Drosophila as a Model Organism to Study Basic Mechanisms of Longevity
Abstract
:1. Introduction
2. Advantages of Drosophila as a Highly Informative Model Organism for Aging Studies
2.1. Advantages and Similarities of Drosophila and Mammals
2.2. Features of the Biology of Drosophila as an Object for the Study of Aging
2.3. Statistical Analysis in Drosophila Aging Studies
2.4. Drosophila as a Model in the Study of Epigenetic Processes
2.5. Limitations in Using Drosophila as a Model in Aging Research
3. Signaling Pathways Regulating Drosophila Lifespan
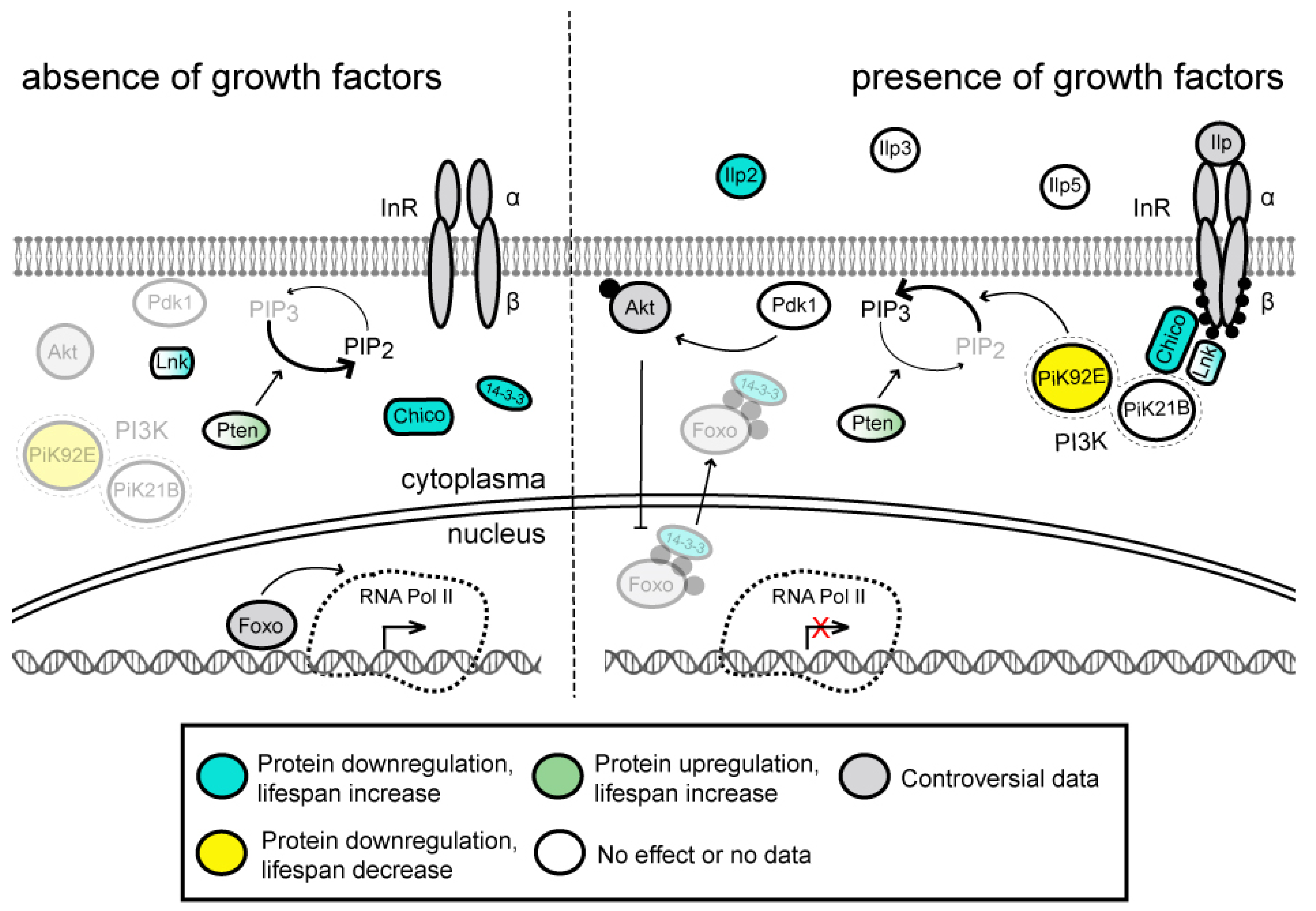
3.1. Insulin/IGF-1 Signaling (IIS) Pathway
3.2. Target of Rapamycin (TOR) Signaling Pathway
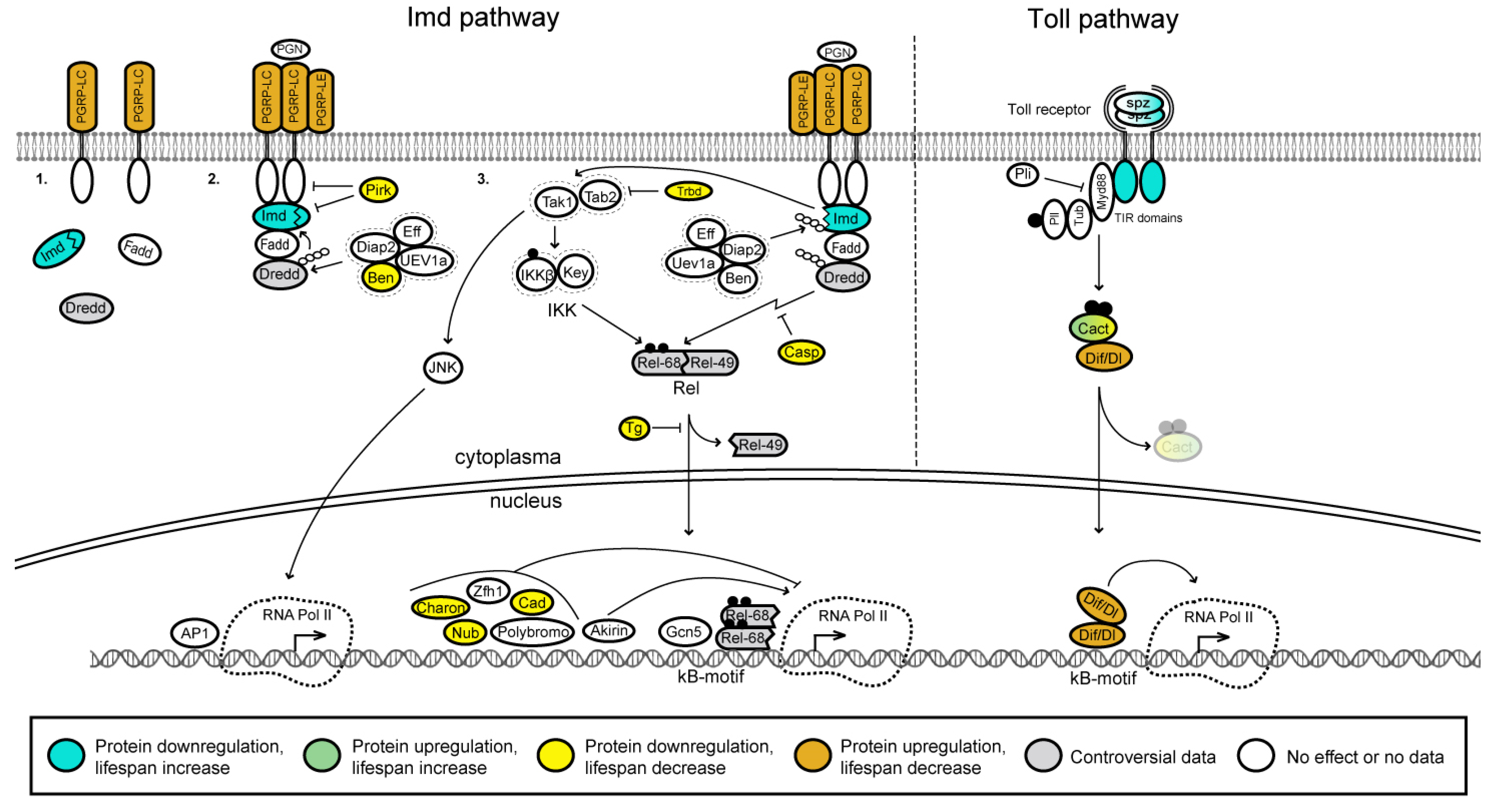
3.3. NF-κB Signaling Pathway
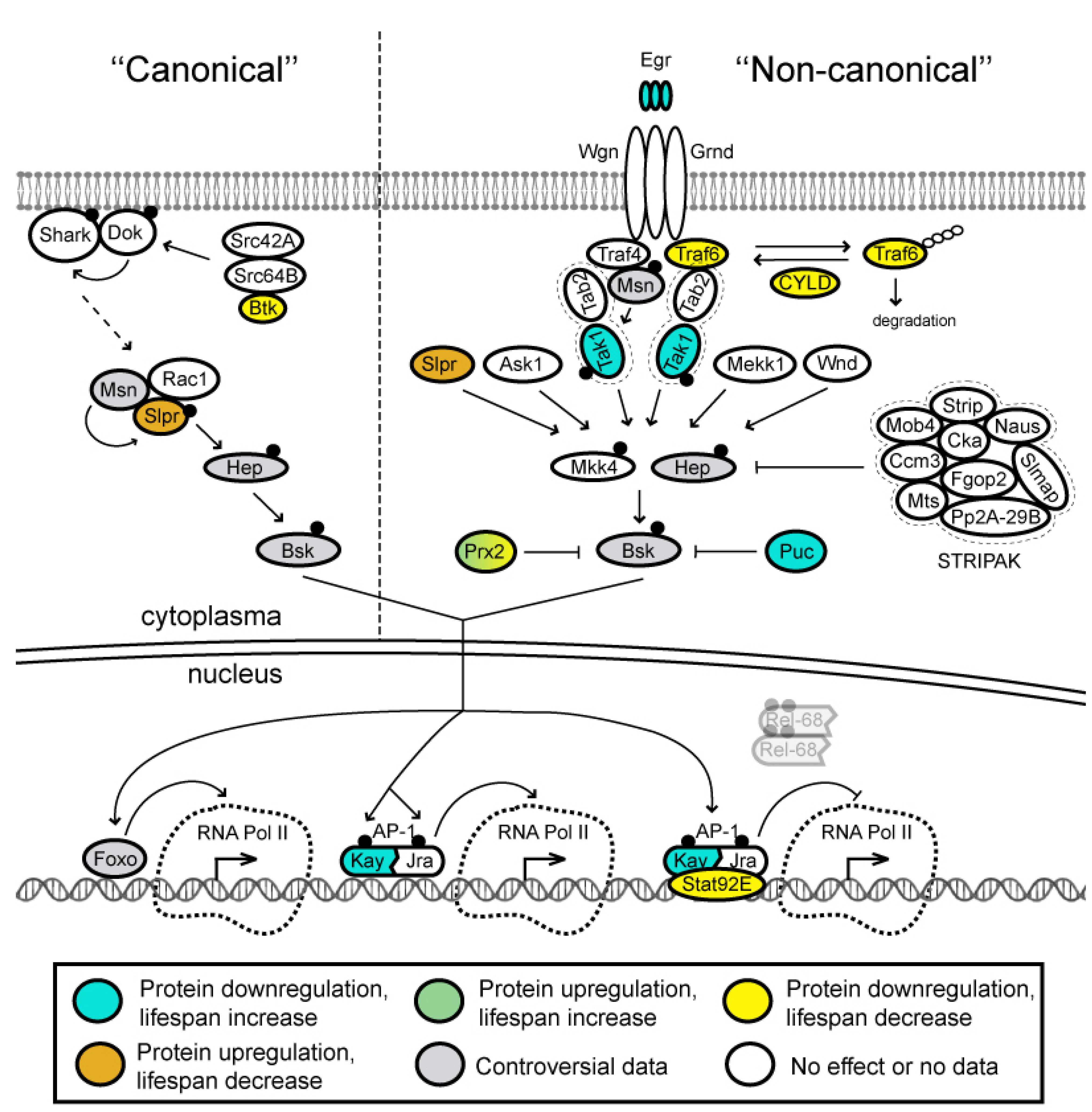
3.4. JNK Signaling Pathway
3.5. Interconnection between IIS, TOR, NF-κB and JNK Pathways
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kennedy, B.K.; Berger, S.L.; Brunet, A.; Campisi, J.; Cuervo, A.M.; Epel, E.S.; Franceschi, C.; Lithgow, G.J.; Morimoto, R.I.; Pessin, J.E.; et al. Geroscience: Linking aging to chronic disease. Cell 2014, 159, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Gan, T.; Fan, L.; Zhao, L.; Misra, M.; Liu, M.; Zhang, M.; Su, Y. JNK Signaling in Drosophila Aging and Longevity. Int. J. Mol. Sci. 2021, 22, 9649. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Lamming, D.W.; Fontana, L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol. 2022, 23, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Miller, H.A.; Dean, E.S.; Pletcher, S.D.; Leiser, S.F. Cell non-autonomous regulation of health and longevity. Elife 2020, 9, e62659. [Google Scholar] [CrossRef]
- Green, M.M. 2010: A century of Drosophila genetics through the prism of the white gene. Genetics 2010, 184, 3–7. [Google Scholar] [CrossRef]
- Rogina, B. For the special issue: Aging studies in Drosophila melanogaster. Exp. Gerontol. 2011, 46, 317–319. [Google Scholar] [CrossRef]
- Promislow, D.E.L.; Flatt, T.; Bonduriansky, R. The Biology of Aging in Insects: From Drosophila to Other Insects and Back. Annu. Rev. Entomol. 2022, 67, 83–103. [Google Scholar] [CrossRef]
- Calissi, G.; Lam, E.W.; Link, W. Therapeutic strategies targeting FOXO transcription factors. Nat. Rev. Drug Discov. 2021, 20, 21–38. [Google Scholar] [CrossRef]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef] [Green Version]
- Rubin, G.M.; Yandell, M.D.; Wortman, J.R.; Gabor Miklos, G.L.; Nelson, C.R.; Hariharan, I.K.; Fortini, M.E.; Li, P.W.; Apweiler, R.; Fleischmann, W.; et al. Comparative genomics of the eukaryotes. Science 2000, 287, 2204–2215. [Google Scholar] [CrossRef] [PubMed]
- Yakovleva, E.U.; Naimark, E.B.; Markov, A.V. Adaptation of Drosophila melanogaster to Unfavorable Growth Medium Affects Lifespan and Age-Related Fecundity. Biochemistry 2016, 81, 1445–1460. [Google Scholar] [CrossRef] [PubMed]
- Shaposhnikov, M.V.; Guvatova, Z.G.; Zemskaya, N.V.; Koval, L.A.; Schegoleva, E.V.; Gorbunova, A.A.; Golubev, D.A.; Pakshina, N.R.; Ulyasheva, N.S.; Solovev, I.A.; et al. Molecular mechanisms of exceptional lifespan increase of Drosophila melanogaster with different genotypes after combinations of pro-longevity interventions. Commun. Biol. 2022, 5, 566. [Google Scholar] [CrossRef] [PubMed]
- Magwere, T.; Chapman, T.; Partridge, L. Sex differences in the effect of dietary restriction on life span and mortality rates in female and male Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 2004, 59, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Gavrilyuk, K.; Burdylyuk, N.; Strilbytska, O.; Storey, K.B.; Kuharskii, V.; Lushchak, O.; Vaiserman, A. Mating status affects Drosophila lifespan, metabolism and antioxidant system. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2020, 246, 110716. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.M.; Promislow, D.E. Sex-specific effects of interventions that extend fly life span. Sci. Aging Knowl. Environ. 2004, 2004, pe30. [Google Scholar] [CrossRef]
- Toivonen, J.M.; Walker, G.A.; Martinez-Diaz, P.; Bjedov, I.; Driege, Y.; Jacobs, H.T.; Gems, D.; Partridge, L. No influence of Indy on lifespan in Drosophila after correction for genetic and cytoplasmic background effects. PLoS Genet. 2007, 3, e95. [Google Scholar] [CrossRef]
- Spencer, C.C.; Howell, C.E.; Wright, A.R.; Promislow, D.E. Testing an ‘aging gene’ in long-lived drosophila strains: Increased longevity depends on sex and genetic background. Aging Cell 2003, 2, 123–130. [Google Scholar] [CrossRef]
- He, Y.; Jasper, H. Studying aging in Drosophila. Methods 2014, 68, 129–133. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Floate, K.D.; Fields, P.G.; Pang, B.-P. Review of treatment methods to remove Wolbachia bacteria from arthropods. Symbiosis 2014, 62, 1–15. [Google Scholar] [CrossRef]
- Yang, J.S.; Nam, H.J.; Seo, M.; Han, S.K.; Choi, Y.; Nam, H.G.; Lee, S.J.; Kim, S. OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 2011, 6, e23525. [Google Scholar] [CrossRef] [PubMed]
- Grandison, R.C.; Piper, M.D.; Partridge, L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 2009, 462, 1061–1064. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; MacLean, A.M.; Sugio, A.; Maqbool, A.; Busscher, M.; Cho, S.T.; Kamoun, S.; Kuo, C.H.; Immink, R.G.H.; Hogenhout, S.A. Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell 2021, 184, 5201–5214.e12. [Google Scholar] [CrossRef]
- Jones, O.R.; Scheuerlein, A.; Salguero-Gómez, R.; Camarda, C.G.; Schaible, R.; Casper, B.B.; Dahlgren, J.P.; Ehrlén, J.; García, M.B.; Menges, E.S.; et al. Diversity of ageing across the tree of life. Nature 2014, 505, 169–173. [Google Scholar] [CrossRef]
- Fernández-Moreno, M.A.; Farr, C.L.; Kaguni, L.S.; Garesse, R. Drosophila melanogaster as a model system to study mitochondrial biology. Methods Mol. Biol. 2007, 372, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.R.; Hanson-Smith, V.; Johnson, A.D. Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science 2013, 342, 104–108. [Google Scholar] [CrossRef]
- Diss, G.; Gagnon-Arsenault, I.; Dion-Coté, A.M.; Vignaud, H.; Ascencio, D.I.; Berger, C.M.; Landry, C.R. Gene duplication can impart fragility, not robustness, in the yeast protein interaction network. Science 2017, 355, 630–634. [Google Scholar] [CrossRef]
- Marlétaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 2018, 564, 64–70. [Google Scholar] [CrossRef]
- Altintas, O.; Park, S.; Lee, S.J. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Rep. 2016, 49, 81–92. [Google Scholar] [CrossRef]
- Nässel, D.R.; Vanden Broeck, J. Insulin/IGF signaling in Drosophila and other insects: Factors that regulate production, release and post-release action of the insulin-like peptides. Cell Mol. Life Sci. 2016, 73, 271–290. [Google Scholar] [CrossRef]
- Mathew, R.; Pal Bhadra, M.; Bhadra, U. Insulin/insulin-like growth factor-1 signalling (IIS) based regulation of lifespan across species. Biogerontology 2017, 18, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Rendón, J.P.; Salceda, R.; Riesgo-Escovar, J.R. Drosophila melanogaster as a Model for Diabetes Type 2 Progression. Biomed. Res. Int. 2018, 2018, 1417528. [Google Scholar] [CrossRef] [PubMed]
- Vinayagam, A.; Kulkarni, M.M.; Sopko, R.; Sun, X.; Hu, Y.; Nand, A.; Villalta, C.; Moghimi, A.; Yang, X.; Mohr, S.E.; et al. An Integrative Analysis of the InR/PI3K/Akt Network Identifies the Dynamic Response to Insulin Signaling. Cell Rep. 2016, 16, 3062–3074. [Google Scholar] [CrossRef] [PubMed]
- Brogiolo, W.; Stocker, H.; Ikeya, T.; Rintelen, F.; Fernandez, R.; Hafen, E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001, 11, 213–221. [Google Scholar] [CrossRef]
