Single-Cell Chromatin Accessibility Data Combined with GWAS Improves Detection of Relevant Cell Types in 59 Complex Phenotypes
Abstract
:1. Introduction
2. Results
2.1. Integration of Bulk Chromatin Accessibility Data and GWAS Data
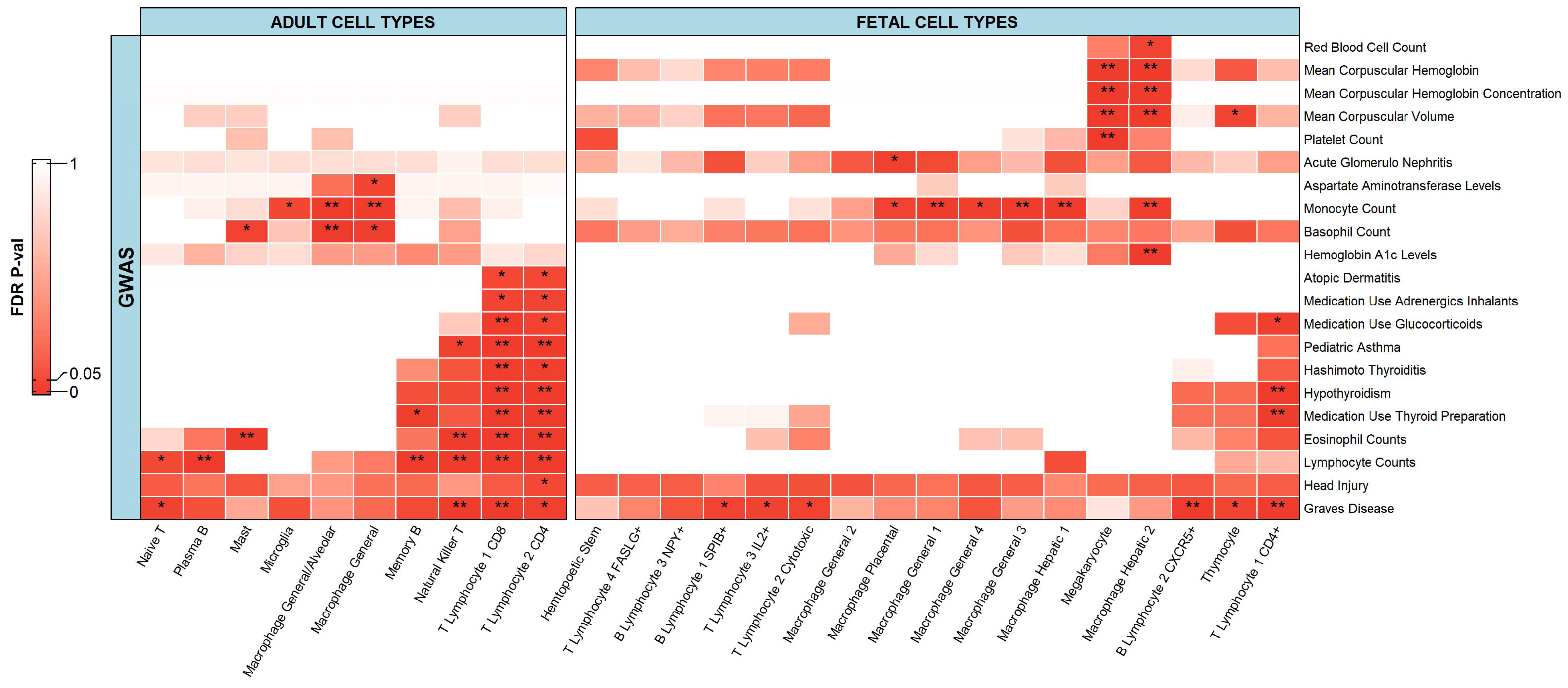
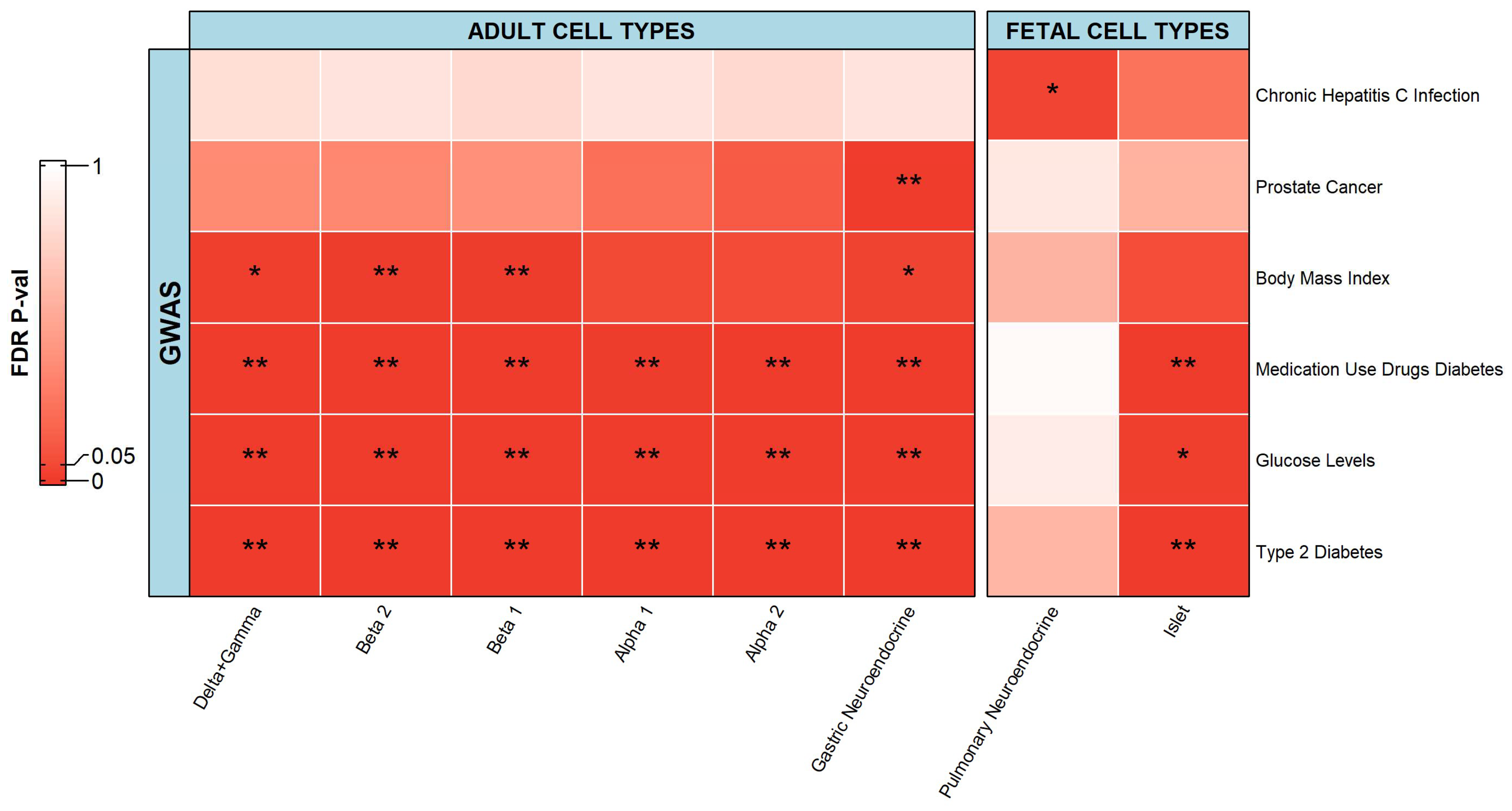
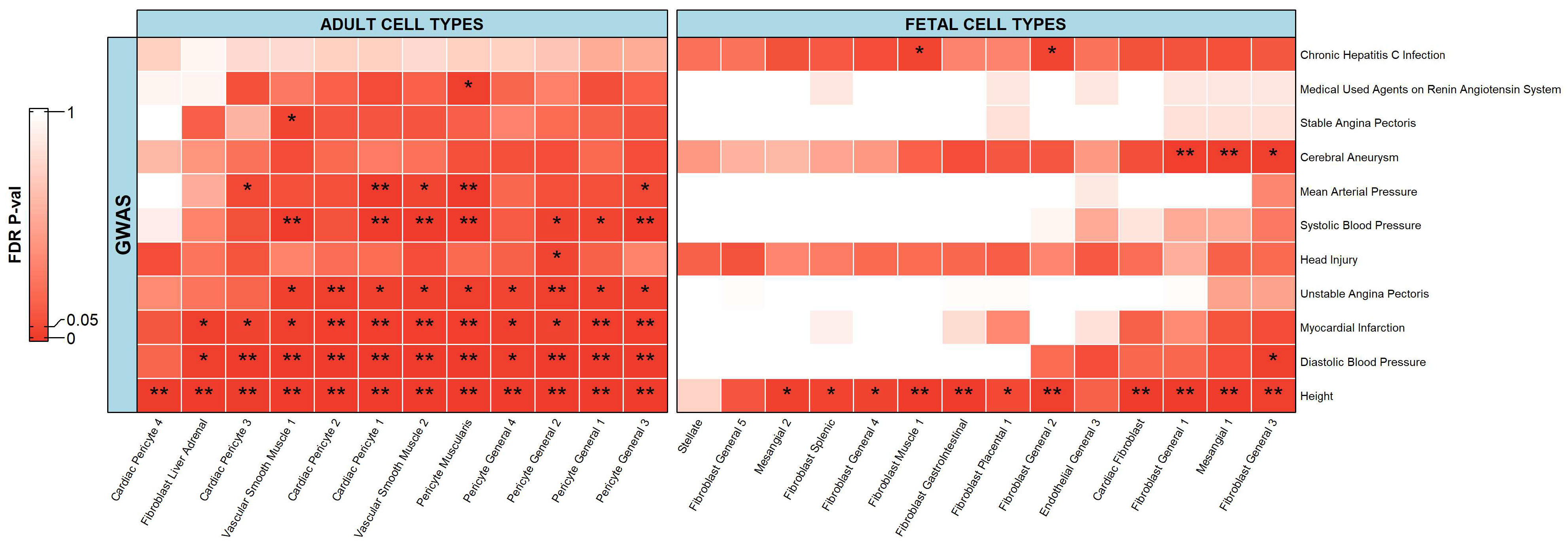
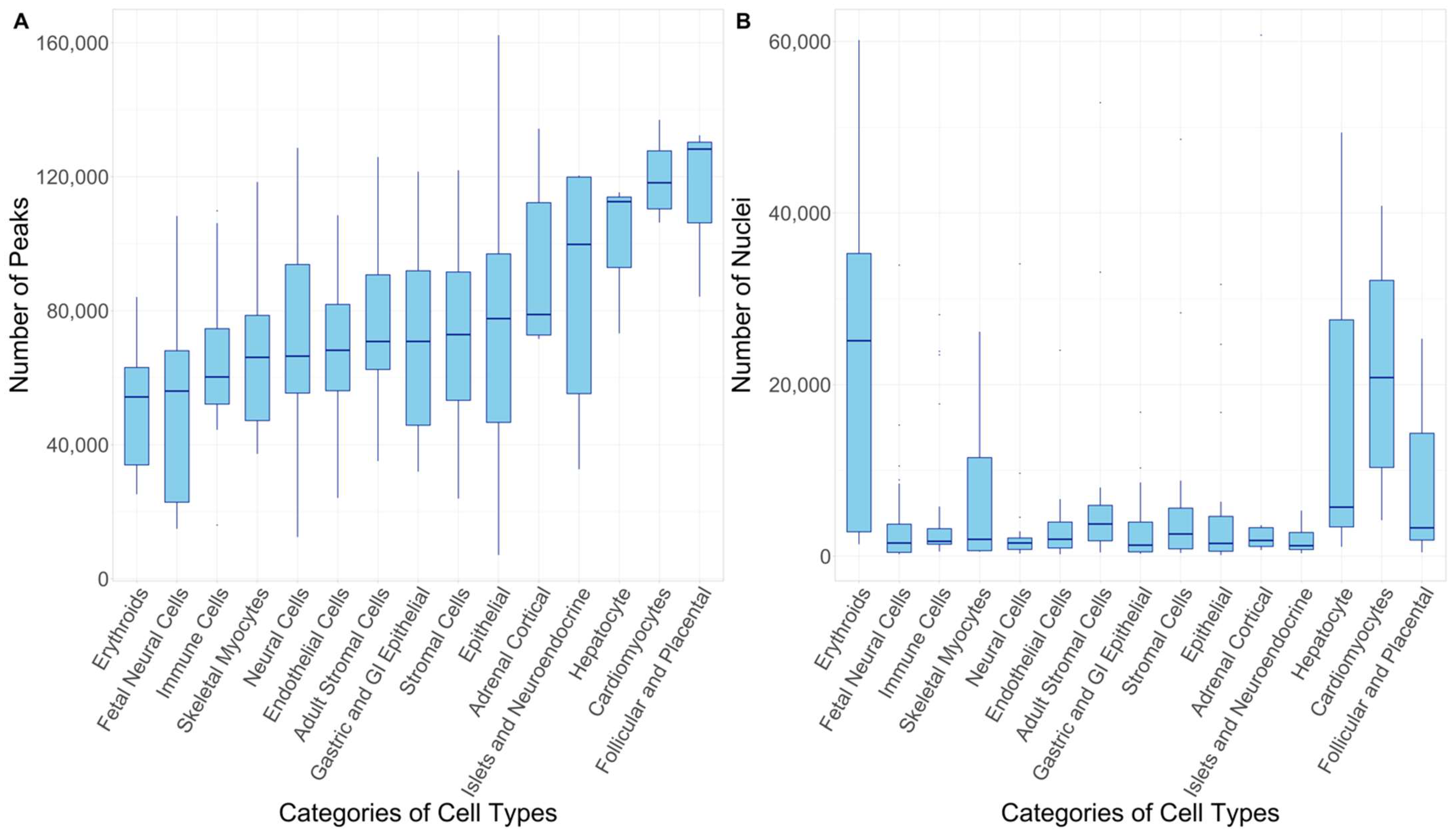
2.2. Integration of Single-Cell Chromatin Accessibility Data and GWAS Data
2.2.1. Immune Cells
2.2.2. Islets and Neuroendocrine Cells
2.2.3. Stromal Cells
2.2.4. Endothelial Cells
3. Discussion
4. Materials and Methods
4.1. GWAS Data
4.2. Chromatin Accessibility Data
4.3. Prioritizing Cell Types Using Linkage Disequilibrium Score Regression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [PubMed]
- Farh, K.K.-H.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.H.; Shishkin, A.A.; et al. Genetic and Epigenetic Fine Mapping of Causal Autoimmune Disease Variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Gusev, A.; Lee, S.H.; Trynka, G.; Finucane, H.; Vilhjálmsson, B.J.; Xu, H.; Zang, C.; Ripke, S.; Bulik-Sullivan, B.; Stahl, E.; et al. Partitioning Heritability of Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. Am. J. Hum. Genet. 2014, 95, 535–552. [Google Scholar] [CrossRef] [PubMed]
- Shooshtari, P.; Huang, H.; Cotsapas, C. Integrative Genetic and Epigenetic Analysis Uncovers Regulatory Mechanisms of Autoimmune Disease. Am. J. Hum. Genet. 2017, 101, 75–86. [Google Scholar] [CrossRef] [PubMed]
- International Multiple Sclerosis Genetics Consortium. Multiple Sclerosis Genomic Map Implicates Peripheral Immune Cells and Microglia in Susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef] [PubMed]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic Studies of Body Mass Index Yield New Insights for Obesity Biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Timshel, P.N.; Thompson, J.J.; Pers, T.H. Genetic Mapping of Etiologic Brain Cell Types for Obesity. Elife 2020, 9, e55851. [Google Scholar] [CrossRef] [PubMed]
