Remodeling of Liver and Plasma Lipidomes in Mice Lacking Cyclophilin D
Abstract
1. Introduction
2. Results
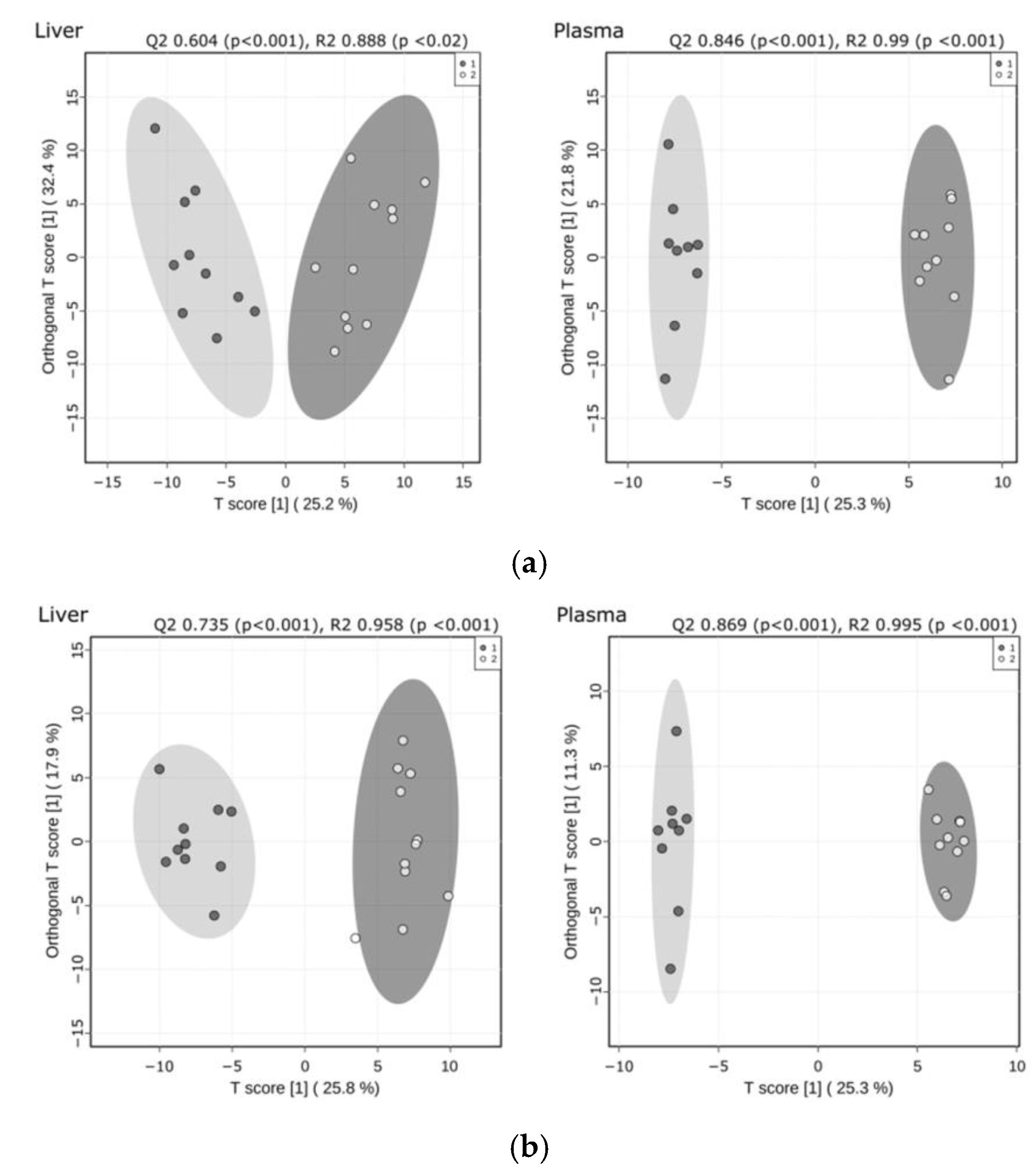
2.1. Significant Remodeling of the Liver and Plasma Lipidomes Occurred in Response to CypD Deletion
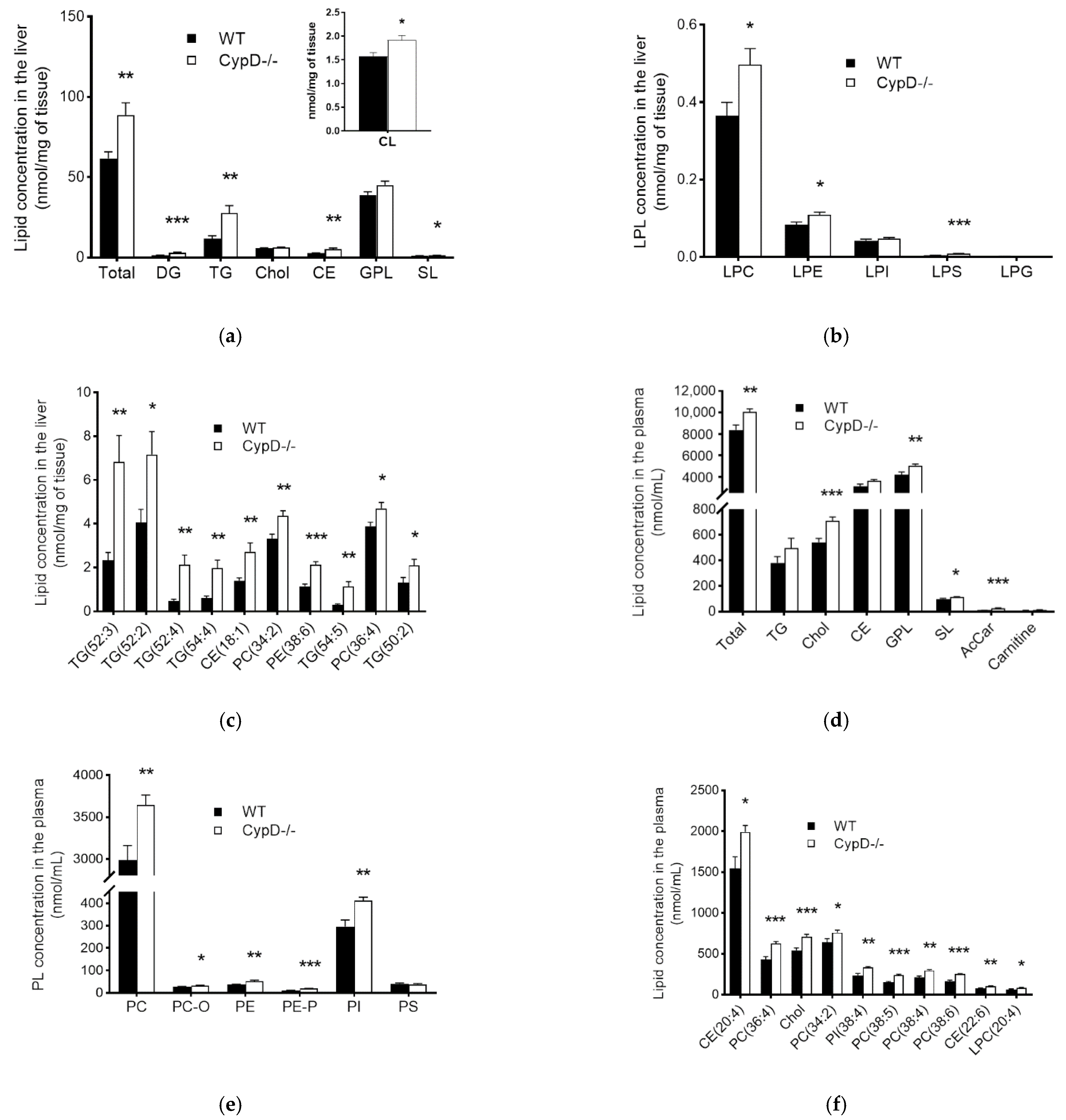
2.2. The Lipid Content Increased Due to CypD Ablation Both in the Liver and Plasma
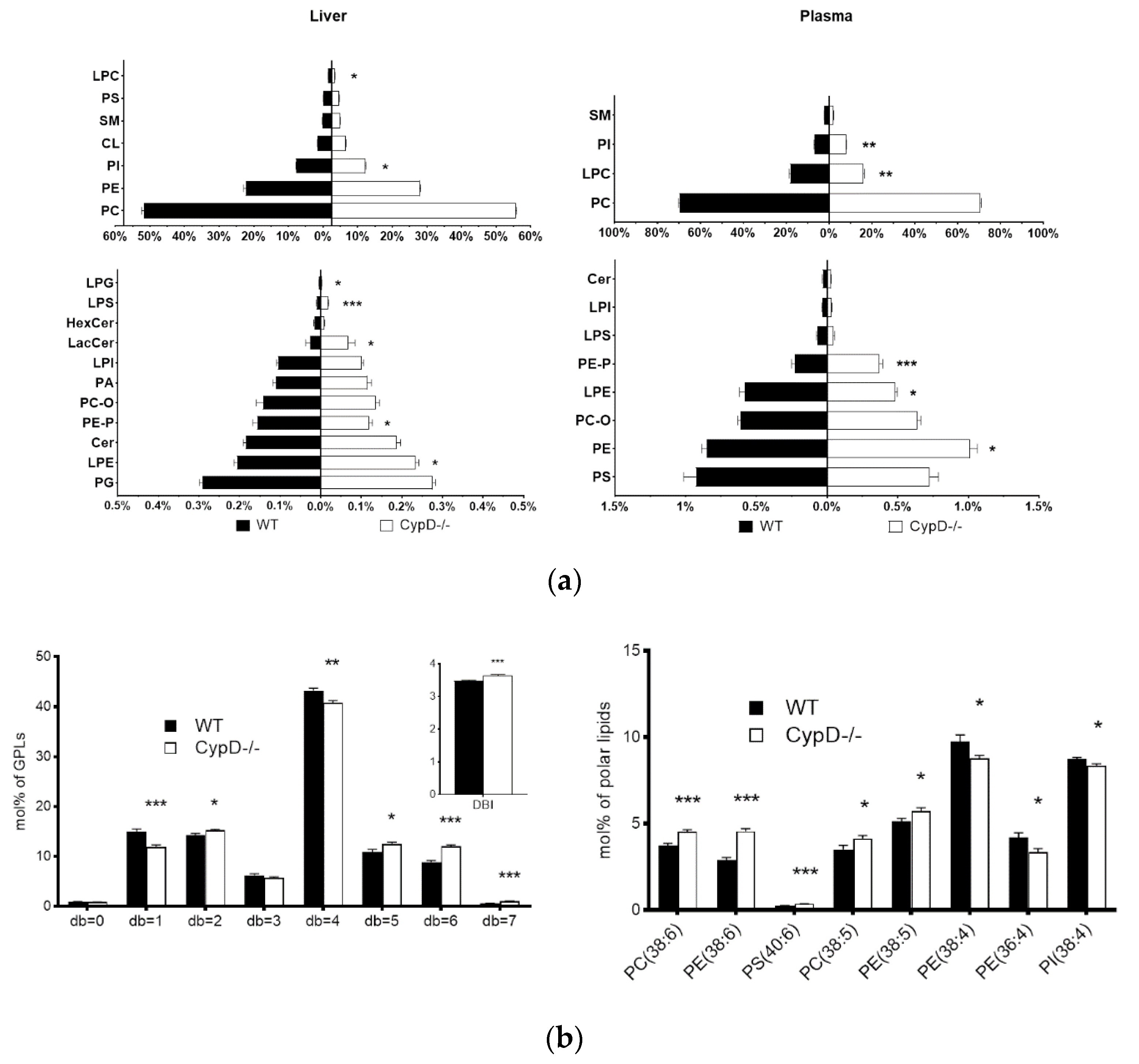
2.3. Lipid Composition Changed Due to CypD Ablation
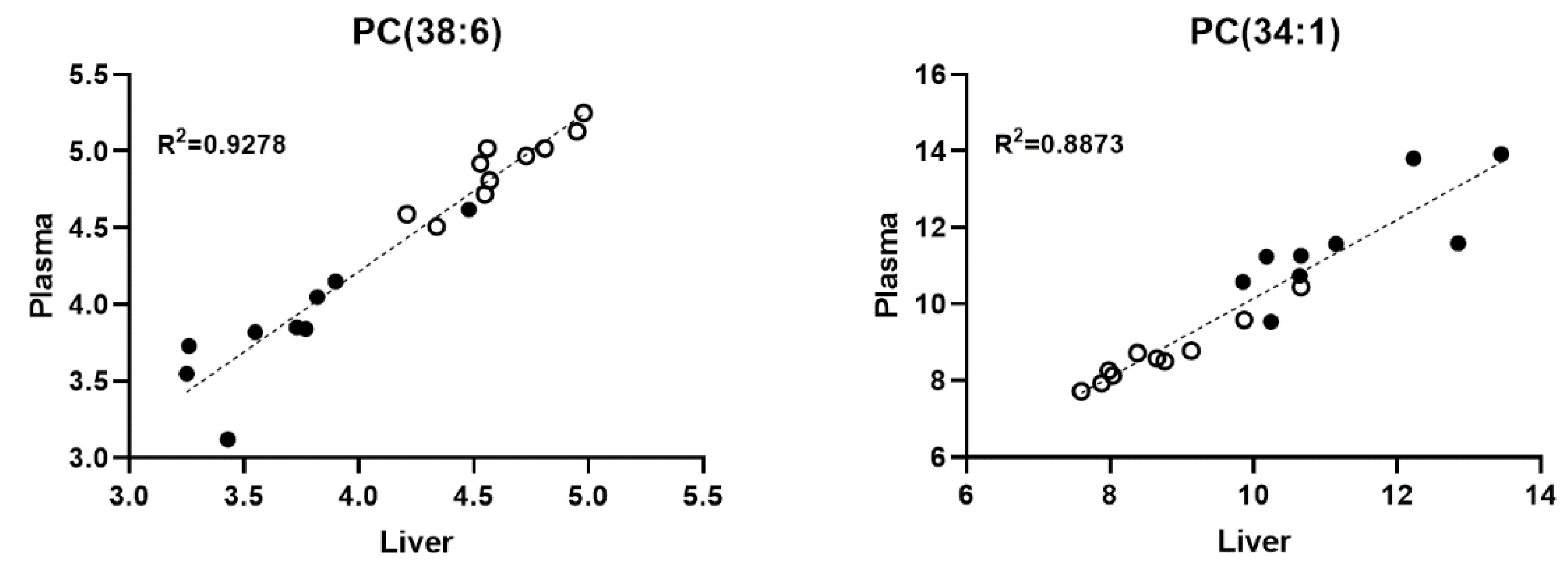
2.4. The Level of Several Liver and Plasma Phospholipid Species Showed a Positive Correlation
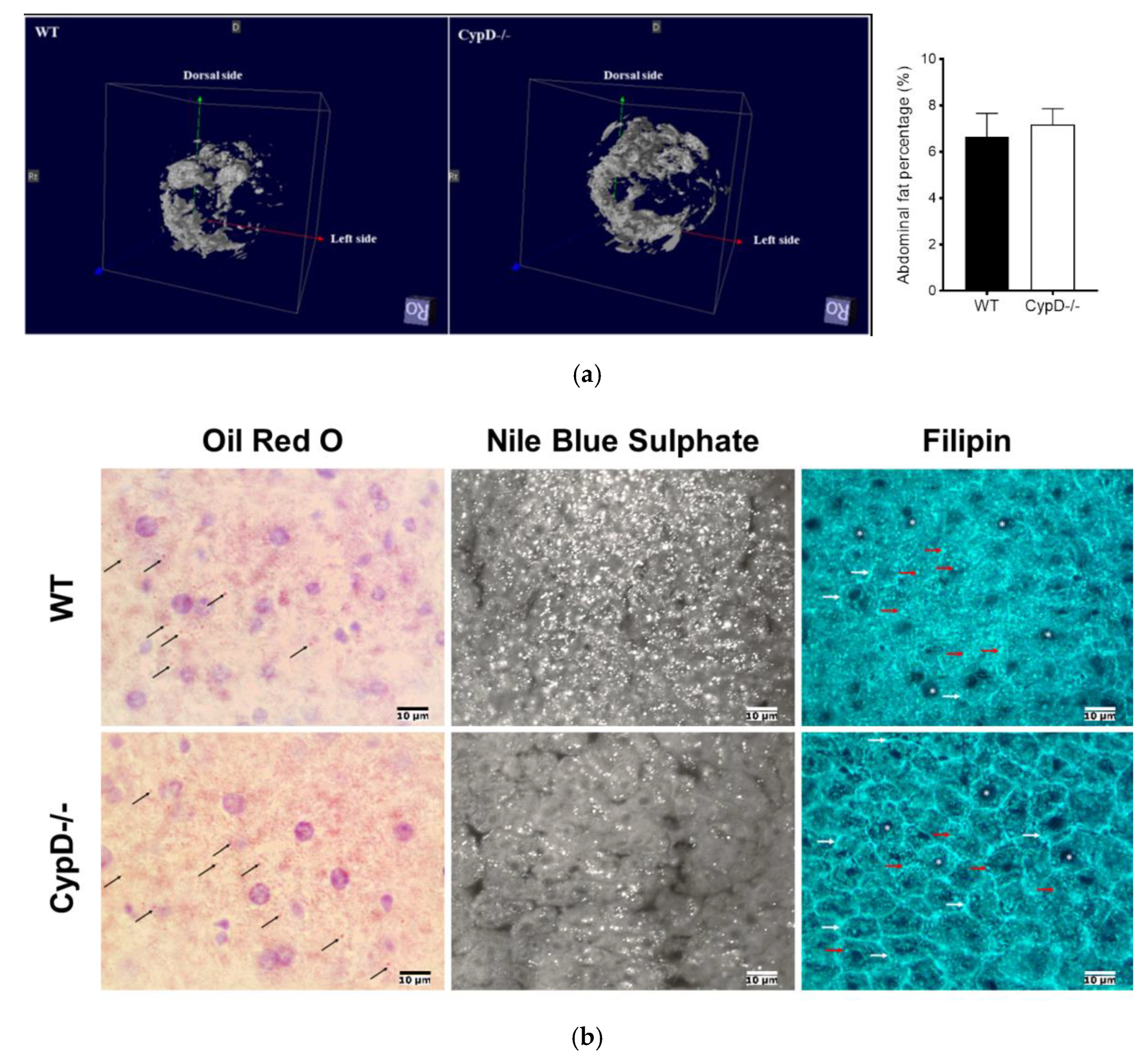
2.5. CypD Ablation Did Not Result in Abdominal Fat Accumulation
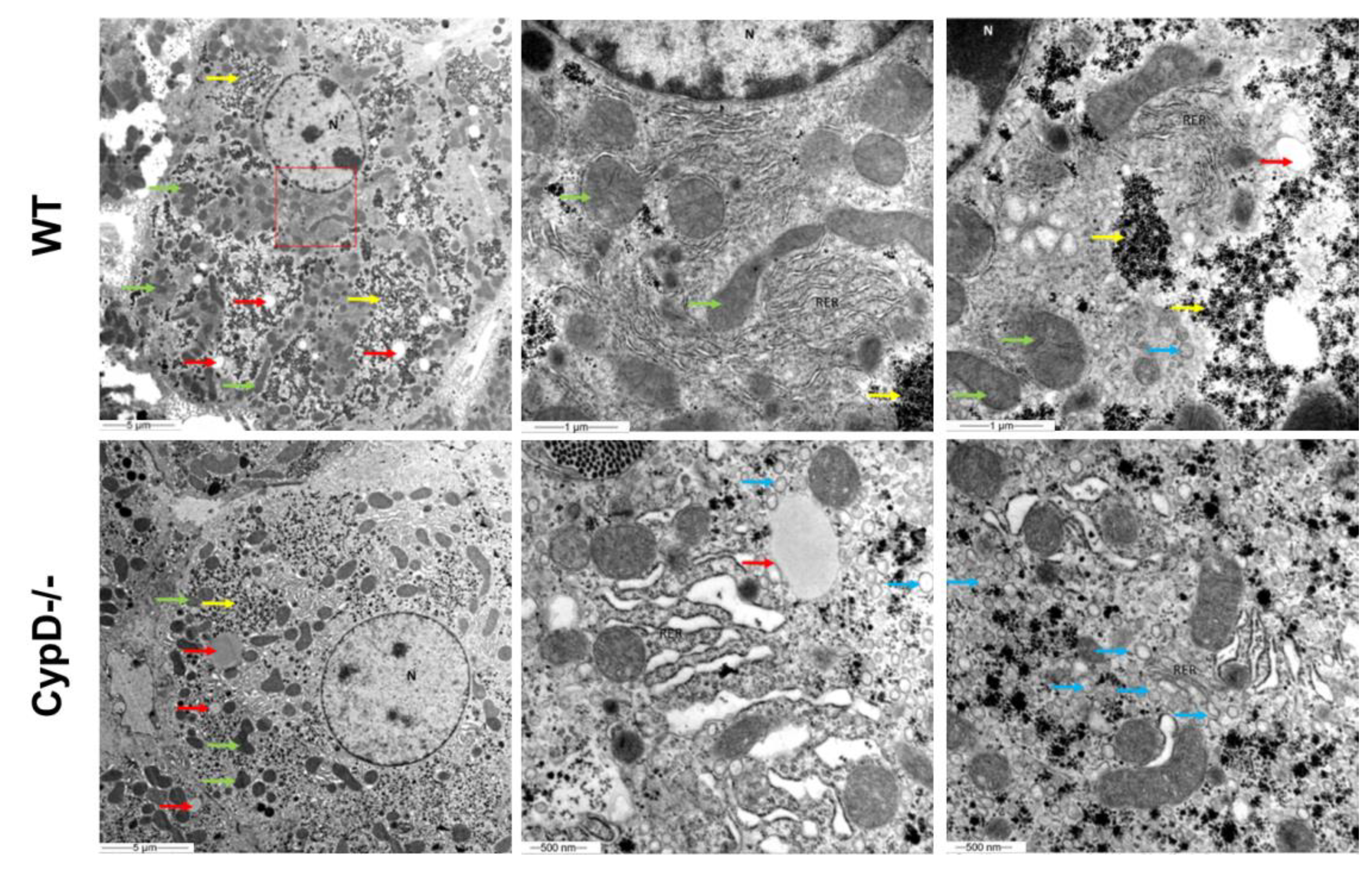
2.6. TEM Investigation Revealed Structural Differences between CypD-/- and WT Hepatocytes
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Lipid Analysis
4.3. MRI Analysis
4.4. Tissue Preparation for Histological and Histochemical Analysis
4.5. Histochemistry
4.6. Electron Microscopy
4.7. Cholesterol Extraction, Sample Preparation and GC-MS Measurements
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Connern, C.P.; Halestrap, A.P. Purification and N-Terminal Sequencing of Peptidyl-Prolyl Cis-Trans-Isomerase from Rat Liver Mitochondrial Matrix Reveals the Existence of a Distinct Mitochondrial Cyclophilin. Biochem. J. 1992, 284, 381–385. [Google Scholar] [CrossRef]
- Nakagawa, T.; Shimizu, S.; Watanabe, T.; Yamaguchi, O.; Otsu, K.; Yamagata, H.; Inohara, H.; Kubo, T.; Tsujimoto, Y. Cyclophilin D-Dependent Mitochondrial Permeability Transition Regulates Some Necrotic but Not Apoptotic Cell Death. Nature 2005, 434, 652–658. [Google Scholar] [CrossRef]
