Two Conserved Amino Acids Characterized in the Island Domain Are Essential for the Biological Functions of Brassinolide Receptors
Abstract
:1. Introduction
2. Results
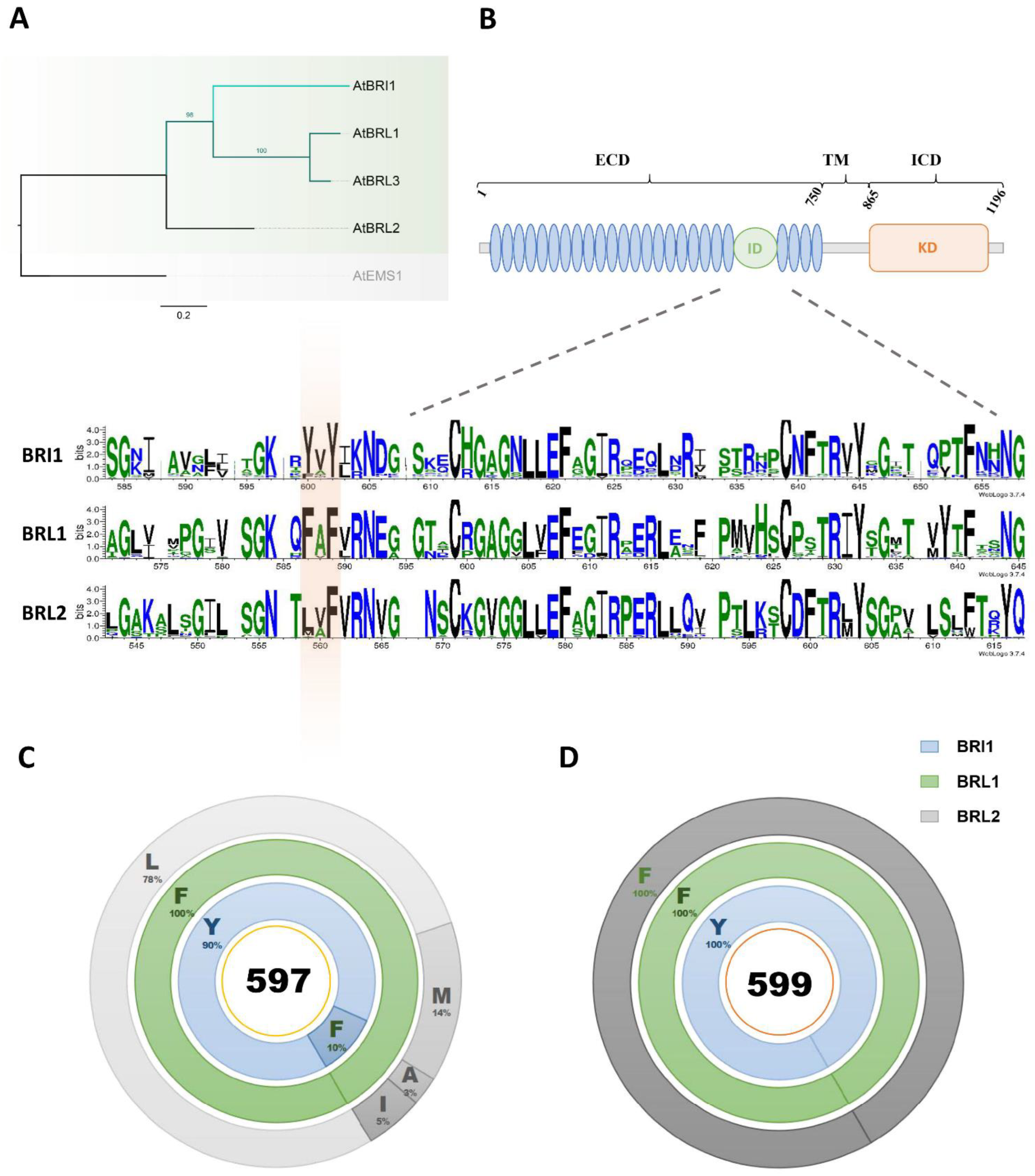
2.1. Tyrosine597 and Tyrosine599 in the BRI1 Island Domain Are Fixed during Plant Evolution
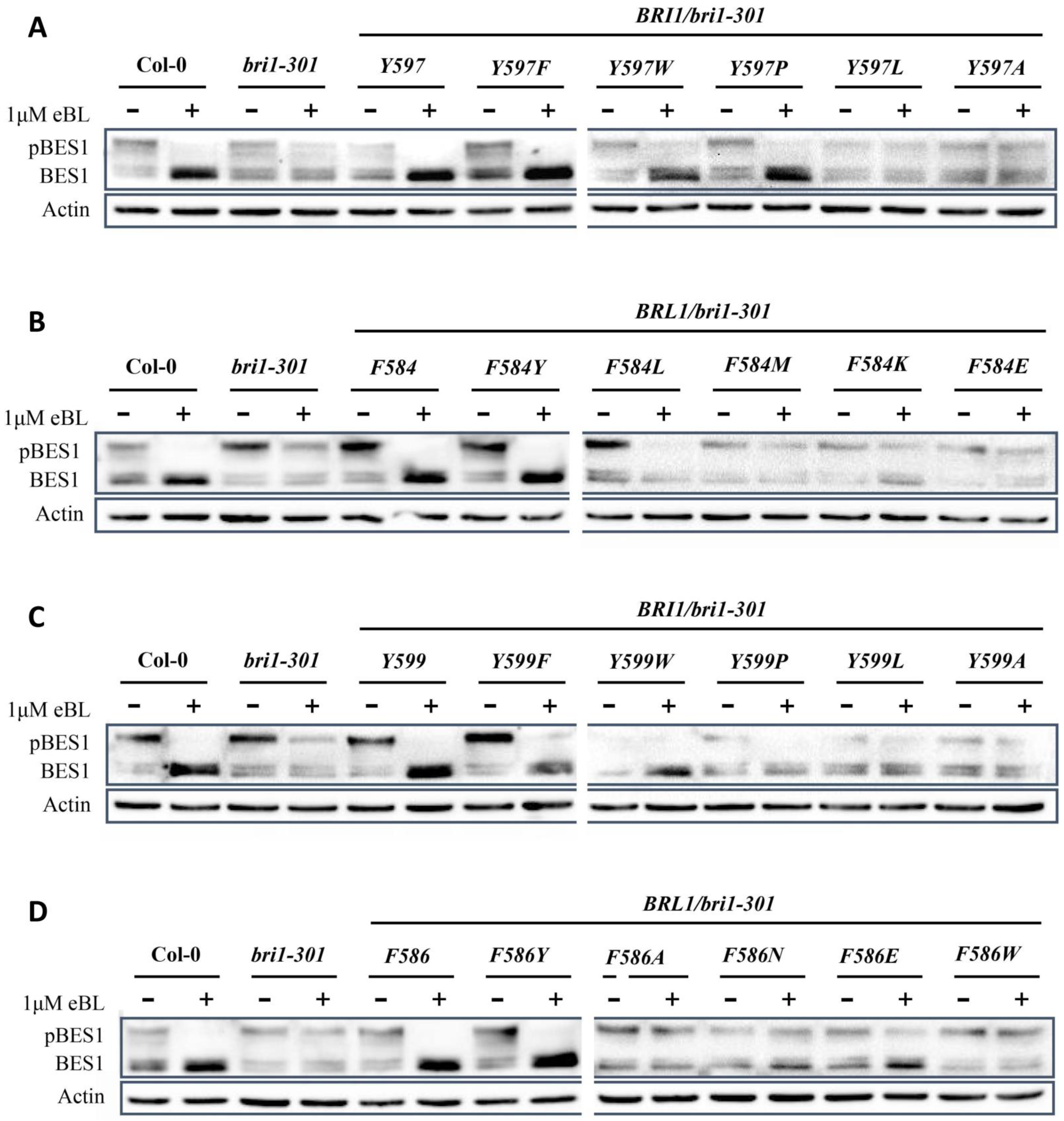
2.2. Tyrosine597 and Phenylalanine597 Differentiate BR Receptors from Non-BR Receptor
2.3. Tyrosine599 Specifies the Essential BR Receptor BRI1 from BRL1/3
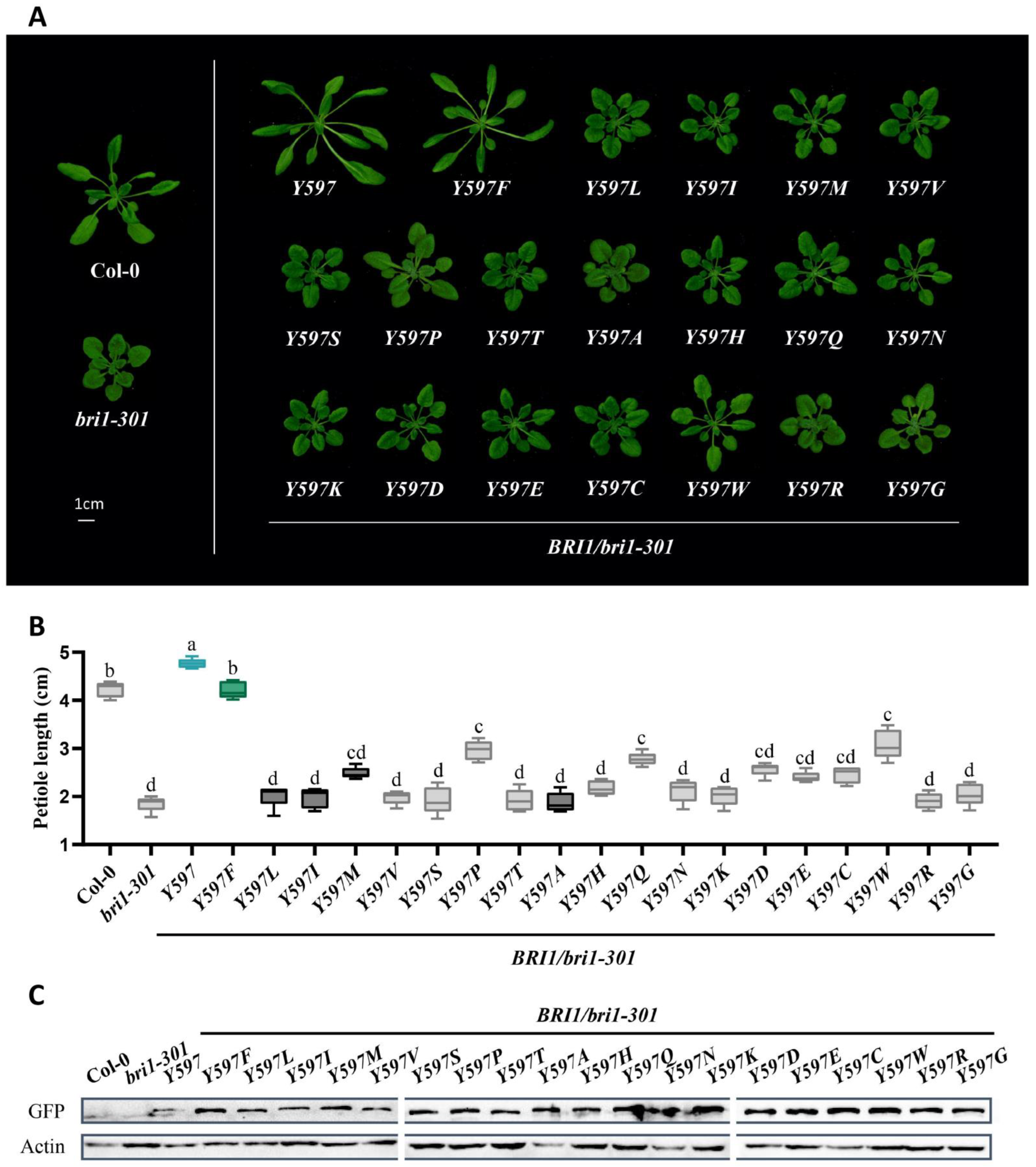
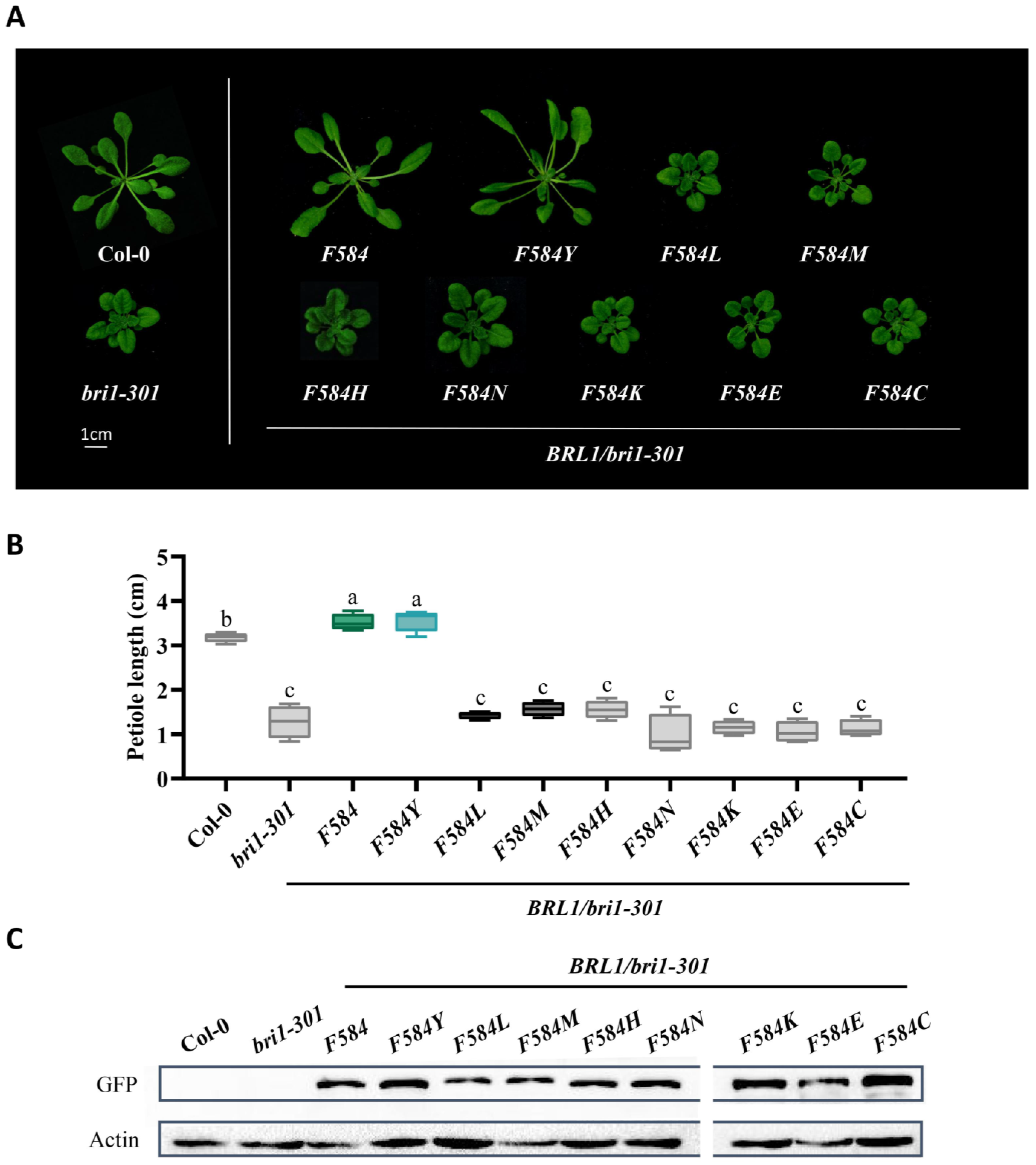
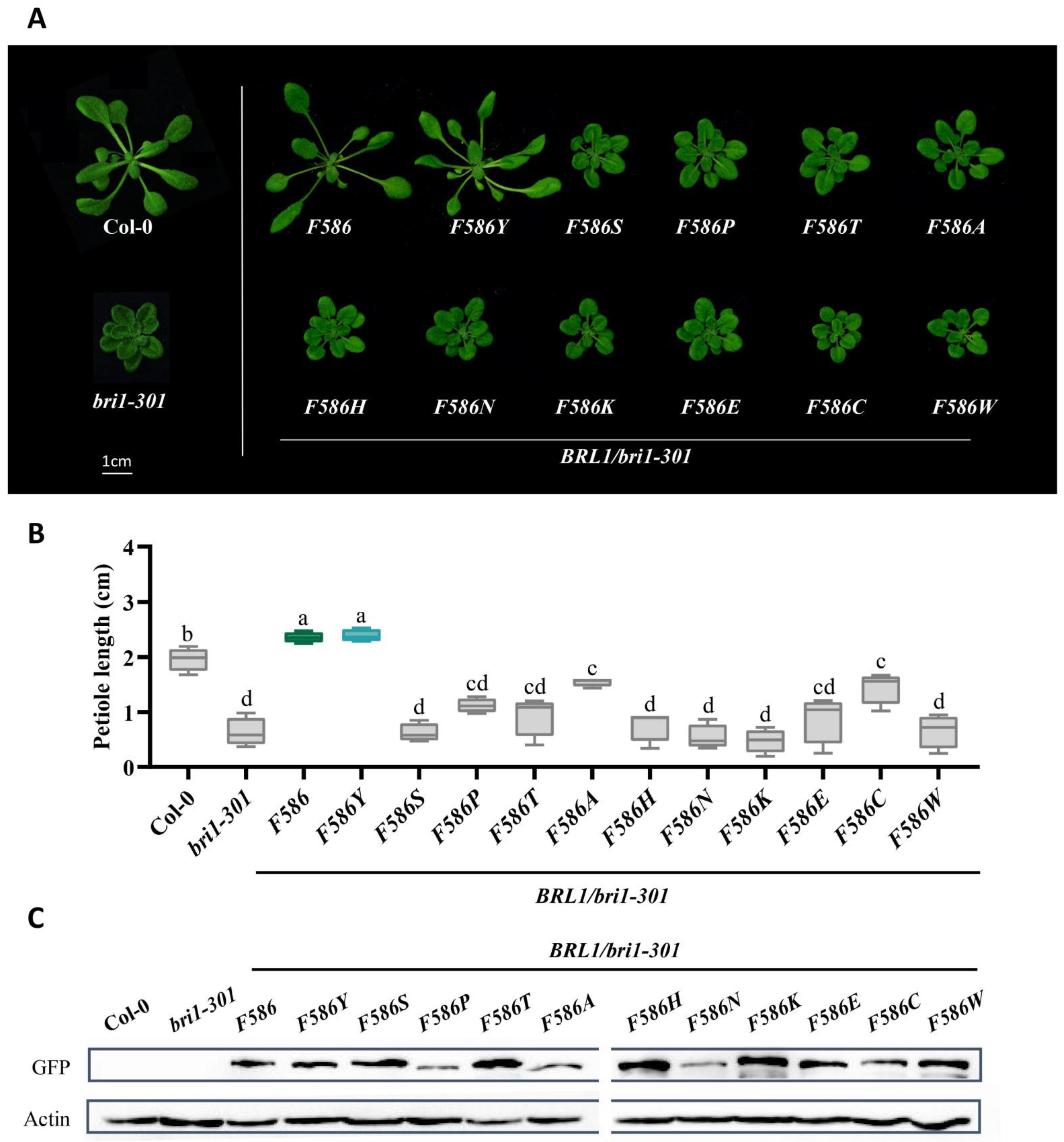
2.4. BR Receptors with Tyrosine597 or Tyrosine599 Showing the Highest Biochemical and Physiological Efficacy in BR Signaling
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Generation of Transgenic Plants
4.3. eBL Treatment of Root
4.4. Immunoblot Analysis
4.5. Phylogenetic Analysis
4.6. Structure Analysis
4.7. Statistics Analysis for Petiole Lengths
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, M.; Németh, K.; Koncz-Kálmán, Z.; Mathur, J.; Kauschmann, A.; Altmann, T.; Rédei, G.P.; Nagy, F.; Schell, J.; Koncz, C. Brassinosteroids Rescue the Deficiency of CYP90, a Cytochrome P450, Controlling Cell Elongation and De-etiolation in Arabidopsis. Cell 1996, 85, 171–182. [Google Scholar] [CrossRef]
