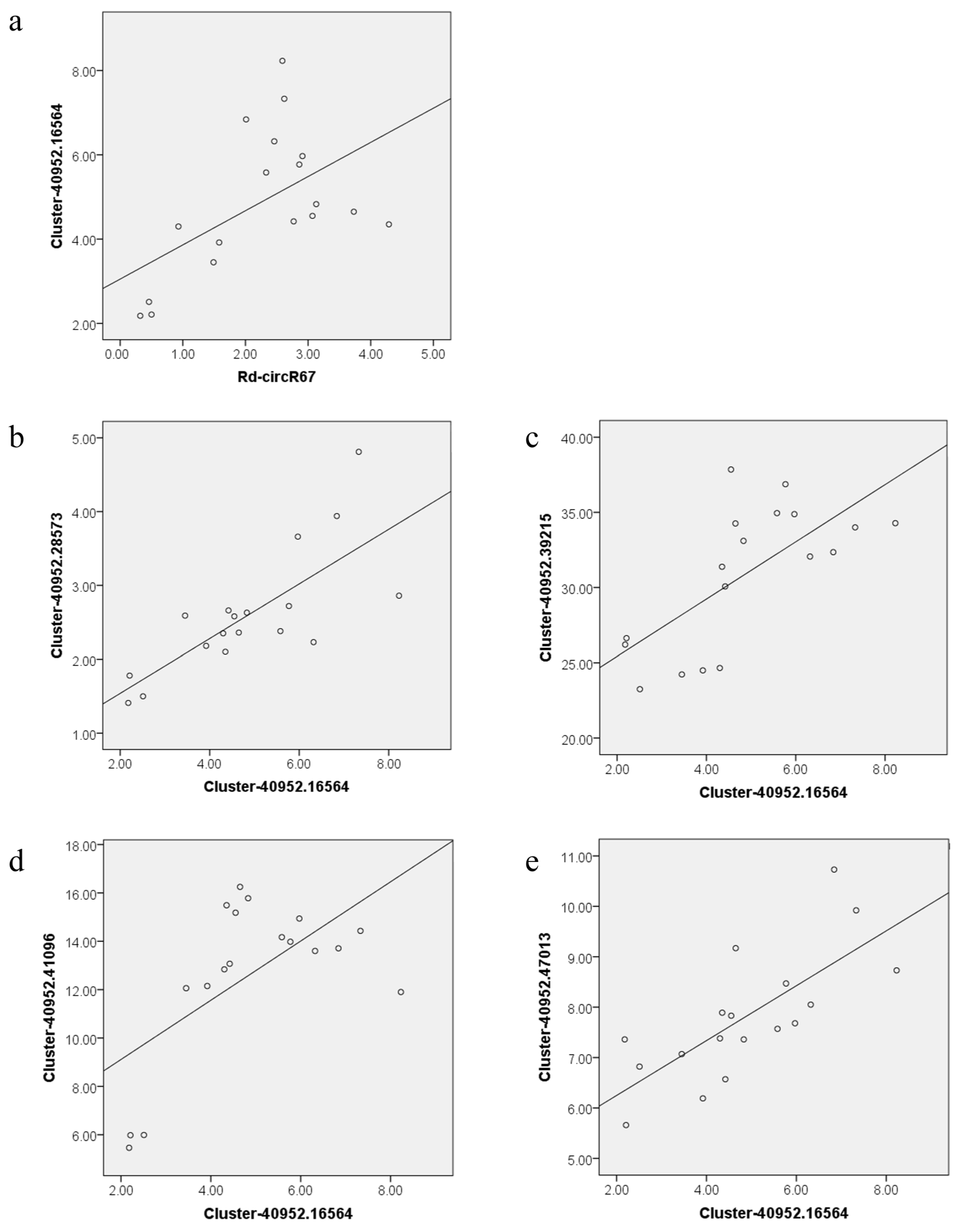
4.7. Correlation Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, H.; Guo, Q.; Li, Q.; Yang, L. Long-reads reveal that Rhododendron delavayi plastid genome contains extensive repeat sequences, and recombination exists among plastid genomes of photosynthetic Ericaceae. PeerJ 2020, 8, e9048. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, H.; Guo, R.; Wang, Q.; Liu, X.; Kuang, W.; Song, H.; Liao, J.; Huang, Y.; Wang, Z. Systematic identification and characterization of circular RNAs involved in flag leaf senescence of rice. Planta 2021, 253, 26. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef]
- Jeck, W.; Sharpless, N. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, P.; Fan, Y.; Lu, Q.; Li, Q.; Yan, J.; Muehlbauer, G.J.; Schnable, P.S.; Dai, M.; Li, L. Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol. 2018, 217, 1292–1306. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.X.; Du, T.Y.; Mao, W.H.; Li, X.H.; Ye, C.Y.; Zhu, Q.H.; Fan, L.J.; Chu, Q.J. PlantcircBase 7.0: Full-length transcripts and conservation of plant circRNAs. Plant Commun. 2022, 3, 2590–3462. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef]
- Hansen, T.; Jensen, T.; Clausen, B.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Quan, W.; Li, G.B.; Hu, X.H.; Wang, Q.; Wang, H.; Li, X.P.; Luo, X.; Feng, Q.; Hu, Z.J.; et al. circRNAs Are Involved in the Rice-Magnaporthe oryzae Interaction. Plant Physiol. 2020, 182, 272–286. [Google Scholar] [CrossRef]
- Tong, W.; Yu, J.; Hou, Y.; Li, F.; Zhou, Q.; Wei, C.; Bennetzen, J.L. Circular RNA architecture and differentiation during leaf bud to young leaf development in tea (Camellia sinensis). Planta 2018, 248, 1417–1429. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Chen, G.; Shi, T. Identifying and Characterizing the Circular RNAs during the Lifespan of Arabidopsis Leaves. Front. Plant Sci. 2017, 8, 1278. [Google Scholar] [CrossRef]
- Zuo, J.; Wang, Y.; Zhu, B.; Luo, Y.; Wang, Q.; Gao, L. Analysis of the Coding and Non-Coding RNA Transcriptomes in Response to Bell Pepper Chilling. Int. J. Mol. Sci. 2018, 19, 2001. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, Y.; Xu, L.; Zhou, D.; Jin, Z.; Zhou, H.; Lin, S.; Cao, J.; Huang, L. CircRNA Expression Pattern and ceRNA and miRNA-mRNA Networks Involved in Anther Development in the CMS Line of Brassica Campestris. Int. J. Mol. Sci. 2019, 20, 4808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.; Chen, L.; Liu, C.; Zhu, Q.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol 2015, 208, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Somoza, I.; Weigel, D.; Franco-Zorilla, J.M.; García, J.A.; Paz-Ares, J. ceRNAs: miRNA target mimic mimics. Cell 2011, 147, 1431–1432. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant. Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Ivashuta, S.; Banks, I.R.; Wiggins, B.E.; Zhang, Y.; Ziegler, T.E.; Roberts, J.K.; Heck, G.R. Regulation of gene expression in plants through miRNA inactivation. PLoS ONE 2011, 6, e21330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, R.; Zhu, Y.; Gong, J.; Yin, S.; Sun, P.; Feng, H.; Wang, Q.; Zhao, S.; Wang, Z.; et al. Identification and Characterization of circRNAs Responsive to Methyl Jasmonate in Arabidopsis Thaliana. Int. J. Mol. Sci. 2020, 21, 792. [Google Scholar] [CrossRef]