- Griffiths, E.J.; Halestrap, A.P. Mitochondrial Non-Specific Pores Remain Closed during Cardiac Ischaemia, but Open upon Reperfusion. Biochem. J. 1995, 307, 93–98. [Google Scholar] [CrossRef]
- Norenberg, M.; Rao, K. The Mitochondrial Permeability Transition in Neurologic Disease. Neurochem. Int. 2007, 50, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Traba, J.; del Arco, A.; Duchen, M.R.; Szabadkai, G.; Satrústegui, J. SCaMC-1 Promotes Cancer Cell Survival by Desensitizing Mitochondrial Permeability Transition via ATP/ADP-Mediated Matrix Ca2+ Buffering. Cell Death Differ. 2012, 19, 650–660. [Google Scholar] [CrossRef]
- Bernardi, P.; von Stockum, S. The Permeability Transition Pore as a Ca2+ Release Channel: New Answers to an Old Question. Cell Calcium 2012, 52, 22–27. [Google Scholar] [CrossRef]
- Lu, X.; Kwong, J.Q.; Molkentin, J.D.; Bers, D.M. Individual Cardiac Mitochondria Undergo Rare Transient Permeability Transition Pore Openings. Circ. Res. 2016, 118, 834–841. [Google Scholar] [CrossRef]
- Agarwal, A.; Wu, P.-H.; Hughes, E.G.; Fukaya, M.; Tischfield, M.A.; Langseth, A.J.; Wirtz, D.; Bergles, D.E. Transient Opening of the Mitochondrial Permeability Transition Pore Induces Microdomain Calcium Transients in Astrocyte Processes. Neuron 2017, 93, 587–605.e7. [Google Scholar] [CrossRef]
- Paillard, M.; Tubbs, E.; Thiebaut, P.-A.; Gomez, L.; Fauconnier, J.; Crola Da Silva, C.; Teixeira, G.; Mewton, N.; Belaidi, E.; Durand, A.; et al. Depressing Mitochondria-Reticulum Interactions Protects Cardiomyocytes From Lethal Hypoxia-Reoxygenation Injury. Circulation 2013, 128, 1555–1565. [Google Scholar] [CrossRef]