- Kim, E.J.; Russinova, E. Brassinosteroid signalling. Curr. Biol. 2020, 30, R294–R298. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Clouse, S.D.; Langford, M.; McMorris, T.C. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996, 111, 671–678. [Google Scholar] [CrossRef]
- Nomura, T.; Kushiro, T.; Yokota, T.; Kamiya, Y.; Bishop, G.J.; Yamaguchi, S. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 2005, 280, 17873–17879. [Google Scholar] [CrossRef]
- Kinoshita, T.; Caño-Delgado, A.; Seto, H.; Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 2005, 433, 167–171. [Google Scholar] [CrossRef]
- Aldukhi, F.; Deb, A.; Zhao, C.; Moffett, A.S.; Shukla, D. Molecular Mechanism of Brassinosteroid Perception by the Plant Growth Receptor BRI1. J. Phys. Chem. B 2020, 124, 355–365. [Google Scholar] [CrossRef]
- He, Z.; Wang, Z.Y.; Li, J.; Zhu, Q.; Lamb, C.; Ronald, P.; Chory, J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 2000, 288, 2360–2363. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef]
- Mora-Garcia, S.; Vert, G.; Yin, Y.; Cano-Delgado, A.; Cheong, H.; Chory, J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004, 18, 448–460. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef]
- Shi, H.; Li, X.; Lv, M.; Li, J. BES1/BZR1 Family Transcription Factors Regulate Plant Development via Brassinosteroid-Dependent and Independent Pathways. Int. J. Mol. Sci. 2022, 23, 10149. [Google Scholar] [CrossRef]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–471. [Google Scholar] [CrossRef]
