Target of Rapamycin Regulates Photosynthesis and Cell Growth in Auxenochlorella pyrenoidosa
Abstract
:1. Introduction
2. Results
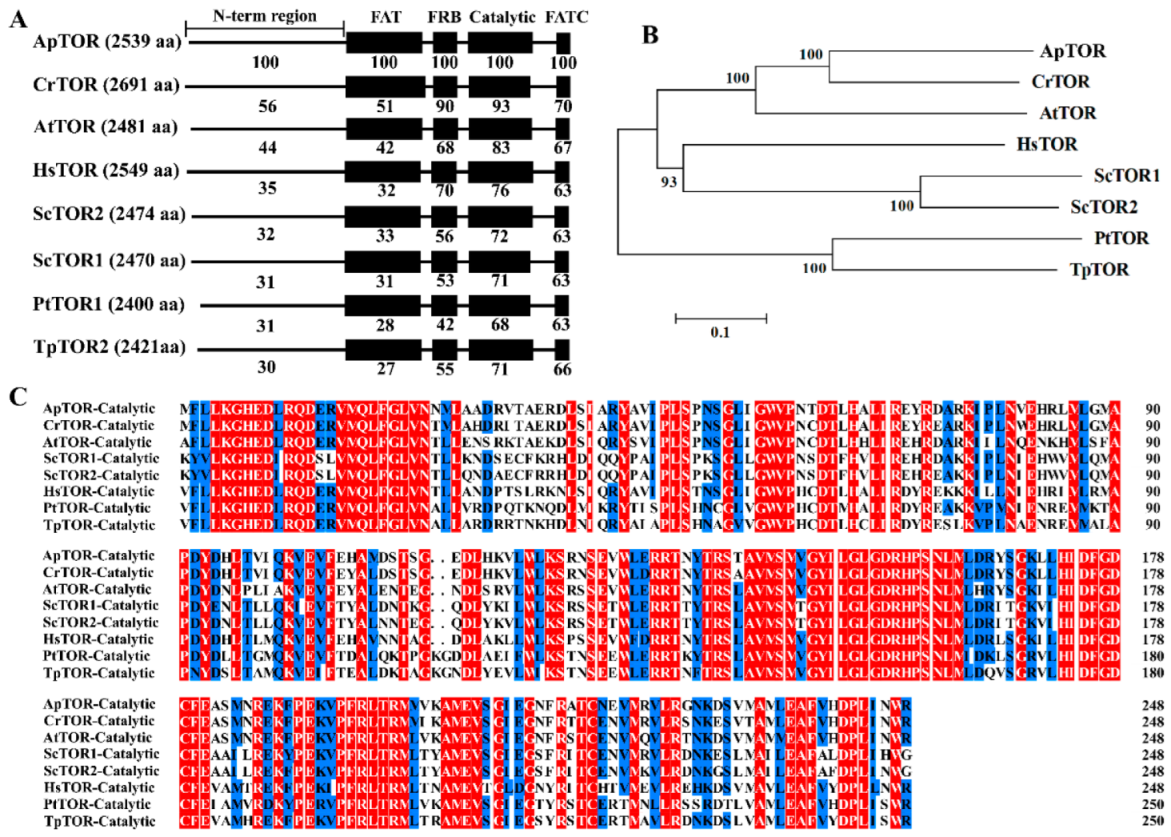
2.1. Conserved TOR Signaling Pathway in Auxenochlorella Pyrenoidosa
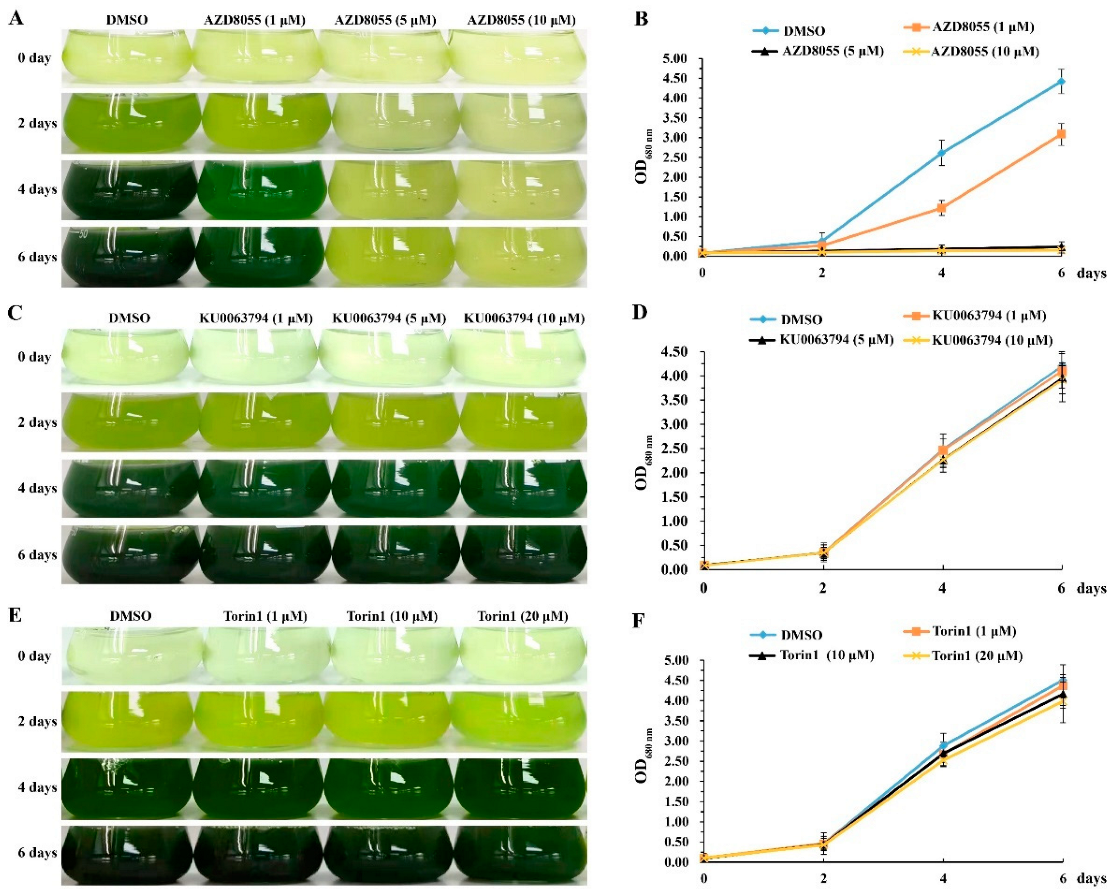
2.2. Effects of TOR Inhibitors on the Growth of Auxenochlorella Pyrenoidosa
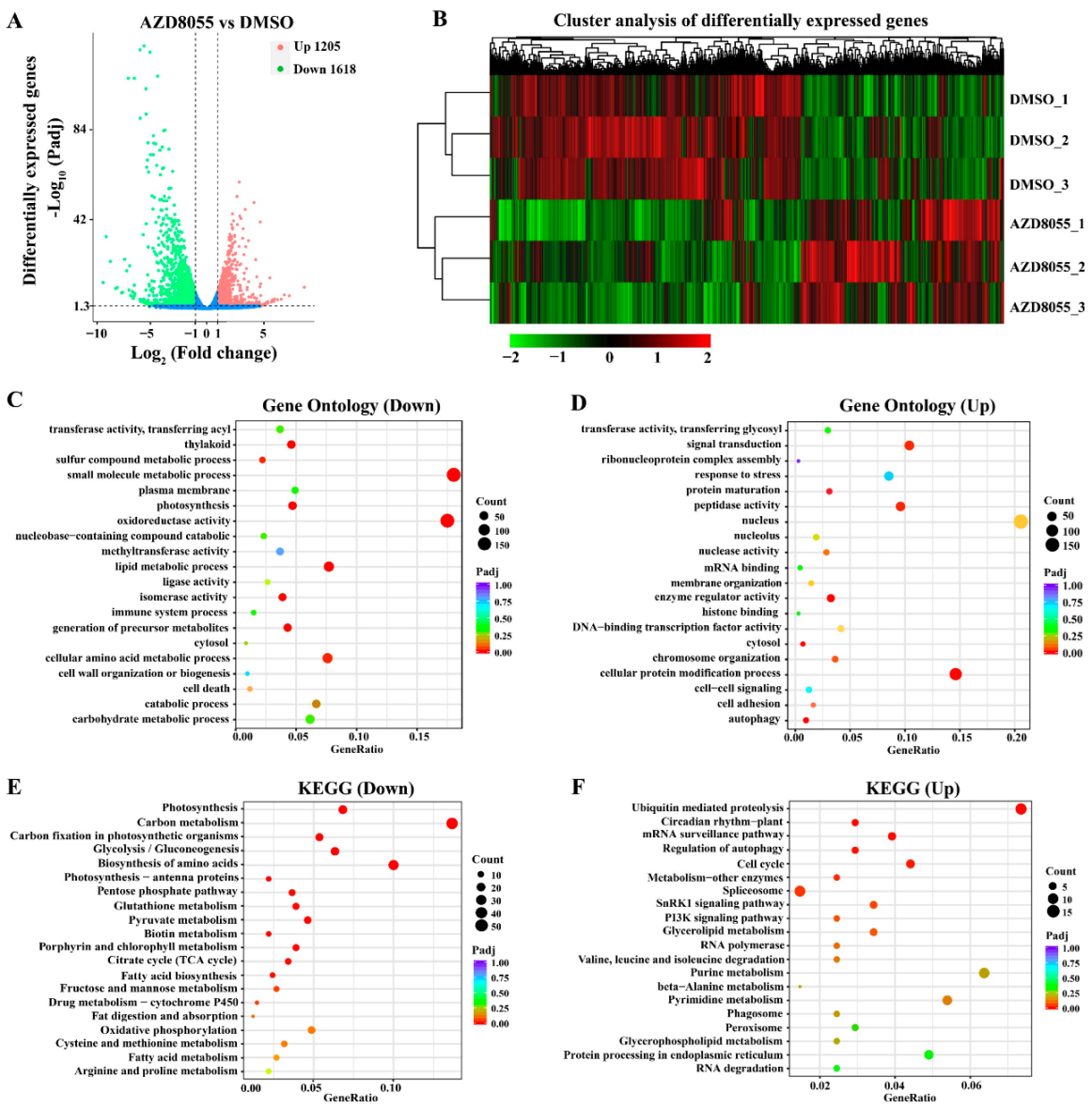
2.3. Analysis of Transcriptome Sequencing under ApTOR Inhibition
2.4. DEGs Involved in Regulating Chloroplast Development and Photosynthesis of Auxenochlorella Pyrenoidosa
2.5. DEGs Involved in Regulating Protein Synthesis and Catabolism of Auxenochlorella Pyrenoidosa
2.6. DEGs Involved in Regulating the Cell Growth of Auxenochlorella Pyrenoidosa
3. Discussion
4. Materials and Methods
4.1. Algae and Growth Condition
4.2. Treatment of Auxenochlorella Pyrenoidosa by TOR Inhibitors
4.3. Phylogenetic Tree Analysis
4.4. Construction of the RNA-seq Library and Transcriptome Sequencing
4.5. Transcriptome Assembly, Annotation and Differential Expression Analysis
4.6. Quantitative Real-Time PCR (qRT-PCR) Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heckman, D.S.; Geiser, D.M.; Eidell, B.R.; Stauffer, R.L.; Kardos, N.L.; Hedges, S.B. Molecular evidence for the early colonization of land by fungi and plants. Science 2001, 293, 1129–1133. [Google Scholar] [CrossRef]
- Spolaore, P.; Joannis-Cassan, C.; Duran, E.; Isambert, A. Commercial applications of microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef]
- Barkia, I.; Saari, N.; Manning, S.R. Microalgae for High-Value Products towards Human Health and Nutrition. Mar. Drugs 2019, 17, 304. [Google Scholar] [CrossRef] [PubMed]
- Torres-Tiji, Y.; Fields, F.J.; Mayfield, S.P. Microalgae as a future food source. Biotechnol. Adv. 2020, 41, 107536. [Google Scholar] [CrossRef]
- Yang, B.; Liu, J.; Jiang, Y.; Chen, F. Chlorella species as hosts for genetic engineering and expression of heterologous proteins: Progress, challenge and perspective. Biotechnol. J. 2016, 11, 1244–1261. [Google Scholar] [CrossRef]
- Krienitz, L.; Huss, V.A.; Bock, C. Chlorella: 125 years of the green survivalist. Trends Plant Sci. 2015, 20, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, J.; Wang, Y.; Feng, Y.; Cui, Q.; Lu, Y. Artificial creation of Chlorella pyrenoidosa mutants for economic sustainable food production. Bioresour. Technol. 2018, 268, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Ning, K.; Zeng, X.; Luo, Y.; Wang, D.; Hu, J.; Li, J.; Xu, H.; Huang, J.; Wan, M.; et al. Genomic Foundation of Starch-to-Lipid Switch in Oleaginous Chlorella spp. Plant Physiol. 2015, 169, 2444–2461. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.C.; Tan, X.B.; Yang, L.B.; Liao, J.Y.; Li, X.Y. Cultivation of Chlorella pyrenoidosa in anaerobic wastewater: The coupled effects of ammonium, temperature and pH conditions on lipids compositions. Bioresour. Technol. 2019, 284, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, Z.; Wei, D. High-yield production of biomass, protein and pigments by mixotrophic Chlorella pyrenoidosa through the bioconversion of high ammonium in wastewater. Bioresour. Technol. 2020, 313, 123499. [Google Scholar] [CrossRef]