- Tubbs, E.; Theurey, P.; Vial, G.; Bendridi, N.; Bravard, A.; Chauvin, M.-A.; Ji-Cao, J.; Zoulim, F.; Bartosch, B.; Ovize, M.; et al. Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Integrity Is Required for Insulin Signaling and Is Implicated in Hepatic Insulin Resistance. Diabetes 2014, 63, 3279–3294. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Connern, C.P.; Griffiths, E.J.; Kerr, P.M. Cyclosporin A Binding to Mitochondrial Cyclophilin Inhibits the Permeability Transition Pore and Protects Hearts from Ischaemia/Reperfusion Injury. In Detection of Mitochondrial Diseases; Gellerich, F.N., Zierz, S., Eds.; Springer: Boston, MA, USA, 1997; pp. 167–172. ISBN 9781461378006. [Google Scholar]
- Matsumoto, S.; Friberg, H.; Ferrand-Drake, M.; Wieloch, T. Blockade of the Mitochondrial Permeability Transition Pore Diminishes Infarct Size in the Rat after Transient Middle Cerebral Artery Occlusion. J. Cereb. Blood Flow Metab. 1999, 19, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Hausenloy, D. Inhibiting Mitochondrial Permeability Transition Pore Opening: A New Paradigm for Myocardial Preconditioning? Cardiovasc. Res. 2002, 55, 534–543. [Google Scholar] [CrossRef]
- Schinzel, A.C.; Takeuchi, O.; Huang, Z.; Fisher, J.K.; Zhou, Z.; Rubens, J.; Hetz, C.; Danial, N.N.; Moskowitz, M.A.; Korsmeyer, S.J. Cyclophilin D Is a Component of Mitochondrial Permeability Transition and Mediates Neuronal Cell Death after Focal Cerebral Ischemia. Proc. Natl. Acad. Sci. USA 2005, 102, 12005–12010. [Google Scholar] [CrossRef] [PubMed]
- Bibli, S.-I.; Papapetropoulos, A.; Iliodromitis, E.K.; Daiber, A.; Randriamboavonjy, V.; Steven, S.; Brouckaert, P.; Chatzianastasiou, A.; Kypreos, K.E.; Hausenloy, D.J.; et al. Nitroglycerine Limits Infarct Size through S-Nitrosation of Cyclophilin D: A Novel Mechanism for an Old Drug. Cardiovasc. Res. 2019, 115, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Fonai, F.; Priber, J.K.; Jakus, P.B.; Kalman, N.; Antus, C.; Pollak, E.; Karsai, G.; Tretter, L.; Sumegi, B.; Veres, B. Lack of Cyclophilin D Protects against the Development of Acute Lung Injury in Endotoxemia. Biochim. Biophys. Acta-Mol. Basis Dis. 2015, 1852, 2563–2573. [Google Scholar] [CrossRef] [PubMed]
- Priber, J.; Fonai, F.; Jakus, P.B.; Racz, B.; Chinopoulos, C.; Tretter, L.; Gallyas, F.; Sumegi, B.; Veres, B. Cyclophilin D Disruption Attenuates Lipopolysaccharide-Induced Inflammatory Response in Primary Mouse Macrophages. Biochem. Cell Biol. 2015, 93, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Luvisetto, S.; Basso, E.; Petronilli, V.; Bernardi, P.; Forte, M. Enhancement of Anxiety, Facilitation of Avoidance Behavior, and Occurrence of Adult-Onset Obesity in Mice Lacking Mitochondrial Cyclophilin D. Neuroscience 2008, 155, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Laker, R.C.; Taddeo, E.P.; Akhtar, Y.N.; Zhang, M.; Hoehn, K.L.; Yan, Z. The Mitochondrial Permeability Transition Pore Regulator Cyclophilin D Exhibits Tissue-Specific Control of Metabolic Homeostasis. PLoS ONE 2016, 11, e0167910. [Google Scholar] [CrossRef]
- Devalaraja-Narashimha, K.; Diener, A.M.; Padanilam, B.J. Cyclophilin D Deficiency Prevents Diet-Induced Obesity in Mice. FEBS Lett. 2011, 585, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Du, H.; Shao, S.; Bo, T.; Yu, C.; Chen, W.; Zhao, L.; Li, Q.; Wang, L.; Liu, X.; et al. Cyclophilin D Deficiency Attenuates Mitochondrial Perturbation and Ameliorates Hepatic Steatosis. Hepatology 2018, 68, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Menazza, S.; Wong, R.; Nguyen, T.; Wang, G.; Gucek, M.; Murphy, E. CypD−/− Hearts Have Altered Levels of Proteins Involved in Krebs Cycle, Branch Chain Amino Acid Degradation and Pyruvate Metabolism. J. Mol. Cell. Cardiol. 