- Trynka, G.; Westra, H.-J.; Slowikowski, K.; Hu, X.; Xu, H.; Stranger, B.E.; Klein, R.J.; Han, B.; Raychaudhuri, S. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-Coding Variants within Complex-Trait Loci. Am. J. Hum. Genet. 2015, 97, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Huang, Y.; Lei, J.; Luo, H.; Zhu, X. The Single-Cell Sequencing: New Developments and Medical Applications. Cell Biosci. 2019, 9, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-Cell RNA-Seq Highlights Intratumoral Heterogeneity in Primary Glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Wagner, J.; Rapsomaniki, M.A.; Chevrier, S.; Anzeneder, T.; Langwieder, C.; Dykgers, A.; Rees, M.; Ramaswamy, A.; Muenst, S.; Soysal, S.D.; et al. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 2019, 177, 1330–1345.e18. [Google Scholar] [CrossRef]
- Sun, D.; Guan, X.; Moran, A.E.; Wu, L.-Y.; Qian, D.Z.; Schedin, P.; Dai, M.-S.; Danilov, A.V.; Alumkal, J.J.; Adey, A.C.; et al. Identifying Phenotype-Associated Subpopulations by Integrating Bulk and Single-Cell Sequencing Data. Nat. Biotechnol. 2022, 40, 527–538. [Google Scholar] [CrossRef]
- Watanabe, K.; Umićević Mirkov, M.; de Leeuw, C.A.; van den Heuvel, M.P.; Posthuma, D. Genetic Mapping of Cell Type Specificity for Complex Traits. Nat. Commun. 2019, 10, 3222. [Google Scholar] [CrossRef]
- Calderon, D.; Bhaskar, A.; Knowles, D.A.; Golan, D.; Raj, T.; Fu, A.Q.; Pritchard, J.K. Inferring Relevant Cell Types for Complex Traits by Using Single-Cell Gene Expression. Am. J. Hum. Genet. 2017, 101, 686–699. [Google Scholar] [CrossRef]
