Structural Basis for the Binding of Allosteric Activators Leucine and ADP to Mammalian Glutamate Dehydrogenase
Abstract
1. Introduction
2. Results
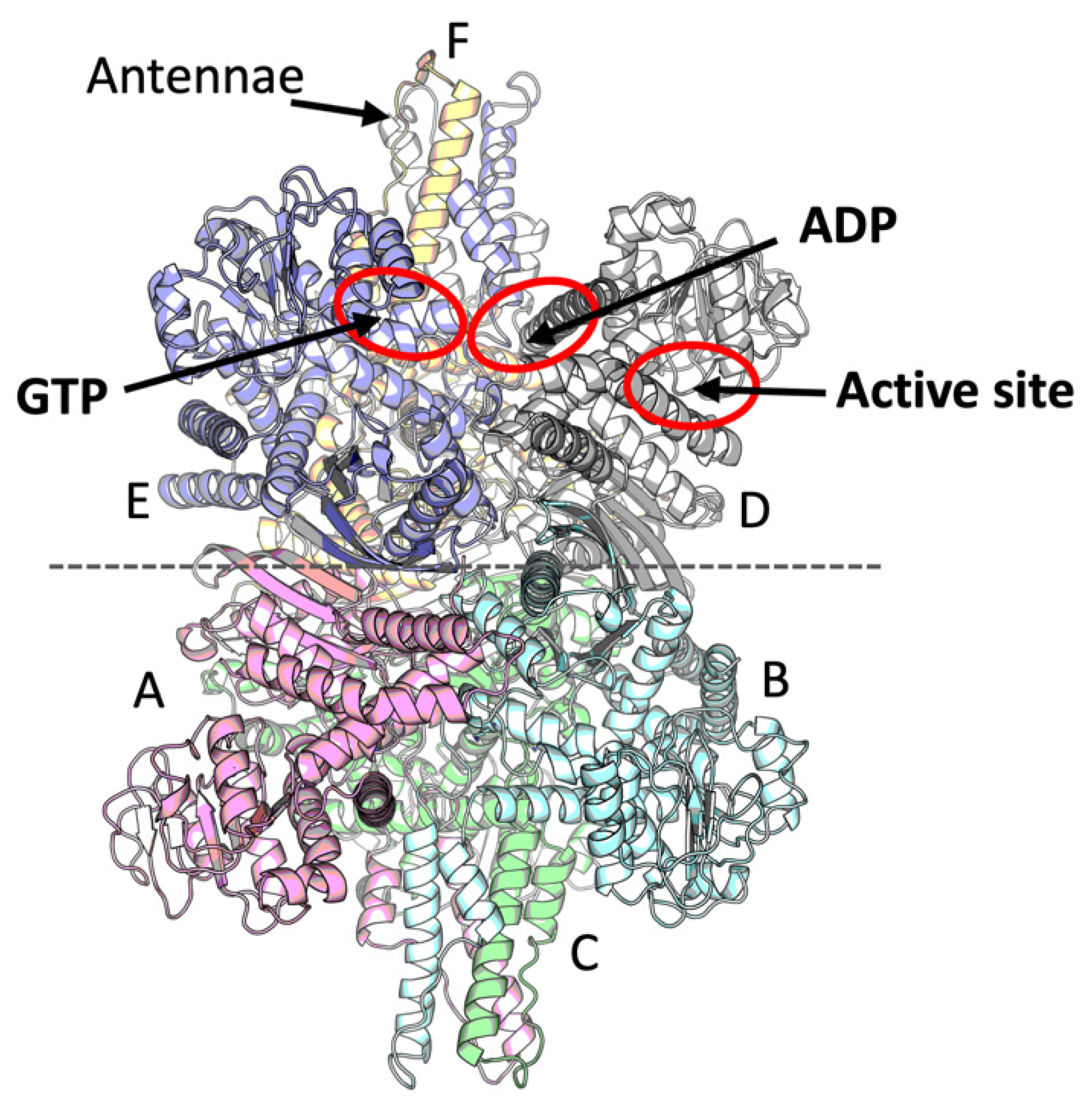
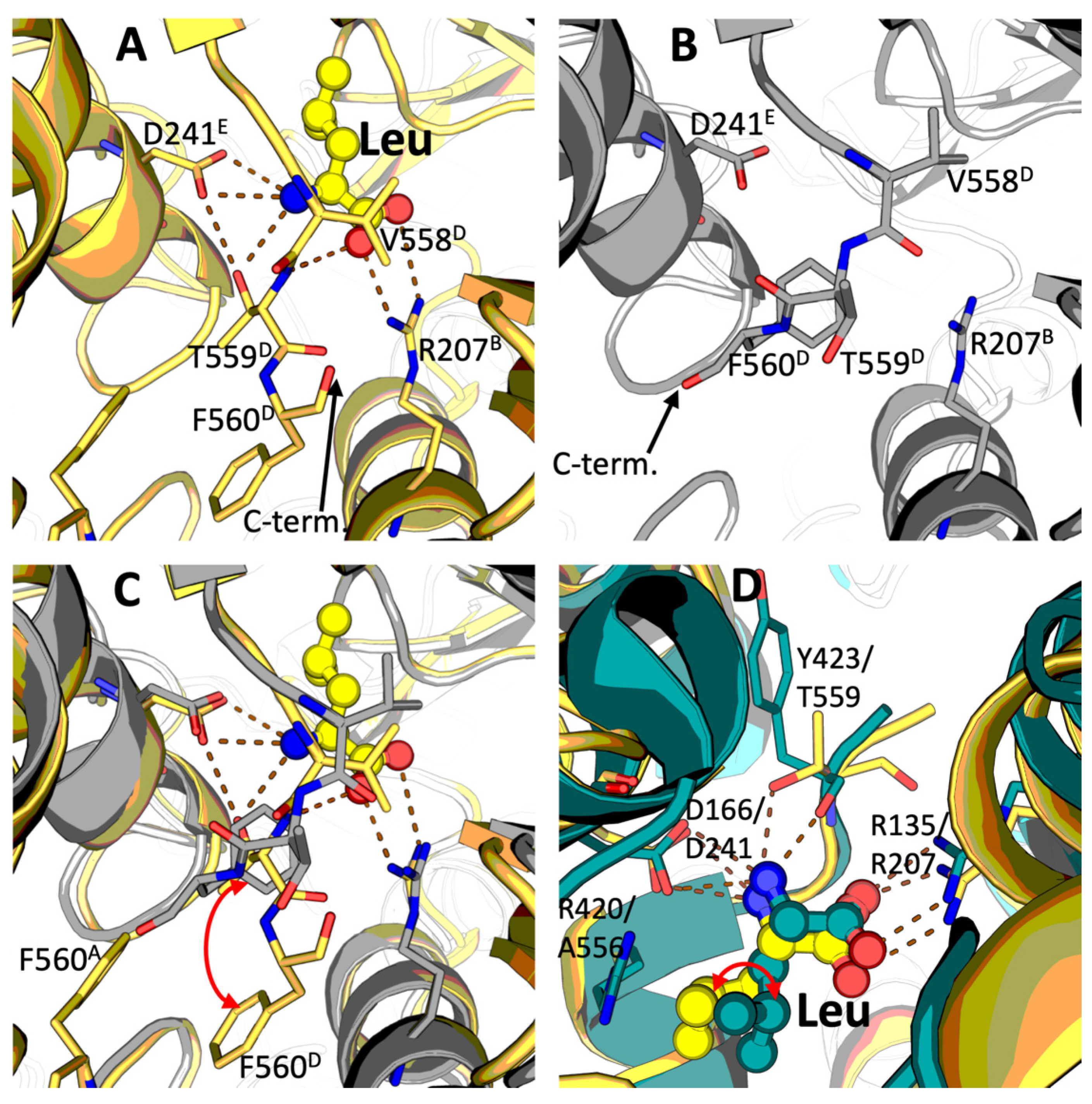
2.1. Identification of the Leucine Binding Site of Mammalian GDH
2.2. Novel Conformation of GDH·ADP Binary Complex at 2.40-angstrom Resolution
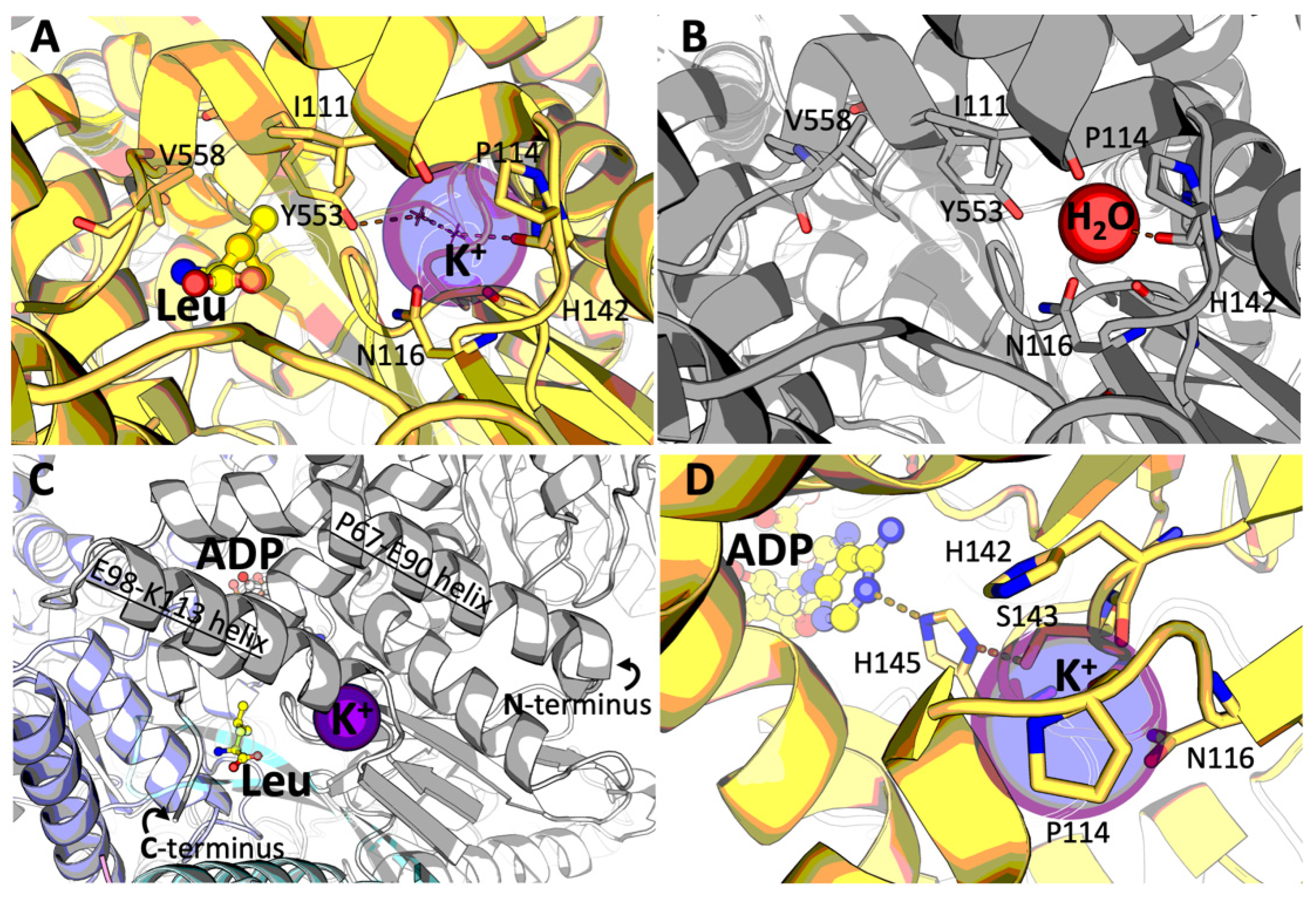
2.3. Identification of the GDH Binding Site for Potassium Ion
3. Discussion
3.1. Leucine Binding Site
3.2. Potassium Ion Binding Site
3.3. Implications of the ADP and Leucine Binding for the GDH Regulation by Acetylation
3.4. Binding of Thiamine Derivatives
4. Materials and Methods
4.1. Reagents
4.2. Crystallization and Data Collection
4.3. Structure Determination and Refinement
4.4. Multiple Sequence Alignment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GDH | glutamate dehydrogenase |
| ThTP | thiamine triphosphate |
| MPD | 2-methyl2,4-pentanediol |
| Cryo-EM | cryo-electron microscopy |
References
- Barratt, R.W.; Strickland, W.N. Purification and characterization of a TPN-specific glutamic acid dehydrogenase from Neurospora crassa. Arch. Biochem. Biophys. 1963, 102, 66–76. [Google Scholar] [CrossRef]
- Veronese, F.M.; Nyc, J.F.; Degani, Y.; Brown, D.M.; Smith, E.L. Nicotinamide Adenine Dinucleotide-specific Glutamate Dehydrogenase of Neurospora. J. Biol. Chem. 1974, 249, 7922–7928. [Google Scholar] [CrossRef]
- Lázaro, M.; Melero, R.; Huet, C.; López-Alonso, J.P.; Delgado, S.; Dodu, A.; Bruch, E.M.; Abriata, L.A.; Alzari, P.M.; Valle, M.; et al. 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme. Commun. Biol. 2021, 4, 684. [Google Scholar] [CrossRef] [PubMed]
- Yielding, K.L.; Tomkins, G.M. An Effect of L-Leucine and Other Essential Amino Acids on the Structure and Activity of Glutamic Dehydrogenase. Proc. Natl. Acad. Sci. USA 1961, 47, 983–989. [Google Scholar] [CrossRef]