- Sachdeva, N.; Kumar, G.D.; Gupta, R.P.; Mathur, A.S.; Manikandan, B.; Basu, B.; Tuli, D.K. Kinetic modeling of growth and lipid body induction in Chlorella pyrenoidosa under heterotrophic conditions. Bioresour. Technol. 2016, 218, 934–943. [Google Scholar] [CrossRef]
- Liu, Z.H.; Li, T.; He, Q.Y.; Sun, Z.; Jiang, Y. Role of Mitochondria in Regulating Lutein and Chlorophyll Biosynthesis in Chlorella pyrenoidosa under Heterotrophic Conditions. Mar. Drugs 2018, 16, 354. [Google Scholar] [CrossRef]
- Burkart, G.M.; Brandizzi, F. A Tour of TOR Complex Signaling in Plants. Trends Biochem. Sci. 2021, 46, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Artins, A.; Caldana, C. The metabolic homeostaTOR: The balance of holding on or letting grow. Curr. Opin. Plant Biol. 2022, 66, 102196. [Google Scholar] [CrossRef]
- Shi, L.; Wu, Y.; Sheen, J. TOR signaling in plants: Conservation and innovation. Development 2018, 145, dev160887. [Google Scholar] [CrossRef] [PubMed]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 2021, 101, 1371–1426. [Google Scholar] [CrossRef] [PubMed]
- Tafur, L.; Kefauver, J.; Loewith, R. Structural Insights into TOR Signaling. Genes 2020, 11, 885. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiong, Y. Plant target of rapamycin signaling network: Complexes, conservations, and specificities. J. Integr. Plant Biol. 2022, 64, 342–370. [Google Scholar]
- Xiong, Y.; Sheen, J. Novel links in the plant TOR kinase signaling network. Curr. Opin. Plant Biol. 2015, 28, 83–91. [Google Scholar] [CrossRef] [PubMed]
- van Dam, T.J.; Zwartkruis, F.J.; Bos, J.L.; Snel, B. Evolution of the TOR pathway. J. Mol. Evol. 2011, 73, 209–220. [Google Scholar] [CrossRef]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR Signaling and Nutrient Sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef]
- Crespo, J.L.; Hall, M.N. Elucidating TOR signaling and rapamycin action: Lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002, 66, 579–591. [Google Scholar] [CrossRef]
- Xu, Q.; Liang, S.; Kudla, J.; Luan, S. Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant. J. 1998, 15, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Sormani, R.; Yao, L.; Menand, B.; Ennar, N.; Lecampion, C.; Meyer, C.; Robaglia, C. Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 2007, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Ishiwata, A.; Watanabe, S.; Yoshikawa, H.; Tanaka, K. Expression of budding yeast FKBP12 confers rapamycin susceptibility to the unicellular red alga Cyanidioschyzon merolae. Biochem. Biophys. Res. Commun. 2013, 439, 264–269. [Google Scholar] [CrossRef]