- Grönke, S.; Clarke, D.F.; Broughton, S.; Andrews, T.D.; Partridge, L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010, 6, e1000857. [Google Scholar] [CrossRef]
- Ruan, Y.; Chen, C.; Cao, Y.; Garofalo, R.S. The Drosophila insulin receptor contains a novel carboxyl-terminal extension likely to play an important role in signal transduction. J. Biol. Chem. 1995, 270, 4236–4243. [Google Scholar] [CrossRef]
- Chen, C.; Jack, J.; Garofalo, R.S. The Drosophila insulin receptor is required for normal growth. Endocrinology 1996, 137, 846–856. [Google Scholar] [CrossRef]
- Yamamoto, R.; Palmer, M.; Koski, H.; Curtis-Joseph, N.; Tatar, M. Aging modulated by the Drosophila insulin receptor through distinct structure-defined mechanisms. Genetics 2021, 217, iyaa037. [Google Scholar] [CrossRef]
- Tatar, M.; Kopelman, A.; Epstein, D.; Tu, M.P.; Yin, C.M.; Garofalo, R.S. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 2001, 292, 107–110. [Google Scholar] [CrossRef]
- Fernandez, R.; Tabarini, D.; Azpiazu, N.; Frasch, M.; Schlessinger, J. The Drosophila insulin receptor homolog: A gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 1995, 14, 3373–3384. [Google Scholar] [CrossRef]
- Weinkove, D.; Leevers, S.J.; MacDougall, L.K.; Waterfield, M.D. p60 is an adaptor for the Drosophila phosphoinositide 3-kinase, Dp110. J. Biol. Chem. 1997, 272, 14606–14610. [Google Scholar] [CrossRef] [PubMed]
- Werz, C.; Köhler, K.; Hafen, E.; Stocker, H. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 2009, 5, e1000596. [Google Scholar] [CrossRef] [PubMed]
- Slack, C.; Werz, C.; Wieser, D.; Alic, N.; Foley, A.; Stocker, H.; Withers, D.J.; Thornton, J.M.; Hafen, E.; Partridge, L. Regulation of lifespan, metabolism, and stress responses by the Drosophila SH2B protein, Lnk. PLoS Genet. 2010, 6, e1000881. [Google Scholar] [CrossRef]
- Leevers, S.J.; Weinkove, D.; MacDougall, L.K.; Hafen, E.; Waterfield, M.D. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996, 15, 6584–6594. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; Deak, M.; Casamayor, A.; Caudwell, F.B.; Morrice, N.; Norman, D.G.; Gaffney, P.; Reese, C.B.; MacDougall, C.N.; Harbison, D.; et al. 3-Phosphoinositide-dependent protein kinase-1 (PDK1): Structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol. 1997, 7, 776–789. [Google Scholar] [CrossRef]
- Verdu, J.; Buratovich, M.A.; Wilder, E.L.; Birnbaum, M.J. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1999, 1, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.S.; Lee, J.H.; Kim, S.; Kim, D.; Koh, H.; Lee, J.; Kim, C.; Kim, J.; Chung, J. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 6144–6149. [Google Scholar] [CrossRef]
- Kim, M.J.; O’Connor, M.B. Drosophila Activin signaling promotes muscle growth through InR/TORC1-dependent and -independent processes. Development 2021, 148, dev190868. [Google Scholar] [CrossRef]
- Webb, A.E.; Kundaje, A.; Brunet, A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell 2016, 15, 673–685. [Google Scholar] [CrossRef]
- Birnbaum, A.; Wu, X.; Tatar, M.; Liu, N.; Bai, H. Age-Dependent Changes in Transcription Factor FOXO Targeting in Female. Front. Genet. 2019, 10, 312. [Google Scholar] [CrossRef] [Green Version]
- Puig, O.; Marr, M.T.; Ruhf, M.L.; Tjian, R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003, 17, 2006–2020. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.D.; Luo, X.; Biteau, B.; Syverson, K.; Jasper, H. 14-3-3 Epsilon antagonizes FoxO to control growth, apoptosis and longevity in Drosophila. Aging Cell 2008, 7, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Tzivion, G.; Dobson, M.; Ramakrishnan, G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim. Biophys. Acta 2011, 1813, 1938–1945. [Google Scholar] [CrossRef]
- van der Horst, A.; Burgering, B.M. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Zheng, H. Role of FoxO transcription factors in aging and age-related metabolic and neurodegenerative diseases. Cell Biosci. 2021, 11, 188. [Google Scholar] [CrossRef]
- Brunet, A.; Kanai, F.; Stehn, J.; Xu, J.; Sarbassova, D.; Frangioni, J.V.; Dalal, S.N.; DeCaprio, J.A.; Greenberg, M.E.; Yaffe, M.B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002, 156, 817–828. [Google Scholar] [CrossRef]
- Sunayama, J.; Tsuruta, F.; Masuyama, N.; Gotoh, Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 2005, 170, 295–304. [Google Scholar] [CrossRef]
- Goberdhan, D.C.; Paricio, N.; Goodman, E.C.; Mlodzik, M.; Wilson, C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 1999, 13, 3244–3258. [Google Scholar] [CrossRef]
- Huang, H.; Potter, C.J.; Tao, W.; Li, D.M.; Brogiolo, W.; Hafen, E.; Sun, H.; Xu, T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development 1999, 126, 5365–5372. [Google Scholar] [CrossRef]
- Gao, X.; Neufeld, T.P.; Pan, D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 2000, 221, 404–418. [Google Scholar] [CrossRef] [Green Version]
- Leslie, N.R.; Batty, I.H.; Maccario, H.; Davidson, L.; Downes, C.P. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene 2008, 27, 5464–5476. [Google Scholar] [CrossRef]
- Bánréti, Á.; Lukácsovich, T.; Csikós, G.; Erdélyi, M.; Sass, M. PP2A regulates autophagy in two alternative ways in Drosophila. Autophagy 2012, 8, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Fischer, P.; Preiss, A.; Nagel, A.C. A triangular connection between Cyclin G, PP2A and Akt1 in the regulation of growth and metabolism in Drosophila. Fly 2016, 10, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Leader, D.P.; Krause, S.A.; Pandit, A.; Davies, S.A.; Dow, J.A.T. FlyAtlas 2: A new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 2018, 46, D809–D815. [Google Scholar] [CrossRef]
- Broughton, S.J.; Piper, M.D.; Ikeya, T.; Bass, T.M.; Jacobson, J.; Driege, Y.; Martinez, P.; Hafen, E.; Withers, D.J.; Leevers, S.J.; et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 2005, 102, 3105–3110. [Google Scholar] [CrossRef]
- Tu, M.P.; Epstein, D.; Tatar, M. The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell 2002, 1, 75–80. [Google Scholar] [CrossRef]
- Clancy, D.J.; Gems, D.; Harshman, L.G.; Oldham, S.; Stocker, H.; Hafen, E.; Leevers, S.J.; Partridge, L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 2001, 292, 104–106. [Google Scholar] [CrossRef]
- Hwangbo, D.S.; Gershman, B.; Gersham, B.; Tu, M.P.; Palmer, M.; Tatar, M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 2004, 429, 562–566. [Google Scholar] [CrossRef]
- Demontis, F.; Perrimon, N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 2010, 143, 813–825. [Google Scholar] [CrossRef] [Green Version]
- Giannakou, M.E.; Goss, M.; Jünger, M.A.; Hafen, E.; Leevers, S.J.; Partridge, L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 2004, 305, 361. [Google Scholar] [CrossRef] [PubMed]
- Wessells, R.J.; Fitzgerald, E.; Cypser, J.R.; Tatar, M.; Bodmer, R. Insulin regulation of heart function in aging fruit flies. Nat. Genet. 2004, 36, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Moya, N.; Niessen, S.; Hoover, H.; Mihaylova, M.M.; Shaw, R.J.; Yates, J.R.; Fischer, W.H.; Thomas, J.B.; Montminy, M. A hormone-dependent module regulating energy balance. Cell 2011, 145, 596–606. [Google Scholar] [CrossRef]
- Lehmann, M. Endocrine and physiological regulation of neutral fat storage in Drosophila. Mol. Cell. Endocrinol. 2018, 461, 165–177. [Google Scholar] [CrossRef]
- Böhni, R.; Riesgo-Escovar, J.; Oldham, S.; Brogiolo, W.; Stocker, H.; Andruss, B.F.; Beckingham, K.; Hafen, E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 1999, 97, 865–875. [Google Scholar] [CrossRef]