- Frieden, C. The Effect of pH and Other Variables on the Dissociation of Beef Liver Glutamic Dehydrogenase. J. Biol. Chem. 1962, 237, 2396–2400. [Google Scholar] [CrossRef]
- Talal, N.; Tomkins, G.M. Allosteric Properties of Glutamate Dehydrogenases from Different Sources. Science 1964, 146, 1309–1311. [Google Scholar] [CrossRef]
- Peterson, P.E.; Smith, T.J. The structure of bovine glutamate dehydrogenase provides insights into the mechanism of allostery. Structure 1999, 7, 769–782. [Google Scholar] [CrossRef]
- Banerjee, S.; Schmidt, T.; Fang, J.; Stanley, C.A.; Smith, T.J. Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation. Biochemistry 2003, 42, 3446–3456. [Google Scholar] [CrossRef]
- Smith, T.J.; Stanley, C.A. Untangling the glutamate dehydrogenase allosteric nightmare. Trends Biochem. Sci. 2008, 33, 557–564. [Google Scholar] [CrossRef]
- Borgnia, M.J.; Banerjee, S.; Merk, A.; Matthies, D.; Bartesaghi, A.; Rao, P.; Pierson, J.; Earl, L.A.; Falconieri, V.; Subramaniam, S.; et al. Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase. Mol. Pharmacol. 2016, 89, 645–651. [Google Scholar] [CrossRef]
- Bailey, J.; Bell, E.T.; Bell, J.E. Regulation of bovine glutamate dehydrogenase. The effects of pH and ADP. J. Biol. Chem. 1982, 257, 5579–5583. [Google Scholar] [CrossRef]
- Couée, I.; Tipton, K.F. Activation of glutamate dehydrogenase by l-leucine. Biochim. Et Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1989, 995, 97–101. [Google Scholar] [CrossRef]
- Mkrtchyan, G.; Aleshin, V.; Parkhomenko, Y.; Kaehne, T.; Di Salvo, M.L.; Parroni, A.; Contestabile, R.; Vovk, A.; Bettendorff, L.; Bunik, V. Molecular mechanisms of the non-coenzyme action of thiamin in brain: Biochemical, structural and pathway analysis. Sci. Rep. 2015, 5, 12583. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wang, B.; Zhang, Y.; Zhu, Y.; Song, B.; Xu, H.; Zhai, Y.; Qiao, M.; Sun, F. A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI. Nat. Commun. 2021, 12, 7257. [Google Scholar] [CrossRef] [PubMed]