2013, 56, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Tavecchio, M.; Lisanti, S.; Bennett, M.J.; Languino, L.R.; Altieri, D.C. Deletion of Cyclophilin D Impairs β-Oxidation and Promotes Glucose Metabolism. Sci. Rep. 2015, 5, 15981. [Google Scholar] [CrossRef] [PubMed]
- Winquist, R.J.; Gribkoff, V.K. Targeting Putative Components of the Mitochondrial Permeability Transition Pore for Novel Therapeutics. Biochem. Pharmacol. 2020, 177, 113995. [Google Scholar] [CrossRef] [PubMed]
- Boyenle, I.D.; Oyedele, A.K.; Ogunlana, A.T.; Adeyemo, A.F.; Oyelere, F.S.; Akinola, O.B.; Adelusi, T.I.; Ehigie, L.O.; Ehigie, A.F. Targeting the Mitochondrial Permeability Transition Pore for Drug Discovery: Challenges and Opportunities. Mitochondrion 2022, 63, 57–71. [Google Scholar] [CrossRef]
- Bernardi, P.; Carraro, M.; Lippe, G. The Mitochondrial Permeability Transition: Recent Progress and Open Questions. FEBS J. 2021, febs.1625. [Google Scholar] [CrossRef]
- Tarasov, K.; Stefanko, A.; Casanovas, A.; Surma, M.A.; Berzina, Z.; Hannibal-Bach, H.K.; Ekroos, K.; Ejsing, C.S. High-Content Screening of Yeast Mutant Libraries by Shotgun Lipidomics. Mol. BioSyst. 2014, 10, 1364–1376. [Google Scholar] [CrossRef]
- Quehenberger, O.; Dennis, E.A. The Human Plasma Lipidome. New Engl. J. Med. 2011, 365, 1812–1823. [Google Scholar] [CrossRef]
- Amanakis, G.; Murphy, E. Cyclophilin D: An Integrator of Mitochondrial Function. Front. Physiol. 2020, 11, 595. [Google Scholar] [CrossRef]
- Elrod, J.W.; Wong, R.; Mishra, S.; Vagnozzi, R.J.; Sakthievel, B.; Goonasekera, S.A.; Karch, J.; Gabel, S.; Farber, J.; Force, T.; et al. Cyclophilin D Controls Mitochondrial Pore–Dependent Ca2+ Exchange, Metabolic Flexibility, and Propensity for Heart Failure in Mice. J. Clin. Investig. 2010, 120, 3680–3687. [Google Scholar] [CrossRef]