- Bryois, J.; Skene, N.G.; Hansen, T.F.; Kogelman, L.J.A.; Watson, H.J.; Brueggeman, L.; Breen, G.; Bulik, C.M.; Arenas, E.; Hjerling-Leffler, J.; et al. Genetic Identification of Cell Types Underlying Brain Complex Traits Yields Novel Insights Into the Etiology of Parkinson’s Disease. Nat. Genet. 2020, 52, 482–493. [Google Scholar] [CrossRef]
- Rotem, A.; Ram, O.; Shoresh, N.; Sperling, R.A.; Goren, A.; Weitz, D.A.; Bernstein, B.E. Single-Cell ChIP-Seq Reveals Cell Subpopulations Defined by Chromatin State. Nat. Biotechnol. 2015, 33, 1165–1172. [Google Scholar] [CrossRef]
- Corces, M.R.; Buenrostro, J.D.; Wu, B.; Greenside, P.G.; Chan, S.M.; Koenig, J.L.; Snyder, M.P.; Pritchard, J.K.; Kundaje, A.; Greenleaf, W.J.; et al. Lineage-Specific and Single-Cell Chromatin Accessibility Charts Human Hematopoiesis and Leukemia Evolution. Nat. Genet. 2016, 48, 1193–1203. [Google Scholar] [CrossRef]
- Litzenburger, U.M.; Buenrostro, J.D.; Wu, B.; Shen, Y.; Sheffield, N.C.; Kathiria, A.; Greenleaf, W.J.; Chang, H.Y. Single-Cell Epigenomic Variability Reveals Functional Cancer Heterogeneity. Genome Biol. 2017, 18, 15. [Google Scholar] [CrossRef]
- Clark, S.J.; Lee, H.J.; Smallwood, S.A.; Kelsey, G.; Reik, W. Single-Cell Epigenomics: Powerful New Methods for Understanding Gene Regulation and Cell Identity. Genome Biol. 2016, 17, 72. [Google Scholar] [CrossRef] [Green Version]
- Schwartzman, O.; Tanay, A. Single-Cell Epigenomics: Techniques and Emerging Applications. Nat. Rev. Genet. 2015, 16, 716–726. [Google Scholar] [CrossRef]
- Macaulay, I.C.; Ponting, C.P.; Voet, T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends Genet. 2017, 33, 155–168. [Google Scholar] [CrossRef]