- Ren, M.; Venglat, P.; Qiu, S.; Feng, L.; Cao, Y.; Wang, E.; Xiang, D.; Wang, J.; Alexander, D.; Chalivendra, S.; et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 2012, 24, 4850–4874. [Google Scholar] [CrossRef]
- Montané, M.H.; Menand, B. TOR inhibitors: From mammalian outcomes to pharmacogenetics in plants and algae. J. Exp. Bot. 2019, 70, 2297–2312. [Google Scholar] [CrossRef] [PubMed]
- Montané, M.H.; Menand, B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 2013, 64, 4361–4374. [Google Scholar] [CrossRef]
- Van Leene, J.; Han, C.; Gadeyne, A.; Eeckhout, D.; Matthijs, C.; Cannoot, B.; De Winne, N.; Persiau, G.; Van De Slijke, E.; Van de Cotte, B.; et al. Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 2019, 5, 316–327. [Google Scholar] [CrossRef]
- Li, L.; Liu, K.H.; Sheen, J. Dynamic Nutrient Signaling Networks in Plants. Annu. Rev. Cell Dev. Biol. 2021, 37, 341–367. [Google Scholar] [CrossRef]
- McCready, K.; Spencer, V.; Kim, M. The Importance of TOR Kinase in Plant Development. Front. Plant Sci. 2020, 11, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Wang, P.; Xiong, Y. Target of Rapamycin Signaling in Plant Stress Responses. Plant Physiol. 2020, 182, 1613–1623. [Google Scholar] [CrossRef]
- Song, Y.; Alyafei, M.S.; Masmoudi, K.; Jaleel, A.; Ren, M. Contributions of TOR Signaling on Photosynthesis. Int. J. Mol. Sci. 2021, 22, 8959. [Google Scholar] [CrossRef] [PubMed]
- Shemi, A.; Ben-Dor, S.; Vardi, A. Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy 2015, 11, 701–715. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.E.; Couso, I.; Crespo, J.L. The TOR Signaling Network in the Model Unicellular Green Alga Chlamydomonas reinhardtii. Biomolecules 2017, 7, 54. [Google Scholar] [CrossRef]
- Mubeen, U.; Jüppner, J.; Alpers, J.; Hincha, D.K.; Giavalisco, P. Target of Rapamycin Inhibition in Chlamydomonas reinhardtii Triggers de Novo Amino Acid Synthesis by Enhancing Nitrogen Assimilation. Plant Cell 2018, 30, 2240–2254. [Google Scholar] [CrossRef]
- Jüppner, J.; Mubeen, U.; Leisse, A.; Caldana, C.; Wiszniewski, A.; Steinhauser, D.; Giavalisco, P. The target of rapamycin kinase affects biomass accumulation and cell cycle progression by altering carbon/nitrogen balance in synchronized Chlamydomonas reinhardtii cells. Plant J. 2018, 93, 355–376. [Google Scholar] [CrossRef]
- Werth, E.G.; McConnell, E.W.; Couso Lianez, I.; Perrine, Z.; Crespo, J.L.; Umen, J.G.; Hicks, L.M. Investigating the effect of target of rapamycin kinase inhibition on the Chlamydomonas reinhardtii phosphoproteome: From known homologs to new targets. New Phytol. 2019, 221, 247–260. [Google Scholar] [CrossRef]
- Benjamin, D.; Colombi, M.; Moroni, C.; Hall, M.N. Rapamycin passes the torch: A new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011, 10, 868–880. [Google Scholar] [CrossRef]