- Moon, S.H.; Jenkins, C.M.; Kiebish, M.A.; Sims, H.F.; Mancuso, D.J.; Gross, R.W. Genetic Ablation of Calcium-Independent Phospholipase A2γ (IPLA2γ) Attenuates Calcium-Induced Opening of the Mitochondrial Permeability Transition Pore and Resultant Cytochrome c Release. J. Biol. Chem. 2012, 287, 29837–29850. [Google Scholar] [CrossRef]
- Moon, S.H.; Jenkins, C.M.; Liu, X.; Guan, S.; Mancuso, D.J.; Gross, R.W. Activation of Mitochondrial Calcium-Independent Phospholipase A2γ (IPLA2γ) by Divalent Cations Mediating Arachidonate Release and Production of Downstream Eicosanoids. J. Biol. Chem. 2012, 287, 14880–14895. [Google Scholar] [CrossRef] [PubMed]
- Paradies, G.; Paradies, V.; De Benedictis, V.; Ruggiero, F.M.; Petrosillo, G. Functional Role of Cardiolipin in Mitochondrial Bioenergetics. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, I.; Sano, R.; d’Azzo, A. Mitochondria-Associated ER Membranes (MAMs) and Lysosomal Storage Diseases. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, M.; Sun, H.; Ferdous, R.U.; Gao, L.; Zhao, J.; Song, Y. Liver Cyclophilin D Deficiency Inhibits the Progression of Early NASH by Ameliorating Steatosis and Inflammation. Biochem. Biophys. Res. Commun. 2022, 594, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Šileikytė, J.; Forte, M. The Mitochondrial Permeability Transition in Mitochondrial Disorders. Oxidative Med. Cell. Longev. 2019, 2019, 1–11. [Google Scholar] [CrossRef]
- Briston, T.; Selwood, D.L.; Szabadkai, G.; Duchen, M.R. Mitochondrial Permeability Transition: A Molecular Lesion with Multiple Drug Targets. Trends Pharmacol. Sci. 2019, 40, 50–70. [Google Scholar] [CrossRef]