- Hellmann, H.; Hobbie, L.; Chapman, A.; Dharmasiri, S.; Dharmasiri, N.; del Pozo, C.; Reinhardt, D.; Estelle, M. Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 2003, 22, 3314–3325. [Google Scholar] [CrossRef] [PubMed]
- Eloy, N.B.; de Freitas Lima, M.; Van Damme, D.; Vanhaeren, H.; Gonzalez, N.; De Milde, L.; Hemerly, A.S.; Beemster, G.T.; Inzé, D.; Ferreira, P.C. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 2011, 68, 351–363. [Google Scholar] [CrossRef]
- Dobritsa, A.A.; Shrestha, J.; Morant, M.; Pinot, F.; Matsuno, M.; Swanson, R.; Møller, B.L.; Preuss, D. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant. Physiol. 2009, 151, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Luccioni, L.; Krzymuski, M.; Sánchez-Lamas, M.; Karayekov, E.; Cerdán, P.D.; Casal, J.J. CONSTANS delays Arabidopsis flowering under short days. Plant J. 2019, 97, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ma, Y.; Guo, T.; Li, G. Identification, biogenesis, function, and mechanism of action of circular RNAs in plants. Plant. Commun. 2022, 100430. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Jia, J.H.; Yang, L.; Xia, Y.C.; Zhang, H.L.; Jia, J.B.; Zhou, R.; Nie, P.Y.; Yin, J.L.; Ma, D.F.; et al. Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol. 2019, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Li, X.; Yan, Y.; Duan, M.H.; Xu, J.H. Identification, characterization, and functional prediction of circular RNAs in maize. Mol. Genet. Genom. 2020, 295, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ren, Y.; Lin, T.; Cui, D. Identification and characterization of CircRNAs involved in the regulation of wheat root length. Biol. Res. 2019, 52, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Diao, S.; Zhang, T.; Chen, D.; He, C.; Zhang, J. Identification and characterization of circular RNAs during the sea buckthorn fruit development. RNA Biol. 2019, 16, 354–361. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Gao, H.; Zhang, X.; Zhuang, N. Comparative transcriptome analysis reveals differential gene expression in sterile and fertile rubber tree varieties during flower bud differentiation. J. Plant Physiol. 2021, 265, 153506. [Google Scholar] [CrossRef]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arab. Thaliana. Sci. 2013, 339, 704–707. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Liu, Y.; Liu, H. Flowering responses to light and temperature. Sci. China Life Sci. 2016, 59, 403–408. [Google Scholar] [CrossRef]
