PIKE-A Modulates Mitochondrial Metabolism through Increasing SDHA Expression Mediated by STAT3/FTO Axis
Abstract
:1. Introduction
2. Results
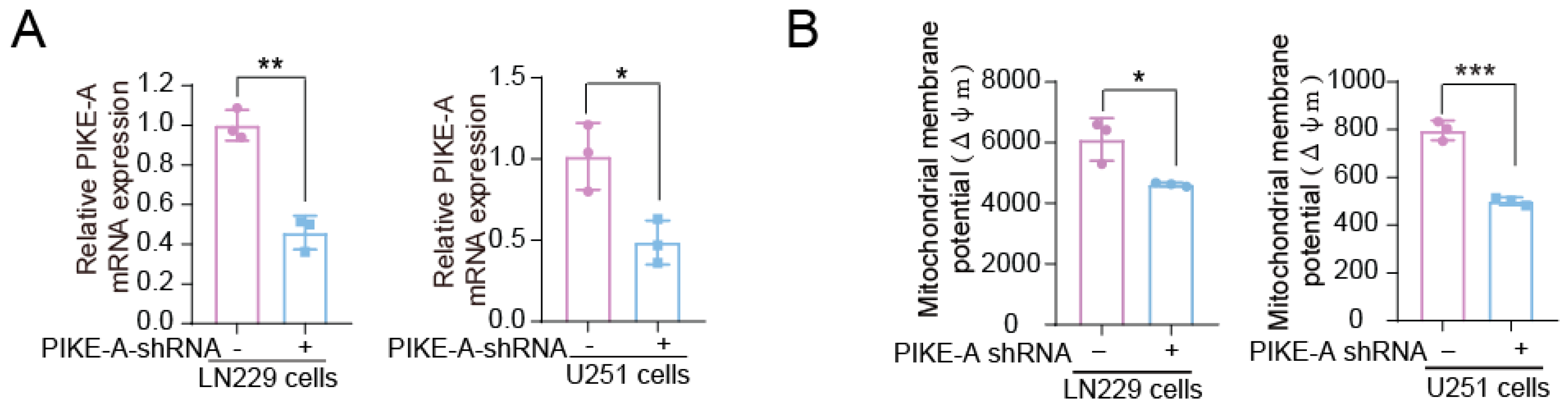
2.1. PIKE-A Promotes Mitochondrial Respiration in Glioblastoma Cells
2.2. PIKE-A Regulates Mitochondrial Respiration through SDHA
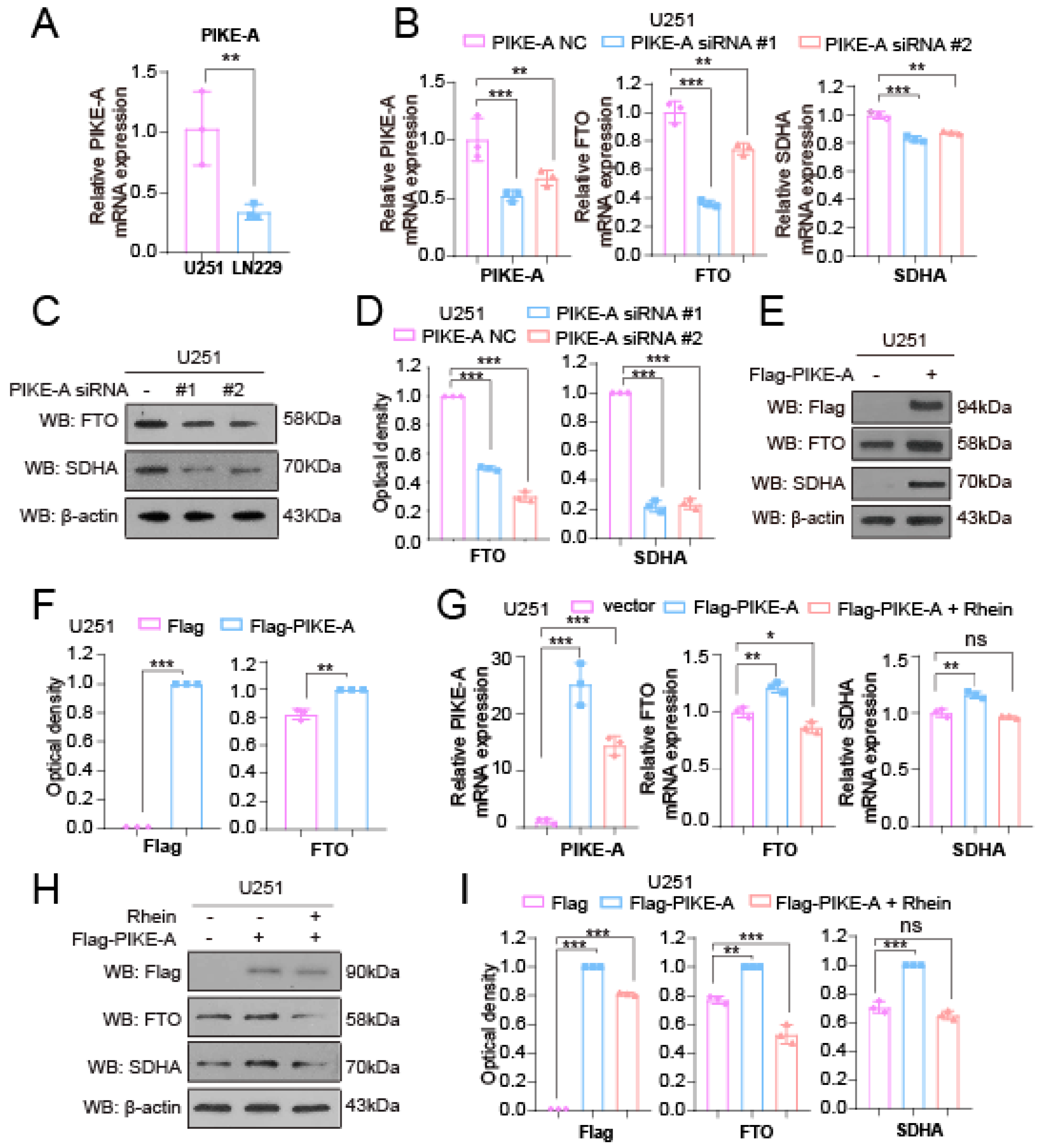
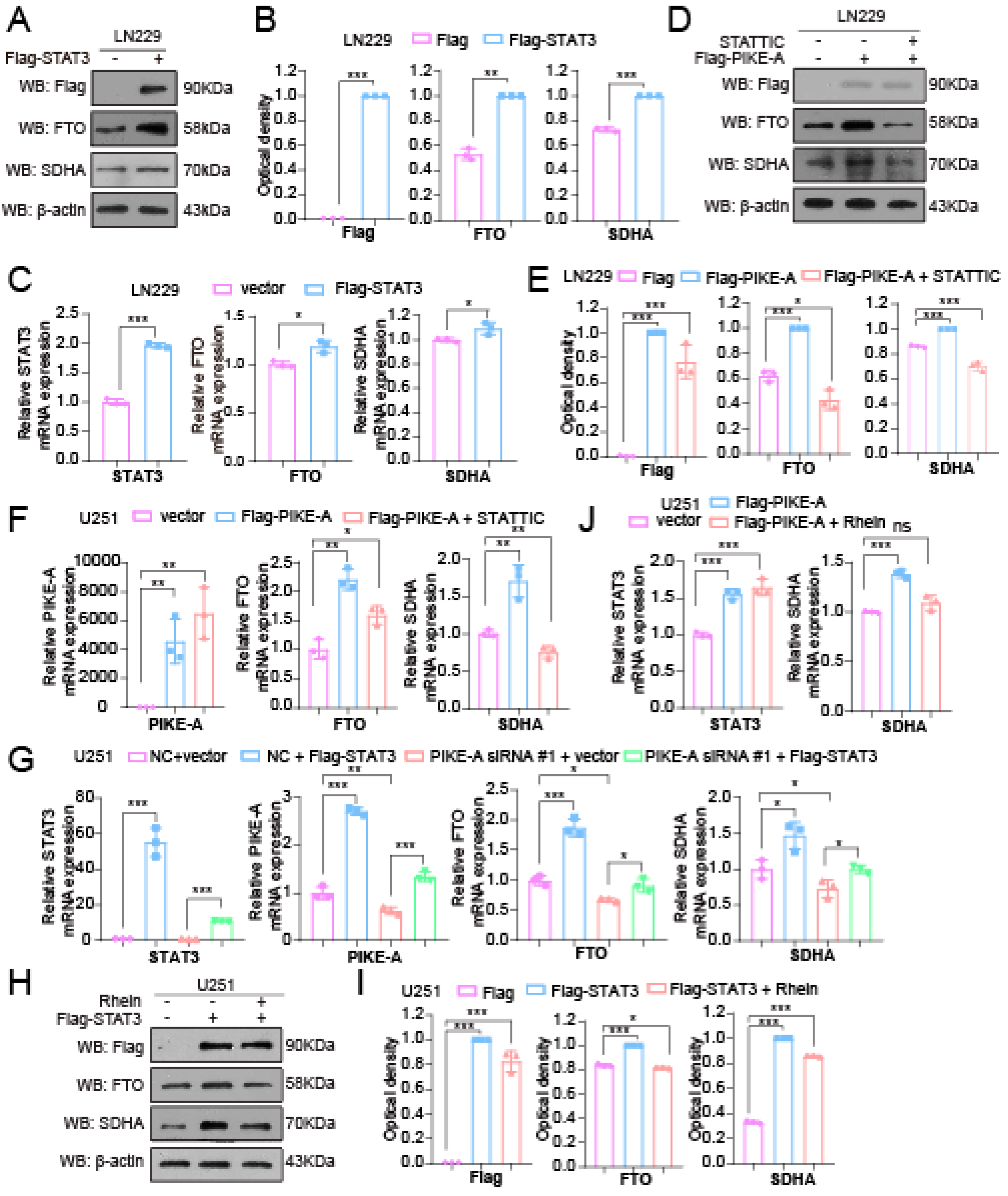
2.3. PIKE-A Increases SDHA Expression through Activating FTO Expression in a STAT3-Dependent Manner
2.4. PIKE-A Regulates Mitochondrial Respiration by Regulating FTO Expression in a STAT3-Dependent Manner
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmid Construction and Lentivirus Packaging
4.3. Cell Proliferation Assay
4.4. Western Blot Analysis
4.5. Mitochondrial Respiratory Chain Complexes Ⅱ (MRCⅡ) Activity Assay
4.6. qRT-PCR
4.7. Mitochondrial Membrane Potential Detection
4.8. RNA-Sequence
4.9. Metabolite Analysis by Gas Chromatography—Mass Spectrometry
4.10. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bleeker, F.E.; Molenaar, R.J.; Leenstra, S. Recent advances in the molecular understanding of glioblastoma. J. Neurooncol. 2012, 108, 11–27. [Google Scholar] [CrossRef]
- Ahn, J.Y.; Rong, R.; Kroll, T.G.; Van Meir, E.G.; Snyder, S.H.; Ye, K. PIKE (phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates Akt activity and mediates cellular invasion. J. Biol. Chem. 2004, 279, 16441–16451. [Google Scholar] [CrossRef]
- Ahn, J.Y.; Hu, Y.; Kroll, T.G.; Allard, P.; Ye, K. PIKE-A is amplified in human cancers and prevents apoptosis by up-regulating Akt. Proc. Natl. Acad. Sci. USA 2004, 101, 6993–6998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sheng, H.; Zhang, X.; Qi, Q.; Chan, C.B.; Li, L.; Shan, C.; Ye, K. Cellular energy stress induces AMPK-mediated regulation of glioblastoma cell proliferation by PIKE-A phosphorylation. Cell Death Dis. 2019, 10, 222. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Y.; Hao, C.; Rempel, S.A.; Ye, K. PIKE-A is a proto-oncogene promoting cell growth, transformation and invasion. Oncogene 2007, 26, 4918–4927. [Google Scholar] [CrossRef]