- Scanga, S.E.; Ruel, L.; Binari, R.C.; Snow, B.; Stambolic, V.; Bouchard, D.; Peters, M.; Calvieri, B.; Mak, T.W.; Woodgett, J.R.; et al. The conserved PI3’K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 2000, 19, 3971–3977. [Google Scholar] [CrossRef] [PubMed]
- Rintelen, F.; Stocker, H.; Thomas, G.; Hafen, E. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 15020–15025. [Google Scholar] [CrossRef] [PubMed]
- Slack, C.; Giannakou, M.E.; Foley, A.; Goss, M.; Partridge, L. dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell 2011, 10, 735–748. [Google Scholar] [CrossRef]
- Partridge, L.; Gems, D.; Withers, D.J. Sex and death: What is the connection? Cell 2005, 120, 461–472. [Google Scholar] [CrossRef]
- Giannakou, M.E.; Partridge, L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem. Sci. 2007, 32, 180–188. [Google Scholar] [CrossRef]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR Signaling and Nutrient Sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, C.S.; Hansen, B.W.; Vang, O. Are invertebrates relevant models in ageing research? Focus on the effects of rapamycin on TOR. Mech. Ageing Dev. 2016, 153, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Antikainen, H.; Driscoll, M.; Haspel, G.; Dobrowolski, R. TOR-mediated regulation of metabolism in aging. Aging Cell 2017, 16, 1219–1233. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.; Rallis, C. The TOR Signaling Pathway in Spatial and Temporal Control of Cell Size and Growth. Front. Cell Dev. Biol. 2017, 5, 61. [Google Scholar] [CrossRef]
- Yao, Y.; Jones, E.; Inoki, K. Lysosomal Regulation of mTORC1 by Amino Acids in Mammalian Cells. Biomolecules 2017, 7, 51. [Google Scholar] [CrossRef]
- Bjedov, I.; Rallis, C. The Target of Rapamycin Signalling Pathway in Ageing and Lifespan Regulation. Genes 2020, 11, 1043. [Google Scholar] [CrossRef]
- Vézina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef]
- Jacinto, E.; Hall, M.N. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 2003, 4, 117–126. [Google Scholar] [CrossRef]
- Bjedov, I.; Toivonen, J.M.; Kerr, F.; Slack, C.; Jacobson, J.; Foley, A.; Partridge, L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010, 11, 35–46. [Google Scholar] [CrossRef]
- Oldham, S.; Montagne, J.; Radimerski, T.; Thomas, G.; Hafen, E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000, 14, 2689–2694. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Billington, C.J.; Pan, D.; Neufeld, T.P. Drosophila target of rapamycin kinase functions as a multimer. Genetics 2006, 172, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, P.T.; Hay, N. The two TORCs and Akt. Dev. Cell 2007, 12, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Soulard, A.; Cohen, A.; Hall, M.N. TOR signaling in invertebrates. Curr. Opin. Cell Biol. 2009, 21, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Glatter, T.; Schittenhelm, R.B.; Rinner, O.; Roguska, K.; Wepf, A.; Jünger, M.A.; Köhler, K.; Jevtov, I.; Choi, H.; Schmidt, A.; et al. Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol. Syst. Biol. 2011, 7, 547. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef]
- Kim, E.; Goraksha-Hicks, P.; Li, L.; Neufeld, T.P.; Guan, K.L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008, 10, 935–945. [Google Scholar] [CrossRef]
- Avruch, J.; Hara, K.; Lin, Y.; Liu, M.; Long, X.; Ortiz-Vega, S.; Yonezawa, K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene 2006, 25, 6361–6372. [Google Scholar] [CrossRef]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef]
- Kim, M.; Lee, J.H. Identification of an AMPK phosphorylation site in Drosophila TSC2 (gigas) that regulate cell growth. Int. J. Mol. Sci. 2015, 16, 7015–7026. [Google Scholar] [CrossRef]
- Cully, M.; Genevet, A.; Warne, P.; Treins, C.; Liu, T.; Bastien, J.; Baum, B.; Tapon, N.; Leevers, S.J.; Downward, J. A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol. Cell. Biol. 2010, 30, 481–495. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Inoki, K.; Kim, E.; Guan, K.L. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc. Natl. Acad. Sci. USA 2006, 103, 6811–6816. [Google Scholar] [CrossRef] [PubMed]
- Jevtov, I.; Zacharogianni, M.; van Oorschot, M.M.; van Zadelhoff, G.; Aguilera-Gomez, A.; Vuillez, I.; Braakman, I.; Hafen, E.; Stocker, H.; Rabouille, C. TORC2 mediates the heat stress response in Drosophila by promoting the formation of stress granules. J. Cell Sci. 2015, 128, 2497–2508. [Google Scholar] [CrossRef] [PubMed]
- Guertin, D.A.; Guntur, K.V.; Bell, G.W.; Thoreen, C.C.; Sabatini, D.M. Functional genomics identifies TOR-regulated genes that control growth and division. Curr. Biol. 2006, 16, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.S.; Evans, J.R.; Edgar, B.A. Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J. Cell Biol. 2007, 179, 1105–1113. [Google Scholar] [CrossRef]
- Teleman, A.A.; Hietakangas, V.; Sayadian, A.C.; Cohen, S.M. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008, 7, 21–32. [Google Scholar] [CrossRef]
- Kuo, Y.; Huang, H.; Cai, T.; Wang, T. Target of Rapamycin Complex 2 regulates cell growth via Myc in Drosophila. Sci. Rep. 2015, 5, 10339. [Google Scholar] [CrossRef]
- Tiebe, M.; Lutz, M.; De La Garza, A.; Buechling, T.; Boutros, M.; Teleman, A.A. REPTOR and REPTOR-BP Regulate Organismal Metabolism and Transcription Downstream of TORC1. Dev. Cell 2015, 33, 272–284. [Google Scholar] [CrossRef]
- Zhang, T.; Zhou, Q.; Ogmundsdottir, M.H.; Möller, K.; Siddaway, R.; Larue, L.; Hsing, M.; Kong, S.W.; Goding, C.R.; Palsson, A.; et al. Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1. J. Cell Sci. 2015, 128, 2938–2950. [Google Scholar] [CrossRef]
- Filer, D.; Thompson, M.A.; Takhaveev, V.; Dobson, A.J.; Kotronaki, I.; Green, J.W.M.; Heinemann, M.; Tullet, J.M.A.; Alic, N. RNA polymerase III limits longevity downstream of TORC1. Nature 2017, 552, 263–267. [Google Scholar] [CrossRef]
- Liu, Y.; Mattila, J.; Ventelä, S.; Yadav, L.; Zhang, W.; Lamichane, N.; Sundström, J.; Kauko, O.; Grénman, R.; Varjosalo, M.; et al. PWP1 Mediates Nutrient-Dependent Growth Control through Nucleolar Regulation of Ribosomal Gene Expression. Dev. Cell 2017, 43, 240–252.e5. [Google Scholar] [CrossRef] [Green Version]
- Demetriades, C.; Doumpas, N.; Teleman, A.A. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 2014, 156, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, Y.; Ting, C.Y.; Bettedi, L.; Kim, K.; Ghaniam, E.; Lilly, M.A. The Rag GTPase Regulates the Dynamic Behavior of TSC Downstream of Both Amino Acid and Growth Factor Restriction. Dev. Cell 2020, 55, 272–288.e5. [Google Scholar] [CrossRef] [PubMed]
- Colombani, J.; Raisin, S.; Pantalacci, S.; Radimerski, T.; Montagne, J.; Léopold, P. A nutrient sensor mechanism controls Drosophila growth. Cell 2003, 114, 739–749. [Google Scholar] [CrossRef]
- Goberdhan, D.C.; Meredith, D.; Boyd, C.A.; Wilson, C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 2005, 132, 2365–2375. [Google Scholar] [CrossRef]
- Ghosh, A.; Rideout, E.J.; Grewal, S.S. TIF-IA-dependent regulation of ribosome synthesis in Drosophila muscle is required to maintain systemic insulin signaling and larval growth. PLoS Genet. 2014, 10, e1004750. [Google Scholar] [CrossRef]
- Kim, J.S.; Ro, S.H.; Kim, M.; Park, H.W.; Semple, I.A.; Park, H.; Cho, U.S.; Wang, W.; Guan, K.L.; Karin, M.; et al. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 2015, 5, 9502. [Google Scholar] [CrossRef]
- Lu, J.; Temp, U.; Müller-Hartmann, A.; Esser, J.; Grönke, S.; Partridge, L. Sestrin is a key regulator of stem cell function and lifespan in response to dietary amino acids. Nat. Aging 2021, 1, 60–72. [Google Scholar] [CrossRef]