- Szűcs, G.; Sója, A.; Péter, M.; Sárközy, M.; Bruszel, B.; Siska, A.; Földesi, I.; Szabó, Z.; Janáky, T.; Vígh, L.; et al. Prediabetes Induced by Fructose-Enriched Diet Influences Cardiac Lipidome and Proteome and Leads to Deterioration of Cardiac Function Prior to the Development of Excessive Oxidative Stress and Cell Damage. Oxidative Med. Cell. Longev. 2019, 2019, 1–21. [Google Scholar] [CrossRef]
- Herzog, R.; Schwudke, D.; Schuhmann, K.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. A Novel Informatics Concept for High-Throughput Shotgun Lipidomics Based on the Molecular Fragmentation Query Language. Genome Biol. 2011, 12, R8. [Google Scholar] [CrossRef]
- Tóth, M.E.; Dukay, B.; Péter, M.; Balogh, G.; Szűcs, G.; Zvara, Á.; Szebeni, G.J.; Hajdu, P.; Sárközy, M.; Puskás, L.G.; et al. Male and Female Animals Respond Differently to High-Fat Diet and Regular Exercise Training in a Mouse Model of Hyperlipidemia. Int. J. Mol. Sci. 2021, 22, 4198. [Google Scholar] [CrossRef]
- Liebisch, G.; Vizcaíno, J.A.; Köfeler, H.; Trötzmüller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J.O. Shorthand Notation for Lipid Structures Derived from Mass Spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef]
- Christie, W.W. Gas Chromatography-Mass Spectrometry Methods for Structural Analysis of Fatty Acids. Lipids 1998, 33, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Kraus, D.; Yang, Q.; Kahn, B. Lipid Extraction from Mouse Feces. BIO-PROTOCOL 2015, 5, e1375. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hu, R.; Yue, J.; Yang, Z.; Zhang, L. Determination of Fecal Sterols by Gas Chromatography–Mass Spectrometry with Solid-Phase Extraction and Injection-Port Derivatization. J. Chromatogr. A 2009, 1216, 1053–1058. [Google Scholar] [CrossRef]