- Finucane, H.K.; Bulik-Sullivan, B.; Gusev, A.; Trynka, G.; Reshef, Y.; Loh, P.-R.; Anttila, V.; Xu, H.; Zang, C.; Farh, K.; et al. Partitioning Heritability by Functional Annotation Using Genome-Wide Association Summary Statistics. Nat. Genet. 2015, 47, 1228–1235. [Google Scholar] [CrossRef]
- Weidinger, S.; Beck, L.A.; Bieber, T.; Kabashima, K.; Irvine, A.D. Atopic Dermatitis. Nat. Rev. Dis. Primers 2018, 4, 1. [Google Scholar] [CrossRef]
- Agrawal, R.; Wisniewski, J.A.; Woodfolk, J.A. The Role of Regulatory T Cells in Atopic Dermatitis. Curr. Probl. Dermatol. 2011, 41, 112–124. [Google Scholar]
- Berroth, A.; Kühnl, J.; Kurschat, N.; Schwarz, A.; Stäb, F.; Schwarz, T.; Wenck, H.; Fölster-Holst, R.; Neufang, G. Role of Fibroblasts in the Pathogenesis of Atopic Dermatitis. J. Allergy Clin. Immunol. 2013, 131, 1547–1554. [Google Scholar] [CrossRef]
- Martin, A.; Davies, T.F. T Cells and Human Autoimmune Thyroid Disease: Emerging Data Show Lack of Need to Invoke Suppressor T Cell Problems. Thyroid 1992, 2, 247–261. [Google Scholar] [CrossRef]
- Rydzewska, M.; Jaromin, M.; Pasierowska, I.E.; Stożek, K.; Bossowski, A. Role of the T and B Lymphocytes in Pathogenesis of Autoimmune Thyroid Diseases. Thyroid Res. 2018, 11, 2. [Google Scholar] [CrossRef]
- McLachlan, S.M.; Rapoport, B. Autoimmune Hypothyroidism: T Cells Caught in the Act. Nat. Med. 2004, 10, 895–896. [Google Scholar] [CrossRef]
- Glick, A.B.; Wodzinski, A.; Fu, P.; Levine, A.D.; Wald, D.N. Impairment of Regulatory T-Cell Function in Autoimmune Thyroid Disease. Thyroid 2013, 23, 871–878. [Google Scholar] [CrossRef]
- Gallo, D.; Piantanida, E.; Gallazzi, M.; Bartalena, L.; Tanda, M.L.; Bruno, A.; Mortara, L. Immunological Drivers in Graves’ Disease: NK Cells as a Master Switcher. Front. Endocrinol. 2020, 11, 406. [Google Scholar] [CrossRef] [PubMed]