- Lee, J.H.; Cho, U.S.; Karin, M. Sestrin regulation of TORC1: Is Sestrin a leucine sensor? Sci. Signal. 2016, 9, re5. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef] [Green Version]
- Bar-Peled, L.; Schweitzer, L.D.; Zoncu, R.; Sabatini, D.M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012, 150, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.H.; Thapar, N.; Guo, L.; Martinez, M.; Maris, J.; Gau, C.L.; Lengyel, J.A.; Tamanoi, F. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 2003, 116, 3601–3610. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Manning, B.D. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem. J. 2008, 412, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Pan, D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001, 15, 1383–1392. [Google Scholar] [CrossRef]
- Tapon, N.; Ito, N.; Dickson, B.J.; Treisman, J.E.; Hariharan, I.K. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 2001, 105, 345–355. [Google Scholar] [CrossRef]
- Potter, C.J.; Huang, H.; Xu, T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 2001, 105, 357–368. [Google Scholar] [CrossRef]
- Potter, C.J.; Pedraza, L.G.; Xu, T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 2002, 4, 658–665. [Google Scholar] [CrossRef]
- Schleich, S.; Teleman, A.A. Akt phosphorylates both Tsc1 and Tsc2 in Drosophila, but neither phosphorylation is required for normal animal growth. PLoS ONE 2009, 4, e6305. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.L.; Tee, A.R.; Short, J.D.; Bergeron, J.M.; Kim, J.; Shen, J.; Guo, R.; Johnson, C.L.; Kiguchi, K.; Walker, C.L. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 2006, 173, 279–289. [Google Scholar] [CrossRef]
- Stocker, H.; Radimerski, T.; Schindelholz, B.; Wittwer, F.; Belawat, P.; Daram, P.; Breuer, S.; Thomas, G.; Hafen, E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 2003, 5, 559–565. [Google Scholar] [CrossRef] [Green Version]
- Saucedo, L.J.; Gao, X.; Chiarelli, D.A.; Li, L.; Pan, D.; Edgar, B.A. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 2003, 5, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Inoki, K.; Ikenoue, T.; Guan, K.L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006, 20, 2820–2832. [Google Scholar] [CrossRef] [PubMed]
- Hietakangas, V.; Cohen, S.M. Re-evaluating AKT regulation: Role of TOR complex 2 in tissue growth. Genes Dev. 2007, 21, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Blumhagen, R.; Lao, U.; Kuo, Y.; Edgar, B.A. LST8 regulates cell growth via target-of-rapamycin complex 2 (TORC2). Mol. Cell. Biol. 2012, 32, 2203–2213. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Pan, D. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 2004, 18, 2479–2484. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.A.; Hardie, D.G. A homologue of AMP-activated protein kinase in Drosophila melanogaster is sensitive to AMP and is activated by ATP depletion. Biochem. J. 2002, 367, 179–186. [Google Scholar] [CrossRef]
- Hindupur, S.K.; González, A.; Hall, M.N. The opposing actions of target of rapamycin and AMP-activated protein kinase in cell growth control. Cold Spring Harb. Perspect. Biol. 2015, 7, a019141. [Google Scholar] [CrossRef]
- Sinnett, S.E.; Brenman, J.E. The Role of AMPK in Drosophila melanogaster. Exp. Suppl. 2016, 107, 389–401. [Google Scholar] [CrossRef]
- Yuan, D.; Zhou, S.; Liu, S.; Li, K.; Zhao, H.; Long, S.; Liu, H.; Xie, Y.; Su, Y.; Yu, F.; et al. The AMPK-PP2A axis in insect fat body is activated by 20-hydroxyecdysone to antagonize insulin/IGF signaling and restrict growth rate. Proc. Natl. Acad. Sci. USA 2020, 117, 9292–9301. [Google Scholar] [CrossRef]
- Lee, J.H.; Budanov, A.V.; Park, E.J.; Birse, R.; Kim, T.E.; Perkins, G.A.; Ocorr, K.; Ellisman, M.H.; Bodmer, R.; Bier, E.; et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 2010, 327, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, M.; Tsuda, M.; Muramatsu, K.; Hatsuda, H.; Morishita, S.; Aigaki, T. A gain-of-function screen identifies wdb and lkb1 as lifespan-extending genes in Drosophila. Biochem. Biophys. Res. Commun. 2011, 405, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chen, J.; Schreiber, S.L.; Clardy, J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 1996, 273, 239–242. [Google Scholar] [CrossRef]
- Albig, A.R.; Decker, C.J. The target of rapamycin signaling pathway regulates mRNA turnover in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2001, 12, 3428–3438. [Google Scholar] [CrossRef]
- Ghartey-Kwansah, G.; Li, Z.; Feng, R.; Wang, L.; Zhou, X.; Chen, F.Z.; Xu, M.M.; Jones, O.; Mu, Y.; Chen, S.; et al. Comparative analysis of FKBP family protein: Evaluation, structure, and function in mammals and Drosophila melanogaster. BMC Dev. Biol. 2018, 18, 7. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.; Tran, T.T.; Taylor, D.; Lee, S.D.; Min, K.J. Effect of rapamycin on lifespan in Drosophila. Geriatr. Gerontol. Int. 2010, 10, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Schinaman, J.M.; Rana, A.; Ja, W.W.; Clark, R.I.; Walker, D.W. Rapamycin modulates tissue aging and lifespan independently of the gut microbiota in Drosophila. Sci. Rep. 2019, 9, 7824. [Google Scholar] [CrossRef]
- Reiling, J.H.; Hafen, E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004, 18, 2879–2892. [Google Scholar] [CrossRef]
- Chang, Y.Y.; Neufeld, T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 2009, 20, 2004–2014. [Google Scholar] [CrossRef]
- Montagne, J.; Stewart, M.J.; Stocker, H.; Hafen, E.; Kozma, S.C.; Thomas, G. Drosophila S6 kinase: A regulator of cell size. Science 1999, 285, 2126–2129. [Google Scholar] [CrossRef]
- Miron, M.; Lasko, P.; Sonenberg, N. Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol. Cell. Biol. 2003, 23, 9117–9126. [Google Scholar] [CrossRef] [PubMed]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Zid, B.M.; Rogers, A.N.; Katewa, S.D.; Vargas, M.A.; Kolipinski, M.C.; Lu, T.A.; Benzer, S.; Kapahi, P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 2009, 139, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Towpik, J.; Graczyk, D.; Gajda, A.; Lefebvre, O.; Boguta, M. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J. Biol. Chem. 2008, 283, 17168–17174. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, C.; Wang, L.; Shetty, M.; Mell, J.C.; Sell, C.; Noguchi, E. Maf1 limits RNA polymerase III-directed transcription to preserve genomic integrity and extend lifespan. Cell Cycle 2021, 20, 247–255. [Google Scholar] [CrossRef]
- Grewal, S.S.; Li, L.; Orian, A.; Eisenman, R.N.; Edgar, B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005, 7, 295–302. [Google Scholar] [CrossRef]
- Casper, A.L.; Baxter, K.; Van Doren, M. no child left behind encodes a novel chromatin factor required for germline stem cell maintenance in males but not females. Development 2011, 138, 3357–3366. [Google Scholar] [CrossRef]
- Gallant, P. Myc function in Drosophila. Cold Spring Harb. Perspect. Med. 2013, 3, a014324. [Google Scholar] [CrossRef]
- Herter, E.K.; Stauch, M.; Gallant, M.; Wolf, E.; Raabe, T.; Gallant, P. snoRNAs are a novel class of biologically relevant Myc targets. BMC Biol. 2015, 13, 25. [Google Scholar] [CrossRef]
- Liu, Y.; Cerejeira Matos, R.; Heino, T.I.; Hietakangas, V. PWP1 promotes nutrient-responsive expression of 5S ribosomal RNA. Biol. Open 2018, 7, bio037911c. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.X.; Regan, J.C.; Eßer, J.; Drews, L.F.; Weinseis, T.; Stinn, J.; Hahn, O.; Miller, R.A.; Grönke, S.; Partridge, L. A TORC1-histone axis regulates chromatin organisation and non-canonical induction of autophagy to ameliorate ageing. eLife 2021, 10, e62233. [Google Scholar] [CrossRef] [PubMed]