- Szucs, S.; Sarvary, A.; Cain, T.; Adany, R. Method Validation for the Simultaneous Determination of Fecal Sterols in Surface Waters by Gas Chromatography-Mass Spectrometry. J. Chromatogr. Sci. 2006, 44, 70–76. [Google Scholar] [CrossRef][Green Version]
- Storey, J.D.; Tibshirani, R. Statistical Significance for Genomewide Studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14-10. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koszegi, B.; Balogh, G.; Berente, Z.; Vranesics, A.; Pollak, E.; Molnar, L.; Takatsy, A.; Poor, V.; Wahr, M.; Antus, C.; et al. Remodeling of Liver and Plasma Lipidomes in Mice Lacking Cyclophilin D. Int. J. Mol. Sci. 2022, 23, 11274. https://doi.org/10.3390/ijms231911274
Koszegi B, Balogh G, Berente Z, Vranesics A, Pollak E, Molnar L, Takatsy A, Poor V, Wahr M, Antus C, et al. Remodeling of Liver and Plasma Lipidomes in Mice Lacking Cyclophilin D. International Journal of Molecular Sciences. 2022; 23(19):11274. https://doi.org/10.3390/ijms231911274
Chicago/Turabian StyleKoszegi, Balazs, Gabor Balogh, Zoltan Berente, Anett Vranesics, Edit Pollak, Laszlo Molnar, Aniko Takatsy, Viktoria Poor, Matyas Wahr, Csenge Antus, and et al. 2022. "Remodeling of Liver and Plasma Lipidomes in Mice Lacking Cyclophilin D" International Journal of Molecular Sciences 23, no. 19: 11274. https://doi.org/10.3390/ijms231911274
APA StyleKoszegi, B., Balogh, G., Berente, Z., Vranesics, A., Pollak, E., Molnar, L., Takatsy, A., Poor, V., Wahr, M., Antus, C., Eros, K., Vigh, L., Gallyas, F., Jr., Peter, M., & Veres, B. (2022). Remodeling of Liver and Plasma Lipidomes in Mice Lacking Cyclophilin D. International Journal of Molecular Sciences, 23(19), 11274. https://doi.org/10.3390/ijms231911274