- Choi, J.; Chen, J.; Schreiber, S.L.; Clardy, J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 1996, 273, 239–242. [Google Scholar] [CrossRef]
- Kaiser, E.; Correa Galvis, V.; Armbruster, U. Efficient photosynthesis in dynamic light environments: A chloroplast’s perspective. Biochem. J. 2019, 476, 2725–2741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, P.; Xiong, F.; Que, Y.; Wang, K.; Yu, L.; Li, Z.; Ren, M. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 2015, 6, 677. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O. Exaptive Evolution of Target of Rapamycin Signaling in Multicellular Eukaryotes. Dev. Cell 2020, 54, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Saba, J.A.; Liakath-Ali, K.; Green, R.; Watt, F.M. Translational control of stem cell function. Nat. Rev. Mol. Cell Biol. 2021, 22, 671–690. [Google Scholar] [CrossRef]
- Mak, T.; Jones, A.W.; Nurse, P. The TOR-dependent phosphoproteome and regulation of cellular protein synthesis. Embo J. 2021, 40, e107911. [Google Scholar] [CrossRef]
- Ganley, I.G.; Lam du, H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Pu, Y.; Luo, X.; Bassham, D.C. TOR-Dependent and -Independent Pathways Regulate Autophagy in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1204. [Google Scholar] [CrossRef]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef]
- Mehrtash, A.B.; Hochstrasser, M. Ubiquitin-dependent protein degradation at the endoplasmic reticulum and nuclear envelope. Semin. Cell Dev. Biol. 2019, 93, 111–124. [Google Scholar] [CrossRef]
- Yuan, H.X.; Xiong, Y.; Guan, K.L. Nutrient sensing, metabolism, and cell growth control. Mol. Cell 2013, 49, 379–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baslam, M.; Mitsui, T.; Sueyoshi, K.; Ohyama, T. Recent Advances in Carbon and Nitrogen Metabolism in C3 Plants. Int. J. Mol. Sci. 2020, 22, 318. [Google Scholar] [CrossRef] [PubMed]
- Caldana, C.; Li, Y.; Leisse, A.; Zhang, Y.; Bartholomaeus, L.; Fernie, A.R.; Willmitzer, L.; Giavalisco, P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013, 73, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, M.E.; Florencio, F.J.; Crespo, J.L. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010, 152, 1874–1888. [Google Scholar] [CrossRef]
- Martin, W.F.; Bryant, D.A.; Beatty, J.T. A physiological perspective on the origin and evolution of photosynthesis. FEMS Microbiol. Rev. 2018, 42, 205–231. [Google Scholar] [CrossRef]
- Stirbet, A.; Lazár, D.; Guo, Y.; Govindjee, G. Photosynthesis: Basics, history and modelling. Ann. Bot. 2020, 126, 511–537. [Google Scholar] [CrossRef]
- Li, L.; Song, Y.; Wang, K.; Dong, P.; Zhang, X.; Li, F.; Li, Z.; Ren, M. TOR-inhibitor insensitive-1 (TRIN1) regulates cotyledons greening in Arabidopsis. Front. Plant Sci. 2015, 6, 861. [Google Scholar] [CrossRef]