- Tomita, T.; Kuzuyama, T.; Nishiyama, M. Structural basis for leucine-induced allosteric activation of glutamate dehydrogenase. J. Biol. Chem. 2011, 286, 37406–37413. [Google Scholar] [CrossRef] [PubMed]
- Tomita, T.; Miyazaki, T.; Miyazaki, J.; Kuzuyama, T.; Nishiyama, M. Hetero-oligomeric glutamate dehydrogenase from Thermus thermophilus. Microbiology 2010, 156, 3801–3813. [Google Scholar] [CrossRef]
- Tomita, T.; Yin, L.; Nakamura, S.; Kosono, S.; Kuzuyama, T.; Nishiyama, M. Crystal structure of the 2-iminoglutarate-bound complex of glutamate dehydrogenase from Corynebacterium glutamicum. FEBS Lett. 2017, 591, 1611–1622. [Google Scholar] [CrossRef]
- Godsora, B.K.J.; Prakash, P.; Punekar, N.S.; Bhaumik, P. Molecular insights into the inhibition of glutamate dehydrogenase by the dicarboxylic acid metabolites. Proteins Struct. Funct. Bioinform. 2021, 90, 810–823. [Google Scholar] [CrossRef]
- Merk, A.; Bartesaghi, A.; Banerjee, S.; Falconieri, V.; Rao, P.; Davis, M.I.; Pragani, R.; Boxer, M.B.; Earl, L.A.; Milne, J.L.S.; et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 2016, 165, 1698–1707. [Google Scholar] [CrossRef]
- Tomita, T.; Matsushita, H.; Yoshida, A.; Kosono, S.; Yoshida, M.; Kuzuyama, T.; Nishiyama, M.; Galperin, M.Y. Glutamate Dehydrogenase from Thermus thermophilus Is Activated by AMP and Leucine as a Complex with Catalytically Inactive Adenine Phosphoribosyltransferase Homolog. J. Bacteriol. 2019, 201, e00710-18. [Google Scholar] [CrossRef]
- Grzechowiak, M.; Sliwiak, J.; Jaskolski, M.; Ruszkowski, M. Structural Studies of Glutamate Dehydrogenase (Isoform 1) From Arabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism. Front. Plant Sci. 2020, 11, 754. [Google Scholar] [CrossRef] [PubMed]