- She, J.; Han, Z.; Kim, T.W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Wang, J.; Yang, M.; Wang, Z.Y.; et al. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, T.; Fujioka, S.; Choe, S.; Takatsuto, S.; Yoshida, S.; Yuan, H.; Feldmann, K.A.; Tax, F.E. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999, 121, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.C.; Chory, J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef] [PubMed]
- She, J.; Han, Z.; Zhou, B.; Chai, J. Structural basis for differential recognition of brassinolide by its receptors. Protein Cell 2013, 4, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Dievart, A.; Hymes, M.J.; Li, J.; Clark, S.E. Brassinosteroid-independent function of BRI1/CLV1 chimeric receptors. Funct. Plant. Biol. 2006, 33, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, D.; Szarejko, I.; Maluszynski, M. New allele of HvBRI1 gene encoding brassinosteroid receptor in barley. J. Appl. Genet. 2011, 52, 257–268. [Google Scholar] [CrossRef]
- Cano-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-Garcia, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef]
- Clay, N.K.; Nelson, T. VH1, a Provascular Cell–Specific Receptor Kinase That Influences Leaf Cell Patterns in Arabidopsis. Plant Cell 2002, 14, 2707–2722. [Google Scholar] [CrossRef]
- Nakamura, A.; Fujioka, S.; Sunohara, H.; Kamiya, N.; Hong, Z.; Inukai, Y.; Miura, K.; Takatsuto, S.; Yoshida, S.; Ueguchi-Tanaka, M.; et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006, 140, 580–590. [Google Scholar] [CrossRef]
- Fàbregas, N.; Lozano-Elena, F.; Blasco-Escámez, D.; Tohge, T.; Martínez-Andújar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 2018, 9, 4680. [Google Scholar] [CrossRef]
- Fàbregas, N.; Li, N.; Boeren, S.; Nash, T.E.; Goshe, M.B.; Clouse, S.D.; de Vries, S.; Caño-Delgado, A.I. The brassinosteroid insensitive1-like3 signalosome complex regulates Arabidopsis root development. Plant Cell 2013, 25, 3377–3388. [Google Scholar] [CrossRef] [Green Version]
- Salazar-Henao, J.E.; Lehner, R.; Betegon-Putze, I.; Vilarrasa-Blasi, J.; Cano-Delgado, A.I. BES1 regulates the localization of the brassinosteroid receptor BRL3 within the provascular tissue of the Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4951–4961. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Wang, H.; Walker, J.C.; Li, J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004, 40, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Dasmeh, P.; Wagner, A. Natural Selection on the Phase-Separation Properties of FUS during 160 My of Mammalian Evolution. Mol. Biol. Evol. 2021, 38, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Gligorijević, V.; Renfrew, P.D.; Kosciolek, T.; Leman, J.K.; Berenberg, D.; Vatanen, T.; Chandler, C.; Taylor, B.C.; Fisk, I.M.; Vlamakis, H.; et al. Structure-based protein function prediction using graph convolutional networks. Nat. Commun. 2021, 12, 3168. [Google Scholar] [CrossRef]
- Konc, J.; Janezic, D. Binding site comparison for function prediction and pharmaceutical discovery. Curr. Opin. Struct. Biol. 2014, 25, 34–39. [Google Scholar] [CrossRef]
- Ali, K.; Li, W.; Qin, Y.; Wang, S.; Feng, L.; Wei, Q.; Bai, Q.; Zheng, B.; Li, G.; Ren, H.; et al. Kinase Function of Brassinosteroid Receptor Specified by Two Allosterically Regulated Subdomains. Front. Plant. Sci. 2021, 12, 802924. [Google Scholar] [CrossRef]
- Liu, L.; Wu, Y.; Liao, Z.; Xiong, J.; Wu, F.; Xu, J.; Lan, H.; Tang, Q.; Zhou, S.; Liu, Y.; et al. Evolutionary conservation and functional divergence of the LFK gene family play important roles in the photoperiodic flowering pathway of land plants. Heredity 2018, 120, 310–328. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.P.; Rosenberg, H.F. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 2002, 30, 411–415. [Google Scholar] [CrossRef]
- Barrett, R.D.H.; Hoekstra, H.E. Molecular spandrels: Tests of adaptation at the genetic level. Nat. Rev. Genet. 2011, 12, 767–780. [Google Scholar] [CrossRef]