- Mallén-Ponce, M.J.; Pérez-Pérez, M.E.; Crespo, J.L. Photosynthetic assimilation of CO2 regulates TOR activity. Proc. Natl. Acad. Sci. USA 2022, 119, e2115261119. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Bio.l 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Siva Sankar, D.; Dengjel, J. Protein complexes and neighborhoods driving autophagy. Autophagy 2021, 17, 2689–2705. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Protein Name | Chlamydomonas reinhardtii | Auxenochlorella pyrenoidosa | Identity (%) |
|---|---|---|---|
| Target of rapamycin (TOR) | CrTOR | ApTOR like | 58 |
| Regulatory associate protein of TOR (RAPTOR) | CrRAPTOR | ApRAPTOR like | 37 |
| Lethal with SEC-13 protein 8 (LST8) | CrLST8 | ApLST8 like | 81 |
| FK506-binding protein 12 kDa (FKBP12) | CrFKBP12 | ApFKBP12 like | 43 |
| Ribosomal protein S6 kinase (S6K) | CrS6K | ApS6K like | 45 |
| Ribosome protein small subunit 6 (RPS6) | CrRPS6 | ApRPS6 like | 78 |
| Transcription factor E2F alpha (E2FA) | CrE2FA | ApE2FA like | 39 |
| Translation initiation factor 2 alpha subunit (eIF2α) | CreIF2α | ApeIF2α like | 72 |
| Type 2A phosphatase associated protein 46 (TAP46) | CrTAP46 | ApTAP46 like | 40 |
| Autophagy protein 1 (ATG1) | CrATG1 | ApATG1 like | 39 |
| Autophagy protein 13 (ATG13) | CrATG13 | ApATG13 like | 32 |
| Gene ID | Log2 (Fold Change) | P-adj | KO Name | KO Description |
|---|---|---|---|---|
| Photosynthesis | ||||
| Cluster-495.6756 | −5.9733 | 5.56 × 10−90 | PETE | Plastocyanin |
| Cluster-498.0 | −5.6390 | 3.47 × 10−5 | PSBE | Photosystem II cytochrome b559 subunit α |
| Cluster-495.7678 | −5.6230 | 6.53 × 10−124 | PSAN | Photosystem I subunit psan |
| Cluster-495.7002 | −5.4544 | 9.90 × 10−104 | PSAH | Photosystem I subunit VI |
| Cluster-495.6324 | −5.3253 | 2.42 × 10−70 | PSBP | Photosystem II oxygen-evolving enhancer 2 |
| Cluster-495.8198 | −5.2049 | 9.45 × 10−3 | ATPF0B | F-type H+ transporting ATPase subunit b |
| Cluster-495.827 | −4.6442 | 6.89 × 10−4 | PETA | Apocytochrome f |
| Cluster-495.10228 | −4.6211 | 4.34 × 10−2 | PSBJ | Photosystem II psbj protein |
| Cluster-495.5726 | −4.0208 | 1.05 × 10−31 | PSBS | Photosystem II 22kda protein |
| Cluster-495.5249 | −4.0063 | 1.02 × 10−46 | PSB27 | Photosystem II Psb27 protein |
| Cluster-495.4105 | −3.9952 | 2.49 × 10−2 | ATPF1A | F-type H+-transporting atpase subunit alpha |
| Cluster-495.6958 | −3.8390 | 5.22 × 10−43 | PSAF | Photosystem I subunit III |
| Cluster-495.7332 | −3.7857 | 2.68 × 10−3 | PSBC | Photosystem II CP43 chlorophyll apoprotein |
| Cluster-495.6505 | −3.4964 | 1.36 × 10−28 | PSAK | Photosystem I subunit X |
| Cluster-495.6979 | −3.3834 | 1.87 × 10−23 | PSBR | Photosystem II 10kda protein |
| Cluster-495.8002 | −3.2884 | 4.79 × 10−6 | ATPF1B | F-type H+-transporting atpase subunit beta |
| Cluster-495.6124 | −3.1305 | 3.85 × 10−3 | PETB | Cytochrome b6 |
| Cluster-495.7894 | −3.0592 | 3.40 × 10−17 | PSAB | Photosystem I P700 chlorophyll a apoprotein |