- Dimovasili, C.; Fadouloglou, V.E.; Kefala, A.; Providaki, M.; Kotsifaki, D.; Kanavouras, K.; Sarrou, I.; Plaitakis, A.; Zaganas, I.; Kokkinidis, M. Crystal structure of glutamate dehydrogenase 2, a positively selected novel human enzyme involved in brain biology and cancer pathophysiology. J. Neurochem. 2021, 157, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Gohara, D.W.; Di Cera, E. Molecular Mechanisms of Enzyme Activation by Monovalent Cations. J. Biol. Chem. 2016, 291, 20840–20848. [Google Scholar] [CrossRef] [PubMed]
- Adelstein, S.J.; Vallee, B.L. Zinc in Beef Liver Glutamic Dehydrogenase. J. Biol. Chem. 1958, 233, 589–593. [Google Scholar] [CrossRef]
- Wolf, G.; Schmidt, W. Zinc and glutamate dehydrogenase in putative glutamatergic brain structures. Acta Histochem. 1983, 72, 15–23. [Google Scholar] [CrossRef]
- Bailey, J.; Powell, L.; Sinanan, L.; Neal, J.; Li, M.; Smith, T.; Bell, E. A novel mechanism of V-type zinc inhibition of glutamate dehydrogenase results from disruption of subunit interactions necessary for efficient catalysis. FEBS J. 2011, 278, 3140–3151. [Google Scholar] [CrossRef] [PubMed]
- Aleshin, V.A.; Mkrtchyan, G.V.; Kaehne, T.; Graf, A.V.; Maslova, M.V.; Bunik, V.I. Diurnal regulation of the function of the rat brain glutamate dehydrogenase by acetylation and its dependence on thiamine administration. J. Neurochem. 2020, 153, 80–102. [Google Scholar] [CrossRef]
- Lombard, D.B.; Alt, F.W.; Cheng, H.L.; Bunkenborg, J.; Streeper, R.S.; Mostoslavsky, R.; Kim, J.; Yancopoulos, G.; Valenzuela, D.; Murphy, A.; et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007, 27, 8807–8814. [Google Scholar] [CrossRef]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef]
- Mkrtchyan, G.V.; Graf, A.; Trofimova, L.; Ksenofontov, A.; Baratova, L.; Bunik, V. Positive correlation between rat brain glutamate concentrations and mitochondrial 2-oxoglutarate dehydrogenase activity. Anal Biochem. 2018, 552, 100–109. [Google Scholar] [CrossRef]