- Cheng, X.; Xin, M.; Xu, R.; Chen, Z.; Cai, W.; Chai, L.; Xu, H.; Jia, L.; Feng, Z.; Wang, Z.; et al. A Single Amino Acid Substitution in STKc_GSK3 Kinase Conferring Semispherical Grains and Its Implications for the Origin of Triticum sphaerococcum. Plant Cell 2020, 32, 923–934. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.; Bai, Q.; Li, C.; Wang, L.; Wei, Q.; Ali, K.; Li, W.; Huang, S.; Xu, H.; Li, G.; et al. Pan-brassinosteroid signaling revealed by functional analysis of NILR1 in land plants. New Phytol. 2022, 235, 1455–1469. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, J.; Shen, Y.; Pu, R.; Hou, M.; Wei, Q.; Zhang, X.; Li, G.; Ren, H.; Wu, G. Brassinosteroids synthesised by CYP85A/A1 but not CYP85A2 function via a BRI1-like receptor but not via BRI1 in Picea abies. J. Exp. Bot. 2021, 72, 1748–1763. [Google Scholar] [CrossRef] [PubMed]
- Todd, A.E.; Orengo, C.A.; Thornton, J.M. Plasticity of enzyme active sites. Trends Biochem. Sci. 2002, 27, 419–426. [Google Scholar] [CrossRef]
- Schrag, J.D.; Winkler, F.K.; Cygler, M. Pancreatic lipases: Evolutionary intermediates in a positional change of catalytic carboxylates? J. Biol. Chem. 1992, 267, 4300–4303. [Google Scholar] [CrossRef]
- Ferreira-Guerra, M.; Marquès-Bueno, M.; Mora-García, S.; Caño-Delgado, A.I. Delving into the evolutionary origin of steroid sensing in plants. Curr. Opin. Plant Biol. 2020, 57, 87–95. [Google Scholar] [CrossRef]