- Steegers, C.; Blok, E.; Lamballais, S.; Jaddoe, V.; Bernardoni, F.; Vernooij, M.; van der Ende, J.; Hillegers, M.; Micali, N.; Ehrlich, S.; et al. The Association between Body Mass Index and Brain Morphology in Children: A Population-Based Study. Brain Struct. Funct. 2021, 226, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Gunstad, J.; Paul, R.H.; Cohen, R.A.; Tate, D.F.; Spitznagel, M.B.; Grieve, S.; Gordon, E. Relationship between Body Mass Index and Brain Volume in Healthy Adults. Int. J. Neurosci. 2008, 118, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Panon, N.; Luangsawang, K.; Rugaber, C.; Tongchit, T.; Thongsepee, N.; Cheaha, D.; Kongjaidee, P.; Changtong, A.; Daradas, A.; Chotimol, P. Correlation between Body Mass Index and Ocular Parameters. Clin. Ophthalmol. 2019, 13, 763–769. [Google Scholar] [CrossRef]
- Guo, M.H.; Hirschhorn, J.N.; Dauber, A. Insights and Implications of Genome-Wide Association Studies of Height. J. Clin. Endocrinol. Metab. 2018, 103, 3155–3168. [Google Scholar] [CrossRef]
- Orakpoghenor, O.; Avazi, D.O.; Markus, T.; Olaolu, O. Lymphocytes: A Brief Review. Sci. J. Immunol. Immunoth. 2019, 3, 4–8. [Google Scholar]
- Duncan, L.; Heathcote, J.; Djurdjev, O.; Levin, A. Screening for Renal Disease Using Serum Creatinine: Who Are We Missing? Nephrol. Dial. Transpl. 2001, 16, 1042–1046. [Google Scholar] [CrossRef]
- Gagnon1, A.L.; Desai, T. Dermatological Diseases in Patients with Chronic Kidney Disease. J. Nephropathol. 2013, 2, 104–109. [Google Scholar] [CrossRef]
- Luecken, M.D.; Theis, F.J. Current Best Practices in Single-Cell RNA-Seq Analysis: A Tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef]
- Single Cell ATAC. Available online: https://www.10xgenomics.com/products/single-cell-atac (accessed on 26 August 2022).