- Bettendorff, L.; Nghiêm, H.-O.; Wins, P.; Lakaye, B. A general method for the chemical synthesis of γ-32P-labeled or unlabeled nucleoside 5′-triphosphates and thiamine triphosphate. Anal. Biochem. 2003, 322, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.; Pissis, C.; Navaza, R.; Mechaly, A.E.; Saul, F.; Alzari, P.M.; Haouz, A. High-Throughput Crystallization Pipeline at the Crystallography Core Facility of the Institut Pasteur. Molecules 2019, 24, 4451. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with theautoPROCtoolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 293–302. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phasercrystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Smart, O.S.; Womack, T.O.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Vonrhein, C.; Bricogne, G. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints inBUSTER. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Moriarty, N.W.; Poon, B.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 148–157. [Google Scholar] [CrossRef]
- Zheng, H.; Cooper, D.R.; Porebski, P.J.; Shabalin, I.G.; Handing, K.B.; Minor, W. CheckMyMetal: A macromolecular metal-binding validation tool. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73 Pt 3, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.S. Global indicators of X-ray data quality. J. Appl. Crystallogr. 2001, 34, 130–135. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment 1 1Edited by J. Thornton. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]

| GDH·ADP·Leu | GDH·ADP | |
|---|---|---|
| Synchrotron beamline | ESRF ID30B | ESRF ID30B |
| Space group | P21 | P1 |
| Unit-cell parameters | ||
| a, b, c (Å) | 90.88, 178.71, 123.88 | 87.51, 92.03, 119.57 |
| α, β, γ (°) | 90, 104.00, 90 | 99.35, 106.73, 109.73 |
| Resolution range (Å) | 120.20–2.45 (2.79–2.45) | 47.46–2.40 (2.48–2.40) |
| Wavelength (Å) | 0.9763 | 0.9763 |
| No. measured reflections | 445,692 | 331,373 |
| No. unique reflections | 82,997 | 96,056 |