- Biteau, B.; Karpac, J.; Supoyo, S.; Degennaro, M.; Lehmann, R.; Jasper, H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010, 6, e1001159. [Google Scholar] [CrossRef] [PubMed]
- Luong, N.; Davies, C.R.; Wessells, R.J.; Graham, S.M.; King, M.T.; Veech, R.; Bodmer, R.; Oldham, S.M. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006, 4, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Endicott, S.J.; Boynton, D.N.; Beckmann, L.J.; Miller, R.A. Long-lived mice with reduced growth hormone signaling have a constitutive upregulation of hepatic chaperone-mediated autophagy. Autophagy 2021, 17, 612–625. [Google Scholar] [CrossRef]
- Bartke, A.; Westbrook, R. Metabolic characteristics of long-lived mice. Front. Genet. 2012, 3, 288. [Google Scholar] [CrossRef]
- Yang, S.; Long, L.H.; Li, D.; Zhang, J.K.; Jin, S.; Wang, F.; Chen, J.G. β-Guanidinopropionic acid extends the lifespan of Drosophila melanogaster via an AMP-activated protein kinase-dependent increase in autophagy. Aging Cell 2015, 14, 1024–1033. [Google Scholar] [CrossRef]
- Xi, J.; Cai, J.; Cheng, Y.; Fu, Y.; Wei, W.; Zhang, Z.; Zhuang, Z.; Hao, Y.; Lilly, M.A.; Wei, Y. The TORC1 inhibitor Nprl2 protects age-related digestive function in. Aging 2019, 11, 9811–9828. [Google Scholar] [CrossRef]
- Proshkina, E.N.; Shaposhnikov, M.V.; Shchegoleva, E.V.; Chernyshova, D.O.; Moskalev, A.A. Influence of Mitf gene overexpression on the life span of Drosophila melanogaster. Proc. Komi Sci. Cent. Ural. Div. Russ. Acad. Sci. 2020, 3, 41–46. [Google Scholar] [CrossRef]
- Greer, C.; Lee, M.; Westerhof, M.; Milholland, B.; Spokony, R.; Vijg, J.; Secombe, J. Myc-dependent genome instability and lifespan in Drosophila. PLoS ONE 2013, 8, e74641. [Google Scholar] [CrossRef]
- Martínez Corrales, G.; Filer, D.; Wenz, K.C.; Rogan, A.; Phillips, G.; Li, M.; Feseha, Y.; Broughton, S.J.; Alic, N. Partial Inhibition of RNA Polymerase I Promotes Animal Health and Longevity. Cell Rep. 2020, 30, 1661–1669.e4. [Google Scholar] [CrossRef] [Green Version]
- Moskalev, A.; Shaposhnikov, M. Pharmacological inhibition of NF-κB prolongs lifespan of Drosophila melanogaster. Aging 2011, 3, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Myllymäki, H.; Valanne, S.; Rämet, M. The Drosophila imd signaling pathway. J. Immunol. 2014, 192, 3455–3462. [Google Scholar] [CrossRef] [PubMed]
- Khor, S.; Cai, D. Control of lifespan and survival by Drosophila NF-κB signaling through neuroendocrine cells and neuroblasts. Aging 2020, 12, 24604–24622. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.H.; Rämet, M. The Drosophila Toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Govind, S. Control of development and immunity by rel transcription factors in Drosophila. Oncogene 1999, 18, 6875–6887. [Google Scholar] [CrossRef] [PubMed]
- Hetru, C.; Hoffmann, J.A. NF-kappaB in the immune response of Drosophila. Cold Spring Harb. Perspect. Biol. 2009, 1, a000232. [Google Scholar] [CrossRef]
- Louradour, I.; Sharma, A.; Morin-Poulard, I.; Letourneau, M.; Vincent, A.; Crozatier, M.; Vanzo, N. Reactive oxygen species-dependent Toll/NF-κB activation in the Drosophila hematopoietic niche confers resistance to wasp parasitism. eLife 2017, 6, e25496. [Google Scholar] [CrossRef]
- Bonnay, F.; Nguyen, X.H.; Cohen-Berros, E.; Troxler, L.; Batsche, E.; Camonis, J.; Takeuchi, O.; Reichhart, J.M.; Matt, N. Akirin specifies NF-κB selectivity of Drosophila innate immune response via chromatin remodeling. EMBO J. 2014, 33, 2349–2362. [Google Scholar] [CrossRef]
- Harnish, J.M.; Link, N.; Yamamoto, S. as a Model for Infectious Diseases. Int. J. Mol. Sci. 2021, 22, 2724. [Google Scholar] [CrossRef]
- Chang, C.I.; Chelliah, Y.; Borek, D.; Mengin-Lecreulx, D.; Deisenhofer, J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 2006, 311, 1761–1764. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, T.; Goldman, W.E.; Mellroth, P.; Steiner, H.; Fukase, K.; Kusumoto, S.; Harley, W.; Fox, A.; Golenbock, D.; Silverman, N. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 2004, 20, 637–649. [Google Scholar] [CrossRef]
- Lim, J.H.; Kim, M.S.; Kim, H.E.; Yano, T.; Oshima, Y.; Aggarwal, K.; Goldman, W.E.; Silverman, N.; Kurata, S.; Oh, B.H. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 2006, 281, 8286–8295. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Huang, X.; Yin, Y. Beyond immunity: The Imd pathway as a coordinator of host defense, organismal physiology and behavior. Dev. Comp. Immunol. 2018, 83, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zaidman-Rémy, A.; Hervé, M.; Poidevin, M.; Pili-Floury, S.; Kim, M.S.; Blanot, D.; Oh, B.H.; Ueda, R.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 2006, 24, 463–473. [Google Scholar] [CrossRef]
- Bischoff, V.; Vignal, C.; Duvic, B.; Boneca, I.G.; Hoffmann, J.A.; Royet, J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006, 2, e14. [Google Scholar] [CrossRef]
- Kleino, A.; Myllymäki, H.; Kallio, J.; Vanha-aho, L.M.; Oksanen, K.; Ulvila, J.; Hultmark, D.; Valanne, S.; Rämet, M. Pirk is a negative regulator of the Drosophila Imd pathway. J. Immunol. 2008, 180, 5413–5422. [Google Scholar] [CrossRef]
- Meinander, A.; Runchel, C.; Tenev, T.; Chen, L.; Kim, C.H.; Ribeiro, P.S.; Broemer, M.; Leulier, F.; Zvelebil, M.; Silverman, N.; et al. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012, 31, 2770–2783. [Google Scholar] [CrossRef]
- Paquette, N.; Broemer, M.; Aggarwal, K.; Chen, L.; Husson, M.; Ertürk-Hasdemir, D.; Reichhart, J.M.; Meier, P.; Silverman, N. Caspase-mediated cleavage, IAP binding, and ubiquitination: Linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol. Cell 2010, 37, 172–182. [Google Scholar] [CrossRef]
- Kim, C.H.; Paik, D.; Rus, F.; Silverman, N. The caspase-8 homolog Dredd cleaves Imd and Relish but is not inhibited by p35. J. Biol. Chem. 2014, 289, 20092–20101. [Google Scholar] [CrossRef]
- Fernando, M.D.; Kounatidis, I.; Ligoxygakis, P. Loss of Trabid, a new negative regulator of the drosophila immune-deficiency pathway at the level of TAK1, reduces life span. PLoS Genet. 2014, 10, e1004117. [Google Scholar] [CrossRef] [Green Version]
- Kounatidis, I.; Chtarbanova, S.; Cao, Y.; Hayne, M.; Jayanth, D.; Ganetzky, B.; Ligoxygakis, P. NF-κB Immunity in the Brain Determines Fly Lifespan in Healthy Aging and Age-Related Neurodegeneration. Cell Rep. 2017, 19, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Stoven, S.; Silverman, N.; Junell, A.; Hedengren-Olcott, M.; Erturk, D.; Engstrom, Y.; Maniatis, T.; Hultmark, D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc. Natl. Acad. Sci. USA 2003, 100, 5991–5996. [Google Scholar] [CrossRef] [PubMed]
- Silverman, N.; Zhou, R.; Stöven, S.; Pandey, N.; Hultmark, D.; Maniatis, T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000, 14, 2461–2471. [Google Scholar] [CrossRef] [PubMed]
- Ertürk-Hasdemir, D.; Broemer, M.; Leulier, F.; Lane, W.S.; Paquette, N.; Hwang, D.; Kim, C.H.; Stöven, S.; Meier, P.; Silverman, N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl. Acad. Sci. USA 2009, 106, 9779–9784. [Google Scholar] [CrossRef]
- Tanji, T.; Yun, E.Y.; Ip, Y.T. Heterodimers of NF-kappaB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 14715–14720. [Google Scholar] [CrossRef]
- Han, Z.S.; Ip, Y.T. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem. 1999, 274, 21355–21361. [Google Scholar] [CrossRef]
- Kunsch, C.; Ruben, S.M.; Rosen, C.A. Selection of optimal kappa B/Rel DNA-binding motifs: Interaction of both subunits of NF-kappa B with DNA is required for transcriptional activation. Mol. Cell. Biol. 1992, 12, 4412–4421. [Google Scholar] [CrossRef]