| Cluster-495.1037 | −2.9414 | 3.35 × 10−20 | PSBP | Photosystem II oxygen-evolving enhancer 2 |
| Cluster-495.4143 | −2.8547 | 8.51 × 10−45 | PSAD | Photosystem I subunit II |
| Cluster-495.6035 | −2.8402 | 3.44 × 10−36 | PSAL | Photosystem I subunit XI |
| Cluster-495.4194 | −2.7017 | 7.15 × 10−18 | PSAO | Photosystem I subunit psao |
| Cluster-495.4418 | −2.3110 | 2.12 × 10−21 | PSAG | Photosystem I subunit V |
| Cluster-495.5609 | −2.2904 | 4.52 × 10−18 | PSBO | Photosystem II oxygen-evolving enhancer 1 |
| Cluster-495.4835 | −1.7041 | 5.65 × 10−12 | PETJ | Cytochrome c6 |
| Cluster-495.6263 | −1.5457 | 9.25 × 10−13 | PETH | Ferredoxin--NADP+ reductase |
| Cluster-495.7190 | −1.4895 | 5.90 × 10−11 | PSBY | Photosystem II psby protein |
| Cluster-495.4933 | −1.2359 | 4.99 × 10−8 | PSB28 | Photosystem II 13kda protein |
| Cluster-495.7871 | 1.1214 | 1.83 × 10−5 | PETF | Ferredoxin |
| Photosynthesis-antenna proteins | ||||
| Cluster-495.6691 | −7.0421 | 1.03 × 10−108 | LHCA4 | Photosystem I chlorophyll a/b binding protein 4 |
| Cluster-495.3640 | −5.9832 | 4.43 × 10−122 | LHCA3 | Photosystem I chlorophyll a/b binding protein 3 |
| Cluster-495.6564 | −5.2717 | 1.81 × 10−78 | LHCA1 | Photosystem I chlorophyll a/b binding protein 1 |
| Cluster-495.5144 | −4.5671 | 6.62 × 10−78 | LHCB4 | Photosystem II chlorophyll a/b binding protein 4 |
| Cluster-495.6386 | −4.1987 | 1.03 × 10−51 | LHCB1 | Photosystem II chlorophyll a/b binding protein 1 |
| Cluster-495.5553 | −3.9840 | 1.96 × 10−76 | LHCA4 | Photosystem I chlorophyll a/b binding protein 4 |
| Cluster-495.6485 | −3.9329 | 1.45 × 10−40 | LHCB5 | Photosystem II chlorophyll a/b binding protein 5 |
| Cluster-495.6349 | −3.4326 | 3.63 × 10−33 | LHCB1 | Photosystem II chlorophyll a/b binding protein 1 |
| Cluster-495.8808 | −2.5721 | 2.26 × 10−2 | LHCB2 | Photosystem II chlorophyll a/b binding protein 2 |
| Carbon fixation in photosynthetic organisms | ||||
| Cluster-495.5099 | −5.0936 | 5.78 × 10−121 | PGK | Phosphoglycerate kinase |
| Cluster-495.2276 | −4.3041 | 1.39 × 10−43 | ALDO | Fructose-bisphosphate aldolase, class I |
| Cluster-495.7895 | −4.0597 | 1.13 × 10−9 | GOT2 | Aspartate aminotransferase |
| Cluster-495.2903 | −3.5833 | 1.72 × 10−36 | MDH2 | Malate dehydrogenase |
| Cluster-495.5815 | −3.0446 | 6.82 × 10−42 | PRKB | Phosphoribulokinase |
| Cluster-495.5677 | −2.9728 | 3.99 × 10−29 | PPDK | Pyruvate, orthophosphate dikinase |
| Cluster-495.5005 | −2.8583 | 3.63 × 10−42 | TPI | Triosephosphate isomerase (TIM) |
| Cluster-495.5217 | −2.7963 | 1.37 × 10−29 | ALDO | Fructose-bisphosphate aldolase, class I |
| Cluster-495.4546 | −2.7251 | 3.02 × 10−38 | TPI | Triosephosphate isomerase (TIM) |
| Cluster-495.5985 | −2.5229 | 6.25 × 10−42 | MDH1 | Malate dehydrogenase |
| Cluster-495.3332 | −2.5073 | 2.29 × 10−19 | RPIA | Ribose 5-phosphate isomerase A |
| Cluster-495.1259 | −2.5056 | 8.33 × 10−6 | PCKA | Phosphoenolpyruvate carboxykinase |