| Multiplicity | 5.4 (5.7) | 3.4 (3.6) |
| Completeness (%) | 92.0 (60.9) | 89.9 (86.0) |
| Average I/σ(I) | 6.8 (1.8) | 5.0 (2.0) |
| Rpima | 0.094 (0.551) | 0.175 (0.398) |
| CC(1/2) | 0.989 (0.407) | 0.925 (0.668) |
| Refinement statistics | ||
| Rworkb (%) | 19.2 | 20.4 |
| Rfreeb (%) | 21.4 | 23.6 |
| No. of non-H atoms | ||
| Macromolecule | 22994 | 22937 |
| Ligands/ions | 222 | 162 |
| Water molecules | 439 | 1019 |
| Average B-factors | 68.9 | 41.4 |
| Rms deviationsc | ||
| Bonds (Å) | 0.010 | 0.010 |
| Angles (°) | 1.37 | 1.27 |
| Molprobity statistics | ||
| Clashscore | 4.26 | 4.30 |
| Ramachandran outliers (%) | 0.07 | 0.00 |
| Ramachandran favored (%) | 97.41 | 98.6 |
| Rotamer outliers (%) | 3.24 | 3.61 |
| C-beta deviations | 0 | 1 |
| PDB entry code | 8AR7 | 8AR8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleshin, V.A.; Bunik, V.I.; Bruch, E.M.; Bellinzoni, M. Structural Basis for the Binding of Allosteric Activators Leucine and ADP to Mammalian Glutamate Dehydrogenase. Int. J. Mol. Sci. 2022, 23, 11306. https://doi.org/10.3390/ijms231911306
Aleshin VA, Bunik VI, Bruch EM, Bellinzoni M. Structural Basis for the Binding of Allosteric Activators Leucine and ADP to Mammalian Glutamate Dehydrogenase. International Journal of Molecular Sciences. 2022; 23(19):11306. https://doi.org/10.3390/ijms231911306
Chicago/Turabian StyleAleshin, Vasily A., Victoria I. Bunik, Eduardo M. Bruch, and Marco Bellinzoni. 2022. "Structural Basis for the Binding of Allosteric Activators Leucine and ADP to Mammalian Glutamate Dehydrogenase" International Journal of Molecular Sciences 23, no. 19: 11306. https://doi.org/10.3390/ijms231911306
APA StyleAleshin, V. A., Bunik, V. I., Bruch, E. M., & Bellinzoni, M. (2022). Structural Basis for the Binding of Allosteric Activators Leucine and ADP to Mammalian Glutamate Dehydrogenase. International Journal of Molecular Sciences, 23(19), 11306. https://doi.org/10.3390/ijms231911306