- Sun, M.; Sheng, H.; Wu, T.; Song, J.; Sun, H.; Wang, Y.; Wang, J.; Li, Z.; Zhao, H.; Tan, J.; et al. PIKE-A promotes glioblastoma growth by driving PPP flux through increasing G6PD expression mediated by phosphorylation of STAT3. Biochem. Pharmacol. 2021, 192, 114736. [Google Scholar] [CrossRef]
- Fernandez-Vizarra, E.; Zeviani, M. Mitochondrial disorders of the OXPHOS system. FEBS Lett. 2021, 595, 1062–1106. [Google Scholar] [CrossRef]
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, R551–R560. [Google Scholar] [CrossRef]
- Sandoval-Acuna, C.; Torrealba, N.; Tomkova, V.; Jadhav, S.B.; Blazkova, K.; Merta, L.; Lettlova, S.; Adamcova, M.K.; Rosel, D.; Brabek, J.; et al. Targeting Mitochondrial Iron Metabolism Suppresses Tumor Growth and Metastasis by Inducing Mitochondrial Dysfunction and Mitophagy. Cancer Res. 2021, 81, 2289–2303. [Google Scholar] [CrossRef]
- Baechler, S.A.; Factor, V.M.; Dalla Rosa, I.; Ravji, A.; Becker, D.; Khiati, S.; Miller Jenkins, L.M.; Lang, M.; Sourbier, C.; Michaels, S.A.; et al. The mitochondrial type IB topoisomerase drives mitochondrial translation and carcinogenesis. Nat. Commun. 2019, 10, 83. [Google Scholar] [CrossRef] [Green Version]
- Bardella, C.; Pollard, P.J.; Tomlinson, I. SDH mutations in cancer. Biochim. Biophys. Acta 2011, 1807, 1432–1443. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.J. Succinate dehydrogenase (SDH) and mitochondrial driven neoplasia. Pathology 2012, 44, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Takenaga, K.; Akimoto, M.; Koshikawa, N.; Yamaguchi, A.; Imanishi, H.; Nakada, K.; Honma, Y.; Hayashi, J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008, 320, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Kashatus, J.A.; Nascimento, A.; Myers, L.J.; Sher, A.; Byrne, F.L.; Hoehn, K.L.; Counter, C.M.; Kashatus, D.F. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 2015, 57, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Payen, V.L.; Perez-Escuredo, J.; De Saedeleer, C.J.; Danhier, P.; Copetti, T.; Dhup, S.; Tardy, M.; Vazeille, T.; Bouzin, C.; et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014, 8, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Rivadeneira, D.B.; Caino, M.C.; Chae, Y.C.; Speicher, D.W.; Tang, H.Y.; Vaira, V.; Bosari, S.; Palleschi, A.; Rampini, P.; et al. The Mitochondrial Unfoldase-Peptidase Complex ClpXP Controls Bioenergetics Stress and Metastasis. PLoS Biol. 2016, 14, e1002507. [Google Scholar] [CrossRef]
- Kang, Y.J.; Bang, B.R.; Han, K.H.; Hong, L.; Shim, E.J.; Ma, J.; Lerner, R.A.; Otsuka, M. Regulation of NKT cell-mediated immune responses to tumours and liver inflammation by mitochondrial PGAM5-Drp1 signalling. Nat. Commun. 2015, 6, 8371. [Google Scholar] [CrossRef]
- Mullen, A.R.; Hu, Z.; Shi, X.; Jiang, L.; Boroughs, L.K.; Kovacs, Z.; Boriack, R.; Rakheja, D.; Sullivan, L.B.; Linehan, W.M.; et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014, 7, 1679–1690. [Google Scholar] [CrossRef]
- Buffet, A.; Burnichon, N.; Favier, J.; Gimenez-Roqueplo, A.P. An overview of 20 years of genetic studies in pheochromocytoma and paraganglioma. Best Pract. Res. Clin. Endocrinol. Metab. 2020, 34, 101416. [Google Scholar] [CrossRef]
- Li, S.T.; Huang, D.; Shen, S.; Cai, Y.; Xing, S.; Wu, G.; Jiang, Z.; Hao, Y.; Yuan, M.; Wang, N.; et al. Myc-mediated SDHA acetylation triggers epigenetic regulation of gene expression and tumorigenesis. Nat. Metab 2020, 2, 256–269. [Google Scholar] [CrossRef]