- Ryu, J.H.; Kim, S.H.; Lee, H.Y.; Bai, J.Y.; Nam, Y.D.; Bae, J.W.; Lee, D.G.; Shin, S.C.; Ha, E.M.; Lee, W.J. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Dantoft, W.; Davis, M.M.; Lindvall, J.M.; Tang, X.; Uvell, H.; Junell, A.; Beskow, A.; Engström, Y. The Oct1 homolog Nubbin is a repressor of NF-κB-dependent immune gene expression that increases the tolerance to gut microbiota. BMC Biol. 2013, 11, 99. [Google Scholar] [CrossRef]
- Myllymäki, H.; Rämet, M. Transcription factor zfh1 downregulates Drosophila Imd pathway. Dev. Comp. Immunol. 2013, 39, 188–197. [Google Scholar] [CrossRef]
- He, X.; Yu, J.; Wang, M.; Cheng, Y.; Han, Y.; Yang, S.; Shi, G.; Sun, L.; Fang, Y.; Gong, S.T.; et al. Bap180/Baf180 is required to maintain homeostasis of intestinal innate immune response in Drosophila and mice. Nat. Microbiol. 2017, 2, 17056. [Google Scholar] [CrossRef] [PubMed]
- Goto, A.; Matsushita, K.; Gesellchen, V.; El Chamy, L.; Kuttenkeuler, D.; Takeuchi, O.; Hoffmann, J.A.; Akira, S.; Boutros, M.; Reichhart, J.M. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in drosophila and mice. Nat. Immunol. 2008, 9, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, B.; Drogaris, P.; Lin, H.H.; Armstrong, H.; Hiragami-Hamada, K.; Imhof, A.; Bonneil, E.; Thibault, P.; Verreault, A.; Festenstein, R.J. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011, 7, e1001354. [Google Scholar] [CrossRef] [PubMed]
- Salminen, T.S.; Rämet, M. Pickle Flavors Relish in Drosophila Immunity. Cell Host Microbe 2016, 20, 273–274. [Google Scholar] [CrossRef]
- Kim, M.; Lee, J.H.; Lee, S.Y.; Kim, E.; Chung, J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc. Natl. Acad. Sci. USA 2006, 103, 16358–16363. [Google Scholar] [CrossRef]
- Park, J.M.; Brady, H.; Ruocco, M.G.; Sun, H.; Williams, D.; Lee, S.J.; Kato, T.; Richards, N.; Chan, K.; Mercurio, F.; et al. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 2004, 18, 584–594. [Google Scholar] [CrossRef]
- Maki, K.; Shibata, T.; Kawabata, S.I. Transglutaminase-catalyzed incorporation of polyamines masks the DNA-binding region of the transcription factor Relish. J. Biol. Chem. 2017, 292, 6369–6380. [Google Scholar] [CrossRef]
- Shibata, T.; Sekihara, S.; Fujikawa, T.; Miyaji, R.; Maki, K.; Ishihara, T.; Koshiba, T.; Kawabata, S. Transglutaminase-catalyzed protein-protein cross-linking suppresses the activity of the NF-κB-like transcription factor relish. Sci. Signal. 2013, 6, ra61. [Google Scholar] [CrossRef]
- Michel, T.; Reichhart, J.M.; Hoffmann, J.A.; Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756–759. [Google Scholar] [CrossRef]
- Gobert, V.; Gottar, M.; Matskevich, A.A.; Rutschmann, S.; Royet, J.; Belvin, M.; Hoffmann, J.A.; Ferrandon, D. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 2003, 302, 2126–2130. [Google Scholar] [CrossRef] [Green Version]
- Jang, I.H.; Chosa, N.; Kim, S.H.; Nam, H.J.; Lemaitre, B.; Ochiai, M.; Kambris, Z.; Brun, S.; Hashimoto, C.; Ashida, M.; et al. A Spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev. Cell 2006, 10, 45–55. [Google Scholar] [CrossRef] [PubMed]
- DeVeale, B.; Brummel, T.; Seroude, L. Immunity and aging: The enemy within? Aging Cell 2004, 3, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Morisato, D.; Anderson, K.V. The spätzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell 1994, 76, 677–688. [Google Scholar] [CrossRef]
- Weber, A.N.; Tauszig-Delamasure, S.; Hoffmann, J.A.; Lelièvre, E.; Gascan, H.; Ray, K.P.; Morse, M.A.; Imler, J.L.; Gay, N.J. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat. Immunol. 2003, 4, 794–800. [Google Scholar] [CrossRef]
- Chowdhury, M.; Li, C.F.; He, Z.; Lu, Y.; Liu, X.S.; Wang, Y.F.; Ip, Y.T.; Strand, M.R.; Yu, X.Q. Toll family members bind multiple Spätzle proteins and activate antimicrobial peptide gene expression in. J. Biol. Chem. 2019, 294, 10172–10181. [Google Scholar] [CrossRef]
- Sun, H.; Bristow, B.N.; Qu, G.; Wasserman, S.A. A heterotrimeric death domain complex in Toll signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 12871–12876. [Google Scholar] [CrossRef]
- Horng, T.; Medzhitov, R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 12654–12658. [Google Scholar] [CrossRef]
- Tauszig-Delamasure, S.; Bilak, H.; Capovilla, M.; Hoffmann, J.A.; Imler, J.L. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 2002, 3, 91–97. [Google Scholar] [CrossRef]
- Xiao, T.; Towb, P.; Wasserman, S.A.; Sprang, S.R. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell 1999, 99, 545–555. [Google Scholar] [CrossRef]
- Moncrieffe, M.C.; Grossmann, J.G.; Gay, N.J. Assembly of oligomeric death domain complexes during Toll receptor signaling. J. Biol. Chem. 2008, 283, 33447–33454. [Google Scholar] [CrossRef] [Green Version]
- Imler, J.L.; Hoffmann, J.A. Signaling mechanisms in the antimicrobial host defense of Drosophila. Curr. Opin. Microbiol. 2000, 3, 16–22. [Google Scholar] [CrossRef]
- Fernandez, N.Q.; Grosshans, J.; Goltz, J.S.; Stein, D. Separable and redundant regulatory determinants in Cactus mediate its dorsal group dependent degradation. Development 2001, 128, 2963–2974. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Khanuja, B.S.; Ip, Y.T. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 1999, 13, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Rutschmann, S.; Jung, A.C.; Hetru, C.; Reichhart, J.M.; Hoffmann, J.A.; Ferrandon, D. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 2000, 12, 569–580. [Google Scholar] [CrossRef]
- Sharma, V.; Mutsuddi, M.; Mukherjee, A. Deltex positively regulates Toll signaling in a JNK independent manner in Drosophila. Genes Cells 2021, 26, 254–263. [Google Scholar] [CrossRef]
- Bai, Z.; Wei, M.; Li, Z.; Xiao, W. Drosophila Uev1a is dually required for Ben-dependent DNA-damage response and fly mobility. Cell. Signal. 2020, 74, 109719. [Google Scholar] [CrossRef]
- Chen, L.; Paquette, N.; Mamoor, S.; Rus, F.; Nandy, A.; Leszyk, J.; Shaffer, S.A.; Silverman, N. Innate immune signaling in Drosophila is regulated by transforming growth factor β (TGF β)-activated kinase (Tak1)-triggered ubiquitin editing. J. Biol. Chem. 2017, 292, 8738–8749. [Google Scholar] [CrossRef]
- Ma, X.; Yang, L.; Yang, Y.; Li, M.; Li, W.; Xue, L. dUev1a modulates TNF-JNK mediated tumor progression and cell death in Drosophila. Dev. Biol. 2013, 380, 211–221. [Google Scholar] [CrossRef]
- Fabian, D.K.; Garschall, K.; Klepsatel, P.; Santos-Matos, G.; Sucena, É.; Kapun, M.; Lemaitre, B.; Schlötterer, C.; Arking, R.; Flatt, T. Evolution of longevity improves immunity in Drosophila. Evol. Lett. 2018, 2, 567–579. [Google Scholar] [CrossRef]
- Lin, Y.R.; Parikh, H.; Park, Y. Stress resistance and lifespan enhanced by downregulation of antimicrobial peptide genes in the Imd pathway. Aging 2018, 10, 622–631. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Li, W.; Yu, H.; Yang, Y.; Li, M.; Xue, L.; Xu, T. Bendless modulates JNK-mediated cell death and migration in Drosophila. Cell Death Differ. 2014, 21, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Kaduskar, B.; Trivedi, D.; Ratnaparkhi, G.S. Caspar SUMOylation regulates Drosophila lifespan. MicroPubl. Biol. 2020. [Google Scholar] [CrossRef]
- Morris, O.; Liu, X.; Domingues, C.; Runchel, C.; Chai, A.; Basith, S.; Tenev, T.; Chen, H.; Choi, S.; Pennetta, G.; et al. Signal Integration by the IκB Protein Pickle Shapes Drosophila Innate Host Defense. Cell Host Microbe 2016, 20, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.A.; Kohn, N.; Spirina, A.; McMillen, A.; Huang, W.; Mackay, T.F.C. Genetic Basis of Increased Lifespan and Postponed Senescence in Drosophila melanogaster. G3 Genes Genomes Genet. 2020, 10, 1087–1098. [Google Scholar] [CrossRef]