- Lloyd, C.M.; Hessel, E.M. Functions of T Cells in Asthma: More than Just T(H)2 Cells. Nat. Rev. Immunol. 2010, 10, 838–848. [Google Scholar] [CrossRef]
- Okajima, M.; Wada, T.; Nishida, M.; Yokoyama, T.; Nakayama, Y.; Hashida, Y.; Shibata, F.; Tone, Y.; Ishizaki, A.; Shimizu, M.; et al. Analysis of T Cell Receptor Vbeta Diversity in Peripheral CD4 and CD8 T Lymphocytes in Patients with Autoimmune Thyroid Diseases. Clin. Exp. Immunol. 2009, 155, 166–172. [Google Scholar] [CrossRef]
- Seaman, W.E. Natural Killer Cells and Natural Killer T Cells. Arthritis Rheum. 2000, 43, 1204–1217. [Google Scholar] [CrossRef]
- Da Silva Xavier, G. The Cells of the Islets of Langerhans. J. Clin. Med. Res. 2018, 7, 54. [Google Scholar] [CrossRef]
- Portha, B.; Chavey, A.; Movassat, J. Early-Life Origins of Type 2 Diabetes: Fetal Programming of the Beta-Cell Mass. Exp. Diabetes Res. 2011, 2011, 105076. [Google Scholar] [CrossRef]
- Gromada, J.; Chabosseau, P.; Rutter, G.A. The α-Cell in Diabetes Mellitus. Nat. Rev. Endocrinol. 2018, 14, 694–704. [Google Scholar] [CrossRef]
- Martínez, M.S.; Manzano, A.; Olivar, L.C.; Nava, M.; Salazar, J.; D’Marco, L.; Ortiz, R.; Chacín, M.; Guerrero-Wyss, M.; Cabrera de Bravo, M.; et al. The Role of the α Cell in the Pathogenesis of Diabetes: A World beyond the Mirror. Int. J. Mol. Sci. 2021, 22, 9504. [Google Scholar] [CrossRef]
- Cerf, M.E. Beta Cell Dysfunction and Insulin Resistance. Front. Endocrinol. 2013, 4, 37. [Google Scholar] [CrossRef]
- Eizirik, D.L.; Pasquali, L.; Cnop, M. Pancreatic β-Cells in Type 1 and Type 2 Diabetes Mellitus: Different Pathways to Failure. Nat. Rev. Endocrinol. 2020, 16, 349–362. [Google Scholar] [CrossRef]
- Gao, R.; Yang, T.; Zhang, Q. δ-Cells: The Neighborhood Watch in the Islet Community. Biology 2021, 10, 74. [Google Scholar] [CrossRef]
- Lawlor, N.; Khetan, S.; Ucar, D.; Stitzel, M.L. Genomics of Islet (Dys)function and Type 2 Diabetes. Trends Genet. 2017, 33, 244–255. [Google Scholar] [CrossRef]
- Ha, J.; Baek, K.-H. Body Mass Index at the Crossroads of Osteoporosis and Type 2 Diabetes. Korean J. Intern. Med. 2020, 35, 1333–1335. [Google Scholar] [CrossRef]
- Dybala, M.P.; Olehnik, S.K.; Fowler, J.L.; Golab, K.; Millis, J.M.; Golebiewska, J.; Bachul, P.; Witkowski, P.; Hara, M. Pancreatic Beta Cell/islet Mass and Body Mass Index. Islets 2019, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Linnemann, A.K.; Baan, M.; Davis, D.B. Pancreatic β-Cell Proliferation in Obesity. Adv. Nutr. 2014, 5, 278–288. [Google Scholar] [CrossRef]
- Thapi, S.; Baeg, K.; Kim, M.K.; Gallagher, E.J. Survival of Patients with Gastroenteropancreatic Neuroendocrine Tumors and Diabetes Mellitus. Pancreas 2021, 50, 1293–1297. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, B.; Joo, J.; Kook, M.-C.; Kim, Y.-I.; Lee, J.Y.; Kim, C.G.; Choi, I.J.; Eom, B.W.; Yoon, H.M.; et al. Body Mass Index and Mortality in Patients with Gastric Cancer: A Large Cohort Study. Gastric Cancer 2018, 21, 913–924. [Google Scholar] [CrossRef]
- Bal, T.; Onlen, Y.; Babayigit, C.; Yumer, Y.; Sahin, S.I. The Impact of Hepatitis C Viremia Status on Lung Functions in Chronic Hepatitis C Patients. Afr. Health Sci. 2019, 19, 1988–1992. [Google Scholar] [CrossRef]
- Segna, D.; Dufour, J.-F. Other Extrahepatic Manifestations of Hepatitis C Virus Infection (Pulmonary, Idiopathic Thrombocytopenic Purpura, Nondiabetes Endocrine Disorders). Clin. Liver Dis. 2017, 21, 607–629. [Google Scholar] [CrossRef] [PubMed]
- Alarcon-Martinez, L.; Yemisci, M.; Dalkara, T. Pericyte Morphology and Function. Histol. Histopathol. 2021, 36, 633–643. [Google Scholar] [PubMed]
- Bergers, G.; Song, S. The Role of Pericytes in Blood-Vessel Formation and Maintenance. Neuro. Oncol. 2005, 7, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Špiranec, K.; Chen, W.; Werner, F.; Nikolaev, V.O.; Naruke, T.; Koch, F.; Werner, A.; Eder-Negrin, P.; Diéguez-Hurtado, R.; Adams, R.H.; et al. Endothelial C-Type Natriuretic Peptide Acts on Pericytes to Regulate Microcirculatory Flow and Blood Pressure. Circulation 2018, 138, 494–508. [Google Scholar] [CrossRef]
- Michael, S.K.; Surks, H.K.; Wang, Y.; Zhu, Y.; Blanton, R.; Jamnongjit, M.; Aronovitz, M.; Baur, W.; Ohtani, K.; Wilkerson, M.K.; et al. High Blood Pressure Arising from a Defect in Vascular Function. Proc. Natl. Acad. Sci. USA 2008, 105, 6702–6707. [Google Scholar] [CrossRef]