| Cluster-495.4145 | −1.8990 | 6.39 × 10−16 | MDH2 | Malate dehydrogenase |
| Cluster-495.4967 | −1.7549 | 3.89 × 10−11 | MAEB | Malate dehydrogenase (NADP+) |
| Cluster-495.3492 | −1.6252 | 7.78 × 10−7 | MAEB | Malate dehydrogenase (NADP+) |
| Cluster-495.5038 | −1.6083 | 3.40 × 10−17 | TKTA | Transketolase |
| Cluster-495.6521 | −1.6040 | 1.68 × 10−14 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| Cluster-495.6227 | −1.2768 | 9.08 × 10−9 | GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| Cluster-495.3372 | −1.1984 | 1.25 × 10−4 | PPC | Phosphoenolpyruvate carboxylase |
| Cluster-495.5601 | −1.0961 | 2.63 × 10−9 | E3.1.3.37 | Sedoheptulose-bisphosphatase |
| Cluster-495.2869 | −1.0914 | 3.29 × 10−7 | FBP | Fructose-1,6-bisphosphatase I |
| Cluster-495.6190 | −1.0435 | 5.51 × 10−5 | GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| Cluster-495.5207 | 1.1296 | 7.49 × 10−7 | E1.1.1.39 | Malate dehydrogenase (decarboxylating) |
| Cluster-495.7215 | 1.4168 | 6.87 × 10−8 | GGAT | Glutamate-glyoxylate aminotransferase |
| Gene ID | Log2 (Fold Change) | P-adj | KO Name | KO Description |
|---|---|---|---|---|
| Regulation of autophagy | ||||
| Cluster-495.165 | 1.6278 | 3.68 × 10−2 | SnRK1α | SNF1-related protein kinase 1 α subunit |
| Cluster-495.6489 | 1.5520 | 9.00 × 10−13 | ATG7 | Autophagy-related protein 7 |
| Cluster-495.4583 | 1.3266 | 5.06 × 10−5 | ATG3 | Autophagy-related protein 3 |
| Cluster-495.7507 | 1.2287 | 1.39 × 10−6 | ATG16L1 | Autophagy-related protein 16-1 |
| Cluster-495.6822 | 1.1679 | 3.45 × 10−6 | SnRK1α | SNF1-related protein kinase 1 α subunit |
| Cluster-495.7877 | 1.1533 | 1.91 × 10−7 | ATG101 | Autophagy-related protein 101 |
| Cluster-495.4705 | 1.0000 | 4.53 × 10−6 | ATG11 | Autophagy-related protein 11 |
| Cluster-495.2673 | −1.6051 | 8.07 × 10−6 | ATG12 | Autophagy-related protein 12 |
| Ubiquitin mediated proteolysis | ||||
| Cluster-495.5469 | 2.9686 | 4.17 × 10−21 | UBLE1B | Ubiquitin-like 1-activating enzyme E1 B |
| Cluster-495.3804 | 2.7399 | 2.91 × 10−53 | UBE2A | Ubiquitin-conjugating enzyme E2 A |
| Cluster-495.4516 | 2.3622 | 8.08 × 10−26 | ERCC8 | DNA excision repair protein ERCC8 |
| Cluster-495.4258 | 1.7941 | 1.47 × 10−13 | CDH1 | Cell division cycle 20-like protein 1 |
| Cluster-495.3283 | 1.7552 | 2.29 × 10−19 | CUL3 | Cullin 3 |
| Cluster-495.5596 | 1.7515 | 2.80 × 10−2 | UBLE1B | Ubiquitin-like 1-activating enzyme E1 B |
| Cluster-495.8038 | 1.4688 | 1.57 × 10−16 | CUL1 | Cullin 1 |
| Cluster-495.4275 | 1.4562 | 6.98 × 10−11 | SKP1 | S-phase kinase-associated protein 1 |
| Cluster-495.6597 | 1.4529 | 1.99 × 10−6 | UBE2E | Ubiquitin-conjugating enzyme E2 E |
| Cluster-495.4573 | 1.2755 | 1.57 × 10−9 | UBE1 | Ubiquitin-activating enzyme E1 |
| Cluster-495.4611 | 1.2633 | 8.88 × 10−9 | CUL4 | Cullin 4 |
| Cluster-495.2951 | 1.1905 | 1.28 × 10−8 | UBE1C | Ubiquitin-activating enzyme E1 C |