- Libert, S.; Chao, Y.; Chu, X.; Pletcher, S.D. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFkappaB signaling. Aging Cell 2006, 5, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Badinloo, M.; Nguyen, E.; Suh, W.; Alzahrani, F.; Castellanos, J.; Klichko, V.I.; Orr, W.C.; Radyuk, S.N. Overexpression of antimicrobial peptides contributes to aging through cytotoxic effects in Drosophila tissues. Arch. Insect Biochem. Physiol. 2018, 98, e21464. [Google Scholar] [CrossRef]
- Herrera, S.C.; Bach, E.A. The Emerging Roles of JNK Signaling in Drosophila Stem Cell Homeostasis. Int. J. Mol. Sci. 2021, 22, 5519. [Google Scholar] [CrossRef]
- Weston, C.R.; Davis, R.J. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 2007, 19, 142–149. [Google Scholar] [CrossRef]
- Davis, R.J. Signal transduction by the JNK group of MAP kinases. Cell 2000, 103, 239–252. [Google Scholar] [CrossRef]
- Ríos-Barrera, L.D.; Riesgo-Escovar, J.R. Regulating cell morphogenesis: The Drosophila Jun N-terminal kinase pathway. Genesis 2013, 51, 147–162. [Google Scholar] [CrossRef]
- Lesch, C.; Jo, J.; Wu, Y.; Fish, G.S.; Galko, M.J. A targeted UAS-RNAi screen in Drosophila larvae identifies wound closure genes regulating distinct cellular processes. Genetics 2010, 186, 943–957. [Google Scholar] [CrossRef]
- Kanda, H.; Igaki, T.; Kanuka, H.; Yagi, T.; Miura, M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J. Biol. Chem. 2002, 277, 28372–28375. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Mutsuddi, M.; Mukherjee, A. Deltex cooperates with TRAF6 to promote apoptosis and cell migration through Eiger-independent JNK activation in Drosophila. Cell Biol. Int. 2021, 45, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Andersen, D.S.; Colombani, J.; Palmerini, V.; Chakrabandhu, K.; Boone, E.; Röthlisberger, M.; Toggweiler, J.; Basler, K.; Mapelli, M.; Hueber, A.O.; et al. The Drosophila TNF receptor Grindelwald couples loss of cell polarity and neoplastic growth. Nature 2015, 522, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Palmerini, V.; Monzani, S.; Laurichesse, Q.; Loudhaief, R.; Mari, S.; Cecatiello, V.; Olieric, V.; Pasqualato, S.; Colombani, J.; Andersen, D.S.; et al. Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions. Nat. Commun. 2021, 12, 2070. [Google Scholar] [CrossRef]
- Igaki, T.; Kanda, H.; Yamamoto-Goto, Y.; Kanuka, H.; Kuranaga, E.; Aigaki, T.; Miura, M. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002, 21, 3009–3018. [Google Scholar] [CrossRef]
- Kodra, A.; de la Cova, C.; Singh, A.S.; Johnston, L.A. The TNF Egr participates in signaling during cell competition in the absence of a requirement for JNK. bioRxiv 2021. [Google Scholar] [CrossRef]
- Grech, A.; Quinn, R.; Srinivasan, D.; Badoux, X.; Brink, R. Complete structural characterisation of the mammalian and Drosophila TRAF genes: Implications for TRAF evolution and the role of RING finger splice variants. Mol. Immunol. 2000, 37, 721–734. [Google Scholar] [CrossRef]
- Liu, H.; Su, Y.C.; Becker, E.; Treisman, J.; Skolnik, E.Y. A Drosophila TNF-receptor-associated factor (TRAF) binds the ste20 kinase Misshapen and activates Jun kinase. Curr. Biol. 1999, 9, 101–104. [Google Scholar] [CrossRef]
- Geuking, P.; Narasimamurthy, R.; Basler, K. A genetic screen targeting the tumor necrosis factor/Eiger signaling pathway: Identification of Drosophila TAB2 as a functionally conserved component. Genetics 2005, 171, 1683–1694. [Google Scholar] [CrossRef] [Green Version]
- Moreno, E.; Yan, M.; Basler, K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr. Biol. 2002, 12, 1263–1268. [Google Scholar] [CrossRef]
- Xue, L.; Igaki, T.; Kuranaga, E.; Kanda, H.; Miura, M.; Xu, T. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev. Cell 2007, 13, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Kauppila, S.; Maaty, W.S.; Chen, P.; Tomar, R.S.; Eby, M.T.; Chapo, J.; Chew, S.; Rathore, N.; Zachariah, S.; Sinha, S.K.; et al. Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene 2003, 22, 4860–4867. [Google Scholar] [CrossRef] [PubMed]
- Igaki, T. Correcting developmental errors by apoptosis: Lessons from Drosophila JNK signaling. Apoptosis 2009, 14, 1021–1028. [Google Scholar] [CrossRef]
- Cha, G.H.; Cho, K.S.; Lee, J.H.; Kim, M.; Kim, E.; Park, J.; Lee, S.B.; Chung, J. Discrete functions of TRAF1 and TRAF2 in Drosophila melanogaster mediated by c-Jun N-terminal kinase and NF-kappaB-dependent signaling pathways. Mol. Cell. Biol. 2003, 23, 7982–7991. [Google Scholar] [CrossRef]
- Takatsu, Y.; Nakamura, M.; Stapleton, M.; Danos, M.C.; Matsumoto, K.; O’Connor, M.B.; Shibuya, H.; Ueno, N. TAK1 participates in c-Jun N-terminal kinase signaling during Drosophila development. Mol. Cell. Biol. 2000, 20, 3015–3026. [Google Scholar] [CrossRef]
- Ryabinina, O.P.; Subbian, E.; Iordanov, M.S. D-MEKK1, the Drosophila orthologue of mammalian MEKK4/MTK1, and Hemipterous/D-MKK7 mediate the activation of D-JNK by cadmium and arsenite in Schneider cells. BMC Cell Biol. 2006, 7, 7. [Google Scholar] [CrossRef]
- Neisch, A.L.; Speck, O.; Stronach, B.; Fehon, R.G. Rho1 regulates apoptosis via activation of the JNK signaling pathway at the plasma membrane. J. Cell Biol. 2010, 189, 311–323. [Google Scholar] [CrossRef]
- Kang, M.J.; Chung, J.; Ryoo, H.D. CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat. Cell Biol. 2012, 14, 409–415. [Google Scholar] [CrossRef]
- La Marca, J.E.; Richardson, H.E. Two-Faced: Roles of JNK Signalling During Tumourigenesis in the. Front. Cell Dev. Biol. 2020, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Kuranaga, E.; Kanuka, H.; Igaki, T.; Sawamoto, K.; Ichijo, H.; Okano, H.; Miura, M. Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat. Cell Biol. 2002, 4, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Tafesh-Edwards, G.; Eleftherianos, I. JNK signaling in Drosophila immunity and homeostasis. Immunol Lett. 2020, 226, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.S.; Moberg, K.H. Drosophila endocytic neoplastic tumor suppressor genes regulate Sav/Wts/Hpo signaling and the c-Jun N-terminal kinase pathway. Cell Cycle 2011, 10, 4110–4118. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Martínez, M.; Stamatakis, K.; Fresno, M. The Dual-Specificity Phosphatase 10 (DUSP10): Its Role in Cancer, Inflammation, and Immunity. Int. J. Mol. Sci. 2019, 20, 1626. [Google Scholar] [CrossRef]
- Pastor-Pareja, J.C.; Wu, M.; Xu, T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis. Model. Mech. 2008, 1, 144–154; discussion 153. [Google Scholar] [CrossRef]
- Uhlirova, M.; Bohmann, D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 2006, 25, 5294–5304. [Google Scholar] [CrossRef]
- Kim, L.K.; Choi, U.Y.; Cho, H.S.; Lee, J.S.; Lee, W.B.; Kim, J.; Jeong, K.; Shim, J.; Kim-Ha, J.; Kim, Y.J. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 2007, 5, e238. [Google Scholar] [CrossRef]
- Biswas, R.; Stein, D.; Stanley, E.R. Drosophila Dok is required for embryonic dorsal closure. Development 2006, 133, 217–227. [Google Scholar] [CrossRef]
- Kučerová, L.; Kubrak, O.I.; Bengtsson, J.M.; Strnad, H.; Nylin, S.; Theopold, U.; Nässel, D.R. Slowed aging during reproductive dormancy is reflected in genome-wide transcriptome changes in Drosophila melanogaster. BMC Genom. 2016, 17, 50. [Google Scholar] [CrossRef]
- Wang, M.C.; Bohmann, D.; Jasper, H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 2005, 121, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.S.; Iijima-Ando, K.; Iijima, K.; Lee, W.J.; Lee, J.H.; Yu, K.; Lee, D.S. JNK/FOXO-mediated neuronal expression of fly homologue of peroxiredoxin II reduces oxidative stress and extends life span. J. Biol. Chem. 2009, 284, 29454–29461. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.W.; Marinissen, M.J.; Oh, S.W.; Chen, X.; Melnick, M.; Perrimon, N.; Gutkind, J.S.; Hou, S.X. CKA, a novel multidomain protein, regulates the JUN N-terminal kinase signal transduction pathway in Drosophila. Mol. Cell. Biol. 2002, 22, 1792–1803. [Google Scholar] [CrossRef] [PubMed]