- Sousa, J.S.; D’Imprima, E.; Vonck, J. Mitochondrial Respiratory Chain Complexes. Subcell Biochem. 2018, 87, 167–227. [Google Scholar] [PubMed]
- Aldera, A.P.; Govender, D. Gene of the month: SDH. J. Clin. Pathol. 2018, 71, 95–97. [Google Scholar] [CrossRef]
- Duan, X.; Yang, L.; Wang, L.; Liu, Q.; Zhang, K.; Liu, S.; Liu, C.; Gao, Q.; Li, L.; Qin, G.; et al. m6A demethylase FTO promotes tumor progression via regulation of lipid metabolism in esophageal cancer. Cell Biosci. 2022, 12, 60. [Google Scholar] [CrossRef]
- Wang, J.; Qiao, Y.; Sun, M.; Sun, H.; Xie, F.; Chang, H.; Wang, Y.; Song, J.; Lai, S.; Yang, C.; et al. FTO promotes colorectal cancer progression and chemotherapy resistance via demethylating G6PD/PARP1. Clin. Transl. Med. 2022, 12, e772. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.; Li, Z.; Su, S.; Sun, W.; Zhang, X.; Li, L.; Li, J.; Liu, S.; Lu, B.; Zhang, S.; et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis 2020, 41, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.B.; Ye, K. PIKE GTPase are phosphoinositide-3-kinase enhancers, suppressing programmed cell death. J. Cell Mol. Med. 2007, 11, 39–53. [Google Scholar] [CrossRef]
- Ma, Y.; Qi, Y.; Wang, L.; Zheng, Z.; Zhang, Y.; Zheng, J. SIRT5-mediated SDHA desuccinylation promotes clear cell renal cell carcinoma tumorigenesis. Free Radic. Biol. Med. 2019, 134, 458–467. [Google Scholar] [CrossRef]
- Gao, H.; Wang, W.; Li, Q. GANT61 suppresses cell survival, invasion and epithelial-mesenchymal transition through inactivating AKT/mTOR and JAK/STAT3 pathways in anaplastic thyroid carcinoma. Cancer Biol. Ther. 2022, 23, 369–377. [Google Scholar] [CrossRef]
- Wu, Q.; Li, J.; Sun, X.; He, D.; Cheng, Z.; Li, J.; Zhang, X.; Xie, Y.; Zhu, Y.; Lai, M. Multi-stage metabolomics and genetic analyses identified metabolite biomarkers of metabolic syndrome and their genetic determinants. EBioMedicine 2021, 74, 103707. [Google Scholar] [CrossRef]