| Cluster-495.8466 | 1.1285 | 6.54 × 10−4 | RBX1 | RING-box protein 1 |
| Cluster-495.1824 | 1.0291 | 3.12 × 10−5 | PPIL2 | Peptidyl-prolyl cis-trans isomerase-like 2 |
| Cluster-495.4193 | 1.0217 | 1.66 × 10−3 | RBX1 | RING-box protein 1 |
| Cluster-495.1695 | −1.1161 | 1.39 × 10−3 | FANCL | E3 ubiquitin-protein ligase FANCL |
| Cluster-495.1057 | −1.2830 | 1.16 × 10−4 | UBE3A | Ubiquitin-protein ligase E3 A |
| Cluster-495.7130 | −1.6390 | 1.07 × 10−10 | UBLE1A | Ubiquitin-like 1-activating enzyme E1 A |
| Cluster-495.9644 | −1.7193 | 5.31 × 10−3 | UBE2S | Ubiquitin-conjugating enzyme E2 S |
| Gene ID | Log2 (Fold Change) | P-adj | KO Name | KO Description |
|---|---|---|---|---|
| Cluster-495.2903 | −3.5833 | 1.72 × 10−36 | MDH2 | Malate dehydrogenase |
| Cluster-495.5985 | −2.5229 | 6.25 × 10−42 | MDH1 | Malate dehydrogenase |
| Cluster-495.1259 | −2.5056 | 8.33 × 10−6 | PCKA | Phosphoenolpyruvate carboxykinase |
| Cluster-495.4145 | −1.8990 | 6.39 × 10−16 | MDH2 | Malate dehydrogenase |
| Cluster-495.5734 | −1.6082 | 5.41 × 10−16 | LSC1 | Succinyl-CoA synthetase alpha subunit |
| Cluster-495.5974 | −1.5814 | 3.82 × 10−15 | ACO | Aconitate hydratase |
| Cluster-495.4348 | −1.4680 | 3.68 × 10−8 | PDHD | Dihydrolipoamide dehydrogenase |
| Cluster-495.4853 | −1.2993 | 1.45 × 10−10 | DLST | α-oxoglutarate dehydrogenase E2 |
| Cluster-495.6949 | −1.2720 | 1.91 × 10−5 | PDHC | Pyruvate dehydrogenase E2 |
| Cluster-495.6523 | −1.2587 | 7.65 × 10−7 | FUMC | Fumarate hydratase, class II |
| Cluster-495.5492 | −1.2187 | 2.33 × 10−8 | IDH1 | Isocitrate dehydrogenase |
| Cluster-495.6105 | −1.2067 | 8.64 × 10−11 | LSC2 | Succinyl-CoA synthetase beta subunit |
| Cluster-495.2778 | −1.1371 | 2.54 × 10−6 | SDHD | Succinate dehydrogenase subunit |
| Cluster-495.5250 | −1.1254 | 1.39 × 10−6 | OGDH | α-oxoglutarate dehydrogenase E1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, T.; Li, L.; Chang, H.; Zhan, J.; Ren, M. Target of Rapamycin Regulates Photosynthesis and Cell Growth in Auxenochlorella pyrenoidosa. Int. J. Mol. Sci. 2022, 23, 11309. https://doi.org/10.3390/ijms231911309
Zhu T, Li L, Chang H, Zhan J, Ren M. Target of Rapamycin Regulates Photosynthesis and Cell Growth in Auxenochlorella pyrenoidosa. International Journal of Molecular Sciences. 2022; 23(19):11309. https://doi.org/10.3390/ijms231911309
Chicago/Turabian StyleZhu, Tingting, Linxuan Li, Huimin Chang, Jiasui Zhan, and Maozhi Ren. 2022. "Target of Rapamycin Regulates Photosynthesis and Cell Growth in Auxenochlorella pyrenoidosa" International Journal of Molecular Sciences 23, no. 19: 11309. https://doi.org/10.3390/ijms231911309
APA StyleZhu, T., Li, L., Chang, H., Zhan, J., & Ren, M. (2022). Target of Rapamycin Regulates Photosynthesis and Cell Growth in Auxenochlorella pyrenoidosa. International Journal of Molecular Sciences, 23(19), 11309. https://doi.org/10.3390/ijms231911309