- Touyz, R.M.; Alves-Lopes, R.; Rios, F.J.; Camargo, L.L.; Anagnostopoulou, A.; Arner, A.; Montezano, A.C. Vascular Smooth Muscle Contraction in Hypertension. Cardiovasc. Res. 2018, 114, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Chalouhi, N.; Hoh, B.L.; Hasan, D. Review of Cerebral Aneurysm Formation, Growth, and Rupture. Stroke 2013, 44, 3613–3622. [Google Scholar] [CrossRef]
- Jiang, B.; Paff, M.; Colby, G.P.; Coon, A.L.; Lin, L.-M. Cerebral Aneurysm Treatment: Modern Neurovascular Techniques. Stroke Vasc. Neurol. 2016, 1, 93–100. [Google Scholar] [CrossRef]
- Levy, M.L.; Levy, D.M.; Manna, B. Pediatric Cerebral Aneurysm. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Avraham, S.; Korin, B.; Chung, J.-J.; Oxburgh, L.; Shaw, A.S. The Mesangial Cell—The Glomerular Stromal Cell. Nat. Rev. Nephrol. 2021, 17, 855–864. [Google Scholar] [CrossRef]
- Pinto, E. Blood Pressure and Ageing. Postgrad. Med. J. 2007, 83, 109–114. [Google Scholar] [CrossRef]
- Zhang, K.; Hocker, J.D.; Miller, M.; Hou, X.; Chiou, J.; Poirion, O.B.; Qiu, Y.; Li, Y.E.; Gaulton, K.J.; Wang, A.; et al. A Single-Cell Atlas of Chromatin Accessibility in the Human Genome. Cell 2021, 184, 5985–6001.e19. [Google Scholar] [CrossRef]
- Cai, W.; Liu, H.; Zhao, J.; Chen, L.Y.; Chen, J.; Lu, Z.; Hu, X. Pericytes in Brain Injury and Repair After Ischemic Stroke. Transl. Stroke Res. 2017, 8, 107–121. [Google Scholar] [CrossRef]
- Nakata, M.; Nakagomi, T.; Maeda, M.; Nakano-Doi, A.; Momota, Y.; Matsuyama, T. Induction of Perivascular Neural Stem Cells and Possible Contribution to Neurogenesis Following Transient Brain Ischemia/Reperfusion Injury. Transl. Stroke Res. 2017, 8, 131–143. [Google Scholar] [CrossRef]
- Bataller, R.; Brenner, D.A. Liver Fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- Sebastiani, G.; Gkouvatsos, K.; Pantopoulos, K. Chronic Hepatitis C and Liver Fibrosis. World J. Gastroenterol. 2014, 20, 11033–11053. [Google Scholar] [CrossRef]
- Sturtzel, C. Endothelial Cells. Adv. Exp. Med. Biol. 2017, 1003, 71–91. [Google Scholar]
- Kondo, Y.; Shimosegawa, T. Direct Effects of Hepatitis C Virus on the Lymphoid Cells. World J. Gastroenterol. 2013, 19, 7889–7895. [Google Scholar] [CrossRef]
- Ghosh, A.; Romani, S.; Kottilil, S.; Poonia, B. Lymphocyte Landscape after Chronic Hepatitis C Virus (HCV) Cure: The New Normal. Int. J. Mol. Sci. 2020, 21, 7473. [Google Scholar] [CrossRef]
- Alonso, J.C. Pleural Effusion in Liver Disease. Semin. Respir. Crit. Care Med. 2010, 31, 698–705. [Google Scholar] [CrossRef]
- Kass, S.M.; Williams, P.M.; Reamy, B.V. Pleurisy. Am. Fam. Physician 2007, 75, 1357–1364. [Google Scholar]
- Vassiliou, A.G.; Kotanidou, A.; Dimopoulou, I.; Orfanos, S.E. Endothelial Damage in Acute Respiratory Distress Syndrome. Int. J. Mol. Sci. 2020, 21, 8793. [Google Scholar] [CrossRef]
- Altmann, T.; Slack, J.; Slatter, M.A.; O’Brien, C.; Cant, A.; Thomas, M.; Brodlie, M.; Annavarapu, S.; Gennery, A.R. Endothelial Cell Damage in Idiopathic Pneumonia Syndrome. Bone Marrow Transplant. 2018, 53, 515–518. [Google Scholar] [CrossRef]
- Loos, R.J.F. 15 Years of Genome-Wide Association Studies and No Signs of Slowing Down. Nat. Commun. 2020, 11, 5900. [Google Scholar] [CrossRef]
- Carter, B.; Zhao, K. The Epigenetic Basis of Cellular Heterogeneity. Nat. Rev. Genet. 2021, 22, 235–250. [Google Scholar] [CrossRef]
- Tak, Y.G.; Farnham, P.J. Making Sense of GWAS: Using Epigenomics and Genome Engineering to Understand the Functional Relevance of SNPs in Non-Coding Regions of the Human Genome. Epigenet. Chromatin 2015, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Sahana, G.; Su, G.; Yu, Y.; Zhang, S.; Lund, M.S.; Sørensen, P. Integrating Sequence-Based GWAS and RNA-Seq Provides Novel Insights into the Genetic Basis of Mastitis and Milk Production in Dairy Cattle. Sci. Rep. 2017, 7, 45560. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Huang, H.; Freebern, E.; Santos, D.J.A.; Dai, D.; Si, J.; Ma, C.; Cao, J.; Guo, G.; Liu, G.E.; et al. Integrating RNA-Seq with GWAS Reveals Novel Insights into the Molecular Mechanism Underpinning Ketosis in Cattle. BMC Genom. 2020, 21, 489. [Google Scholar] [CrossRef] [PubMed]