| Gene | shRNA Sequence |
|---|---|
| PIKE-A | CCGGGCTGTGATCAATAGCCAGGAACTCGAGTTCCTGGCTATTGATCACAGCTTTTTG |
| Gene | siRNA Sequence |
| PIKE-A | #1:GAGAAACGAAGCTTGGATACT |
| #2:TGTGATCAATAGCCAGGAACT |
| Antibodies | Source | Identifier |
|---|---|---|
| FTO Rabbit mAb | proteintech | 27226-1-AP |
| SDHA (D6J9M) XP® Rabbit mAb | CST | 11998 |
| Bata Actin Mouse McAb | proteintech | 66009-1-Ig |
| Flag Rabbit PolyAb | proteintech | 20543-1-AP |
| Gene | Real-Time PCR Primers Sequence |
|---|---|
| PIKE-A | Forward primer: 5′- AGAGGCAGTTCGTTGTAGCT -3′ |
| Reverse primer: 5′- TCTGTCTTCTCCAGCACCTG -3′ | |
| STAT3 | Forward primer: 5′- CAGCAGCTTGACACACGGTA -3′ |
| Reverse primer: 5′- AAACACCAAAGTGGCATGTGA -3′ | |
| FTO | Forward primer: 5′-AACACCAGGCTCTTTACGGTC-3′ |
| Reverse primer: 5′-TGTCCGTTGTAGGATGAACCC-3′ | |
| SDHA | Forward primer: 5′-CAAACAGGAACCCGAGGTTTT-3′ |
| Reverse primer: 5′-CAGCTTGGTAACACATGCTGTAT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, M.; Yan, Q.; Qiao, Y.; Zhao, H.; Wang, Y.; Shan, C.; Zhang, S. PIKE-A Modulates Mitochondrial Metabolism through Increasing SDHA Expression Mediated by STAT3/FTO Axis. Int. J. Mol. Sci. 2022, 23, 11304. https://doi.org/10.3390/ijms231911304
Sun M, Yan Q, Qiao Y, Zhao H, Wang Y, Shan C, Zhang S. PIKE-A Modulates Mitochondrial Metabolism through Increasing SDHA Expression Mediated by STAT3/FTO Axis. International Journal of Molecular Sciences. 2022; 23(19):11304. https://doi.org/10.3390/ijms231911304
Chicago/Turabian StyleSun, Mingming, Qi Yan, Yaya Qiao, Huifang Zhao, Yingzhi Wang, Changliang Shan, and Shuai Zhang. 2022. "PIKE-A Modulates Mitochondrial Metabolism through Increasing SDHA Expression Mediated by STAT3/FTO Axis" International Journal of Molecular Sciences 23, no. 19: 11304. https://doi.org/10.3390/ijms231911304