- Ashton-Beaucage, D.; Udell, C.M.; Gendron, P.; Sahmi, M.; Lefrançois, M.; Baril, C.; Guenier, A.S.; Duchaine, J.; Lamarre, D.; Lemieux, S.; et al. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 2014, 12, e1001809. [Google Scholar] [CrossRef] [PubMed]
- Bond, D.; Foley, E. A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 2009, 5, e1000655. [Google Scholar] [CrossRef] [PubMed]
- Goudreault, M.; D’Ambrosio, L.M.; Kean, M.J.; Mullin, M.J.; Larsen, B.G.; Sanchez, A.; Chaudhry, S.; Chen, G.I.; Sicheri, F.; Nesvizhskii, A.I.; et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteom. 2009, 8, 157–171. [Google Scholar] [CrossRef]
- Ribeiro, P.S.; Josué, F.; Wepf, A.; Wehr, M.C.; Rinner, O.; Kelly, G.; Tapon, N.; Gstaiger, M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 2010, 39, 521–534. [Google Scholar] [CrossRef]
- La Marca, J.E.; Diepstraten, S.T.; Hodge, A.L.; Wang, H.; Hart, A.H.; Richardson, H.E.; Somers, W.G. Strip and Cka negatively regulate JNK signalling during Drosophila spermatogenesis. Development 2019, 146, dev174292. [Google Scholar] [CrossRef]
- Delaney, J.R.; Stöven, S.; Uvell, H.; Anderson, K.V.; Engström, Y.; Mlodzik, M. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. EMBO J. 2006, 25, 3068–3077. [Google Scholar] [CrossRef]
- Nandipati, K.C.; Subramanian, S.; Agrawal, D.K. Protein kinases: Mechanisms and downstream targets in inflammation-mediated obesity and insulin resistance. Mol. Cell. Biochem. 2017, 426, 27–45. [Google Scholar] [CrossRef]
- Weina, T.; Ying, L.; Yiwen, W.; Huan-Huan, Q. What we have learnt from Drosophila model organism: The coordination between insulin signaling pathway and tumor cells. Heliyon 2022, 8, e09957. [Google Scholar] [CrossRef]
- Dantzer, R. Neuroimmune Interactions: From the Brain to the Immune System and Vice Versa. Physiol. Rev. 2018, 98, 477–504. [Google Scholar] [CrossRef] [PubMed]
- Gernert, M.; Feja, M. Bypassing the Blood-Brain Barrier: Direct Intracranial Drug Delivery in Epilepsies. Pharmaceutics 2020, 12, 1134. [Google Scholar] [CrossRef] [PubMed]
- Dulac, A.; Issa, A.R.; Sun, J.; Matassi, G.; Jonas, C.; Chérif-Zahar, B.; Cattaert, D.; Birman, S. A Novel Neuron-Specific Regulator of the V-ATPase in Drosophila. eNeuro 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Yu, G.; Li, K.; Du, Y.; Yuan, Z.; Gao, Y.; Fan, X.; Yang, D.; Mao, X.; Yang, M. VhaAC39-1 regulates gut homeostasis and affects the health span in Drosophila. Mech. Ageing Dev. 2022, 204, 111673. [Google Scholar] [CrossRef] [PubMed]
- Strilbytska, O.M.; Semaniuk, U.V.; Storey, K.B.; Edgar, B.A.; Lushchak, O.V. Activation of the Tor/Myc signaling axis in intestinal stem and progenitor cells affects longevity, stress resistance and metabolism in Drosophila. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 203, 92–99. [Google Scholar] [CrossRef]
- Hatfield, I.; Harvey, I.; Yates, E.R.; Redd, J.R.; Reiter, L.T.; Bridges, D. The role of TORC1 in muscle development in Drosophila. Sci. Rep. 2015, 5, 9676. [Google Scholar] [CrossRef]
- Chang, K.; Kang, P.; Liu, Y.; Huang, K.; Miao, T.; Sagona, A.P.; Nezis, I.P.; Bodmer, R.; Ocorr, K.; Bai, H. TGFB-INHB/activin signaling regulates age-dependent autophagy and cardiac health through inhibition of MTORC2. Autophagy 2020, 16, 1807–1822. [Google Scholar] [CrossRef]
- Zhan, L.; Xie, Q.; Tibbetts, R.S. Opposing roles of p38 and JNK in a Drosophila model of TDP-43 proteinopathy reveal oxidative stress and innate immunity as pathogenic components of neurodegeneration. Hum. Mol. Genet. 2015, 24, 757–772. [Google Scholar] [CrossRef]
- Ji, S.; Luo, Y.; Cai, Q.; Cao, Z.; Zhao, Y.; Mei, J.; Li, C.; Xia, P.; Xie, Z.; Xia, Z.; et al. LC Domain-Mediated Coalescence Is Essential for Otu Enzymatic Activity to Extend Drosophila Lifespan. Mol. Cell 2019, 74, 363–377. [Google Scholar] [CrossRef]
- Dantoft, W.; Lundin, D.; Esfahani, S.S.; Engström, Y. The POU/Oct Transcription Factor Pdm1/nub Is Necessary for a Beneficial Gut Microbiota and Normal Lifespan of Drosophila. J. Innate Immun. 2016, 8, 412–426. [Google Scholar] [CrossRef]
- Wu, K.; Tang, Y.; Zhang, Q.; Zhuo, Z.; Sheng, X.; Huang, J.; Ye, J.; Li, X.; Liu, Z.; Chen, H. Aging-related upregulation of the homeobox gene caudal represses intestinal stem cell differentiation in Drosophila. PLoS Genet. 2021, 17, e1009649. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.S.; Ayres, J.S.; Brandt, S.M.; Costa, A.; Dionne, M.S.; Gordon, M.D.; Mabery, E.M.; Moule, M.G.; Pham, L.N.; Shirasu-Hiza, M.M. Drosophila eiger mutants are sensitive to extracellular pathogens. PLoS Pathog. 2007, 3, e41. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Sachan, N.; Mutsuddi, M.; Mukherjee, A. Kinase active Misshapen regulates Notch signaling in Drosophila melanogaster. Exp. Cell. Res. 2015, 339, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Ishibashi, T.; Ishikawa, H.O.; Sato, H.; Usui, T.; Okuda, T.; Yashiro, H.; Ishikawa, H.; Taikou, Y.; Minami, A.; et al. A gain-of-function screen to identify genes that reduce lifespan in the adult of Drosophila melanogaster. BMC Genet. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.C.; Bohmann, D.; Jasper, H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell 2003, 5, 811–816. [Google Scholar] [CrossRef]
- Biteau, B.; Hochmuth, C.E.; Jasper, H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 2008, 3, 442–455. [Google Scholar] [CrossRef]
- Hamada, N.; Bäckesjö, C.M.; Smith, C.I.; Yamamoto, D. Functional replacement of Drosophila Btk29A with human Btk in male genital development and survival. FEBS Lett. 2005, 579, 4131–4137. [Google Scholar] [CrossRef]
- Kikuchi, M.; Sekiya, M.; Hara, N.; Miyashita, A.; Kuwano, R.; Ikeuchi, T.; Iijima, K.M.; Nakaya, A. Disruption of a RAC1-centred network is associated with Alzheimer’s disease pathology and causes age-dependent neurodegeneration. Hum. Mol. Genet. 2020, 29, 817–833. [Google Scholar] [CrossRef]
- Magwire, M.M.; Yamamoto, A.; Carbone, M.A.; Roshina, N.V.; Symonenko, A.V.; Pasyukova, E.G.; Morozova, T.V.; Mackay, T.F. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 2010, 6, e1001037. [Google Scholar] [CrossRef]
- Soory, A.; Ratnaparkhi, G.S. SUMOylation of Jun fine-tunes the Drosophila gut immune response. PLoS Pathog. 2022, 18, e1010356. [Google Scholar] [CrossRef]
- Larson, K.; Yan, S.J.; Tsurumi, A.; Liu, J.; Zhou, J.; Gaur, K.; Guo, D.; Eickbush, T.H.; Li, W.X. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012, 8, e1002473. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogienko, A.A.; Omelina, E.S.; Bylino, O.V.; Batin, M.A.; Georgiev, P.G.; Pindyurin, A.V. Drosophila as a Model Organism to Study Basic Mechanisms of Longevity. Int. J. Mol. Sci. 2022, 23, 11244. https://doi.org/10.3390/ijms231911244
Ogienko AA, Omelina ES, Bylino OV, Batin MA, Georgiev PG, Pindyurin AV. Drosophila as a Model Organism to Study Basic Mechanisms of Longevity. International Journal of Molecular Sciences. 2022; 23(19):11244. https://doi.org/10.3390/ijms231911244
Chicago/Turabian StyleOgienko, Anna A., Evgeniya S. Omelina, Oleg V. Bylino, Mikhail A. Batin, Pavel G. Georgiev, and Alexey V. Pindyurin. 2022. "Drosophila as a Model Organism to Study Basic Mechanisms of Longevity" International Journal of Molecular Sciences 23, no. 19: 11244. https://doi.org/10.3390/ijms231911244