- Zheng, B.; Xing, K.; Zhang, J.; Liu, H.; Ali, K.; Li, W.; Bai, Q.; Ren, H. Evolutionary Analysis and Functional Identification of Ancient Brassinosteroid Receptors in Ceratopteris richardii. Int. J. Mol. Sci. 2022, 23, 6795. [Google Scholar] [CrossRef]
- Wang, H.; Mao, H. On the origin and evolution of plant brassinosteroid receptor kinases. J. Mol. Evol. 2014, 78, 118–129. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Zhang, J.; Tian, X.; Liu, H.; Ali, K.; Bai, Q.; Zheng, B.; Wu, G.; Ren, H. Two Conserved Amino Acids Characterized in the Island Domain Are Essential for the Biological Functions of Brassinolide Receptors. Int. J. Mol. Sci. 2022, 23, 11454. https://doi.org/10.3390/ijms231911454
Li W, Zhang J, Tian X, Liu H, Ali K, Bai Q, Zheng B, Wu G, Ren H. Two Conserved Amino Acids Characterized in the Island Domain Are Essential for the Biological Functions of Brassinolide Receptors. International Journal of Molecular Sciences. 2022; 23(19):11454. https://doi.org/10.3390/ijms231911454
Chicago/Turabian StyleLi, Wenjuan, Jiaojiao Zhang, Xiaoyi Tian, Hui Liu, Khawar Ali, Qunwei Bai, Bowen Zheng, Guang Wu, and Hongyan Ren. 2022. "Two Conserved Amino Acids Characterized in the Island Domain Are Essential for the Biological Functions of Brassinolide Receptors" International Journal of Molecular Sciences 23, no. 19: 11454. https://doi.org/10.3390/ijms231911454