- Xu, L.H.; Liu, F.Q.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.F.; Xie, D.X. The SCF(COI1) Ubiquitin-Ligase Complexes Are Required for Jasmonate Response in Arabidopsis. Plant Cell 2002, 14, 1919–1935. [Google Scholar] [CrossRef]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Dotson, B.; Rey, C.; Lindsey, J.; Bleecker, A.B.; Binder, B.M.; Patterson, S.E. New Clothes for the Jasmonic Acid Receptor COI1: Delayed Abscission, Meristem Arrest and Apical Dominance. PLoS ONE 2013, 8, e60505. [Google Scholar] [CrossRef]
- Zhao, W.; Cheng, Y.; Zhang, C.; You, Q.; Shen, X.; Guo, W.; Jiao, Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017, 7, 5636. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Qin, Y.; Xu, T.; Lin, W.; Jia, Q.; He, Q.; Liu, K.; Du, J.; Chen, L.; Yang, X.; Du, F.; et al. Reference-Free and De Novo Identification of Circular Rnas. bioRxiv 2020. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Meng, X.; Chen, H.; Liu, Y.; Xue, J.; Zhou, Y.; Chen, M. PlantCircNet: A database for plant circRNA-miRNA-mRNA regulatory networks. Database 2017, 2017, bax089. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Fan, Y.; Han, Z.; Lu, X.; Arbab, A.; Nazar, M.; Yang, Y.; Yang, Z. Short Time-Series Expression Transcriptome Data Reveal the Gene Expression Patterns of Dairy Cow Mammary Gland as Milk Yield Decreased Process. Genes 2021, 12, 942. [Google Scholar] [CrossRef]
- Li, T.; Chen, B.; Yang, P.; Wang, D.; Du, B.; Kang, L. Long Non-coding RNA Derived from lncRNA-mRNA Co-expression Networks Modulates the Locust Phase Change. Genom. Proteom. Bioinform. 2020, 18, 664–678. [Google Scholar] [CrossRef]
- Chang, T.H.; Huang, H.Y.; Hsu, J.B.; Weng, S.L.; Horng, J.T.; Huang, H.D. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinform. 2013, 14, S4. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, E.; He, Y.; Billiau, K.; Van de Peer, Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 2010, 26, 1566–1568. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Xiao, Y.; Zhang, X.; Tang, M.; Tang, J. Identification and Characterization of Circular RNAs Involved in the Flower Development and Senescence of Rhododendron delavayi Franch. Int. J. Mol. Sci. 2022, 23, 11214. https://doi.org/10.3390/ijms231911214
Xu X, Xiao Y, Zhang X, Tang M, Tang J. Identification and Characterization of Circular RNAs Involved in the Flower Development and Senescence of Rhododendron delavayi Franch. International Journal of Molecular Sciences. 2022; 23(19):11214. https://doi.org/10.3390/ijms231911214
Chicago/Turabian StyleXu, Xiaorong, Yufeng Xiao, Ximin Zhang, Ming Tang, and Jing Tang. 2022. "Identification and Characterization of Circular RNAs Involved in the Flower Development and Senescence of Rhododendron delavayi Franch" International Journal of Molecular Sciences 23, no. 19: 11214. https://doi.org/10.3390/ijms231911214
APA StyleXu, X., Xiao, Y., Zhang, X., Tang, M., & Tang, J. (2022). Identification and Characterization of Circular RNAs Involved in the Flower Development and Senescence of Rhododendron delavayi Franch. International Journal of Molecular Sciences, 23(19), 11214. https://doi.org/10.3390/ijms231911214