- Gòdia, M.; Reverter, A.; González-Prendes, R.; Ramayo-Caldas, Y.; Castelló, A.; Rodríguez-Gil, J.-E.; Sánchez, A.; Clop, A. A Systems Biology Framework Integrating GWAS and RNA-Seq to Shed Light on the Molecular Basis of Sperm Quality in Swine. Genet. Sel. Evol. 2020, 52, 72. [Google Scholar] [CrossRef]
- Perzel Mandell, K.A.; Eagles, N.J.; Wilton, R.; Price, A.J.; Semick, S.A.; Collado-Torres, L.; Ulrich, W.S.; Tao, R.; Han, S.; Szalay, A.S.; et al. Genome-Wide Sequencing-Based Identification of Methylation Quantitative Trait Loci and Their Role in Schizophrenia Risk. Nat. Commun. 2021, 12, 5251. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, A.; Xu, T.; Mo, X.; Zhang, Y. Promoter DNA Methylation in GWAS-Identified Genes as Potential Functional Elements for Blood Pressure: An Observational and Mendelian Randomization Study. Front. Genet. 2021, 12, 791146. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of Native Chromatin for Fast and Sensitive Epigenomic Profiling of Open Chromatin, DNA-Binding Proteins and Nucleosome Position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Kawakatsu, T. Whole-Genome Bisulfite Sequencing and Epigenetic Variation in Cereal Methylomes. Methods Mol. Biol. 2020, 2072, 119–128. [Google Scholar]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A Cross-Population Atlas of Genetic Associations for 220 Human Phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef]
- Shooshtari, P.; Feng, S.; Nelakuditi, V.; Foong, J.; Brudno, M.; Cotsapas, C. OCHROdb: A Comprehensive, Quality Checked Database of Open Chromatin Regions from Sequencing Data. bioRxiv 2018. [Google Scholar] [CrossRef]
- Domcke, S.; Hill, A.J.; Daza, R.M.; Cao, J.; O’Day, D.R.; Pliner, H.A.; Aldinger, K.A.; Pokholok, D.; Zhang, F.; Milbank, J.H.; et al. A Human Cell Atlas of Fetal Chromatin Accessibility. Science 2020, 370, eaba7612. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Wu, Y.; Byrne, E.M.; Zheng, Z.; Kemper, K.E.; Yengo, L.; Mallett, A.J.; Yang, J.; Visscher, P.M.; Wray, N.R. Genome-Wide Association Study of Medication-Use and Associated Disease in the UK Biobank. Nat. Commun. 2019, 10, 1891. [Google Scholar] [CrossRef]
- Slatkin, M. Linkage Disequilibrium—Understanding the Evolutionary Past and Mapping the Medical Future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef] [Green Version]
- Bulik-Sullivan, B.K.; Loh, P.-R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score Regression Distinguishes Confounding from Polygenicity in Genome-Wide Association Studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- ComplexHeatmap. Available online: https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html (accessed on 26 August 2022).



| Category of Cell Types | Number of Adult Cell Types | Number of Fetal Cell Types |
|---|---|---|
| Immune Cells | 10 | 17 |
| Endothelial Cells | 9 | 8 |
| Erythroids | 0 | 5 |
| Cardiomyocytes | 2 | 2 |
| Stromal Cells | 12 | 14 |
| Adult Stromal Cells | 22 | 0 |
| Skeletal Myocyte | 2 | 5 |
| Follicular and Placental | 1 | 2 |
| Epithelial | 19 | 9 |
| Gastric and GI Epithelial | 12 | 7 |
| Islets and Neuroendocrine | 6 | 2 |
| Fetal Neural Cells | 0 | 25 |
| Neural Cells | 11 | 11 |
| Adrenal Cortical Cells | 4 | 2 |
| Hepatocytes | 2 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, A.C.; Foroutan, A.; Qian, B.; Hosseini Naghavi, N.; Shabani, K.; Shooshtari, P. Single-Cell Chromatin Accessibility Data Combined with GWAS Improves Detection of Relevant Cell Types in 59 Complex Phenotypes. Int. J. Mol. Sci. 2022, 23, 11456. https://doi.org/10.3390/ijms231911456
Das AC, Foroutan A, Qian B, Hosseini Naghavi N, Shabani K, Shooshtari P. Single-Cell Chromatin Accessibility Data Combined with GWAS Improves Detection of Relevant Cell Types in 59 Complex Phenotypes. International Journal of Molecular Sciences. 2022; 23(19):11456. https://doi.org/10.3390/ijms231911456
Chicago/Turabian StyleDas, Akash Chandra, Aidin Foroutan, Brian Qian, Nader Hosseini Naghavi, Kayvan Shabani, and Parisa Shooshtari. 2022. "Single-Cell Chromatin Accessibility Data Combined with GWAS Improves Detection of Relevant Cell Types in 59 Complex Phenotypes" International Journal of Molecular Sciences 23, no. 19: 11456. https://doi.org/10.3390/ijms231911456







