Characterization of New Defensin Antimicrobial Peptides and Their Expression in Bed Bugs in Response to Bacterial Ingestion and Injection
Abstract
:1. Introduction
2. Results
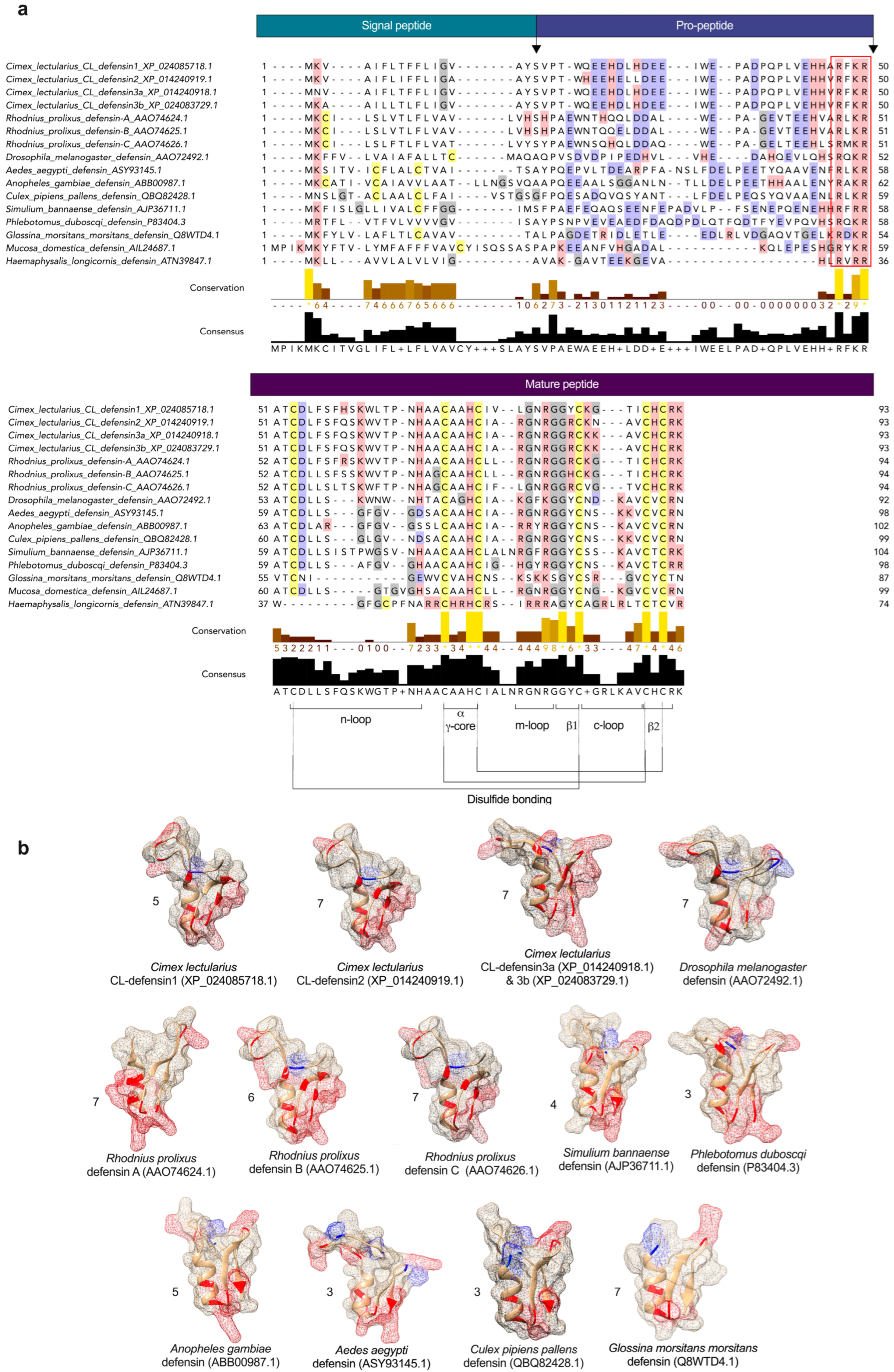
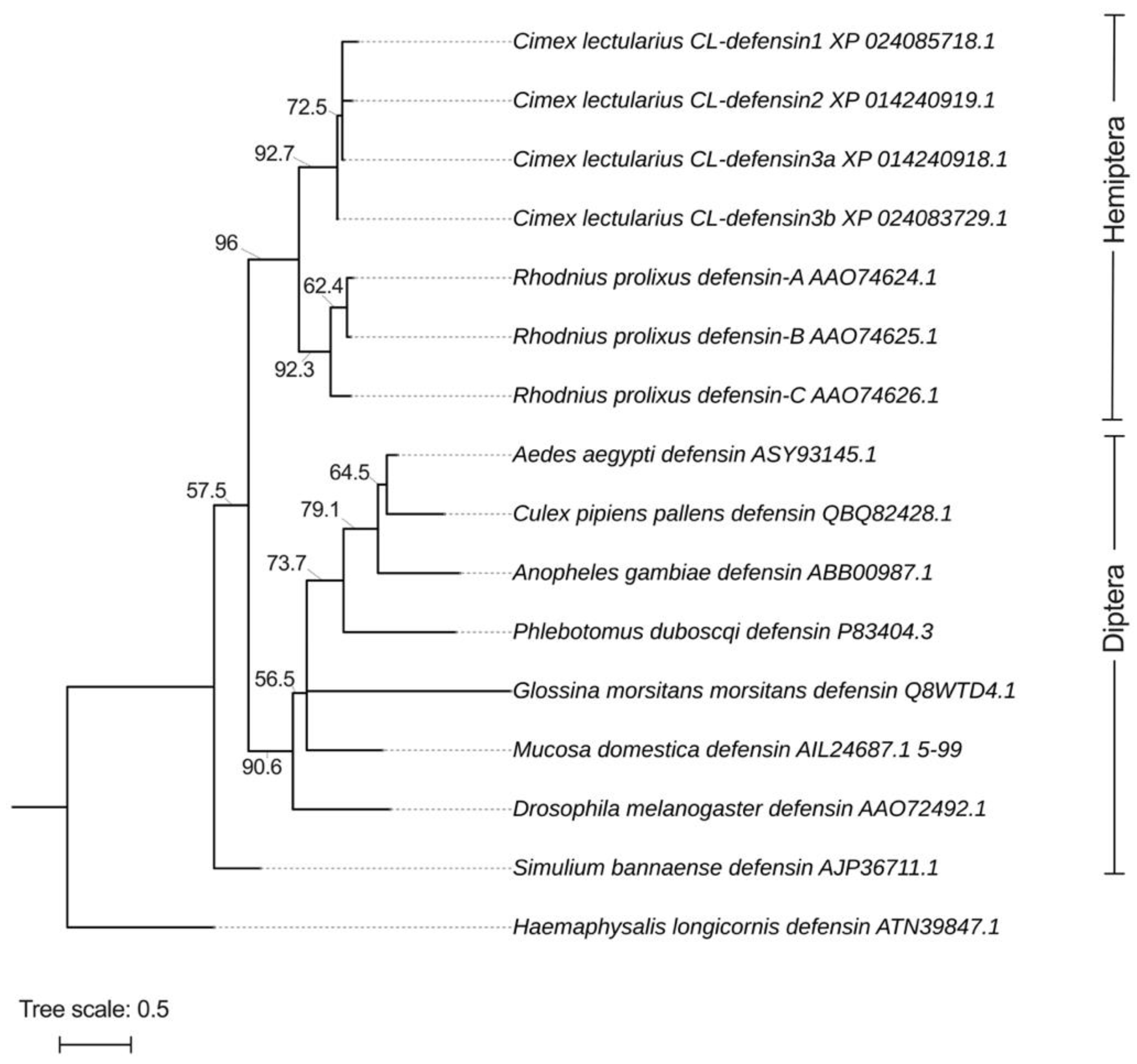
2.1. Structural and Phylogenetic Analyses of CL-Defensins (Figure 1 and Figure 2)
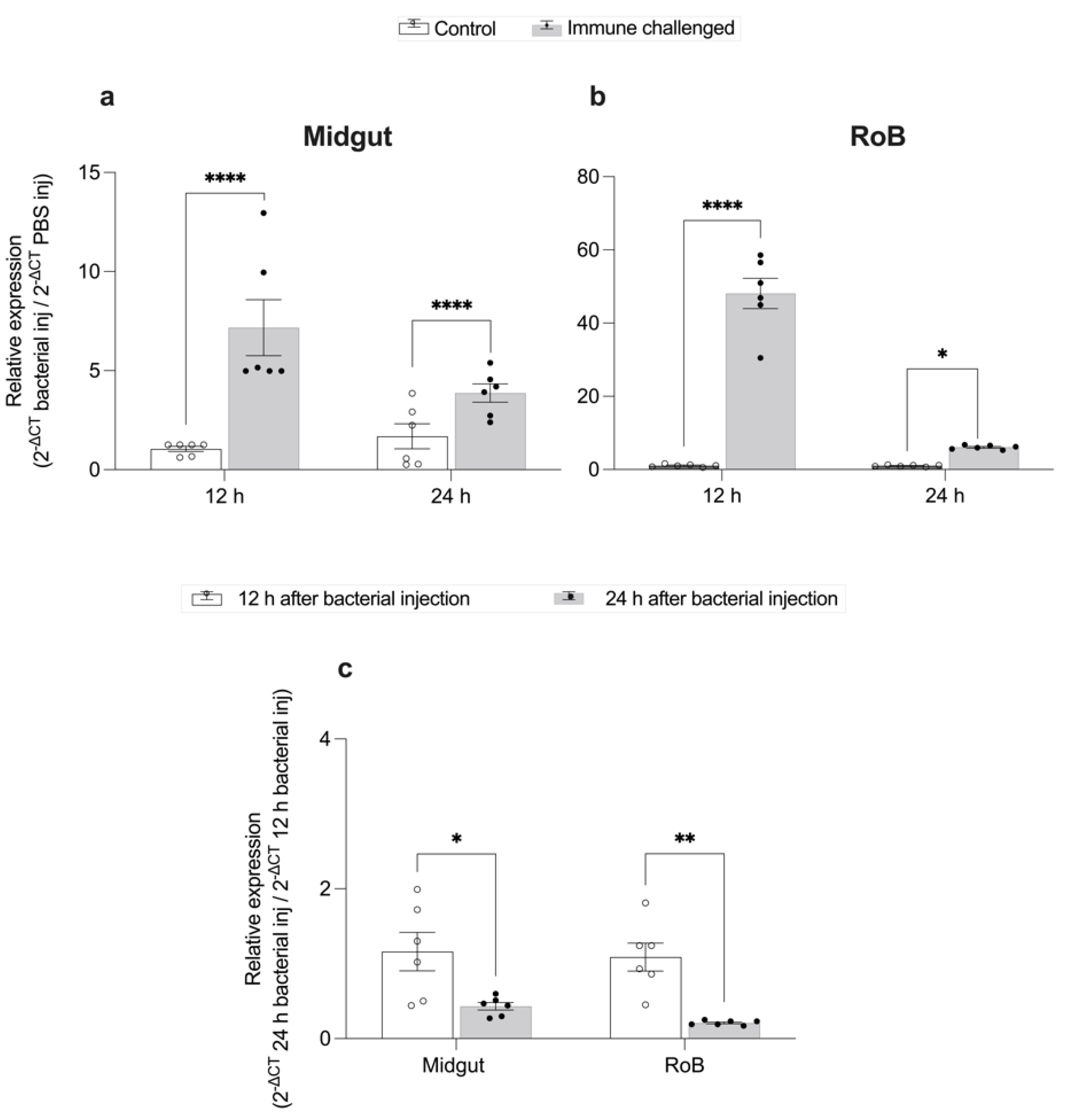
2.2. Time-Dependent Defensin Expression Prompted by Bacterial Injection (Figure 3)
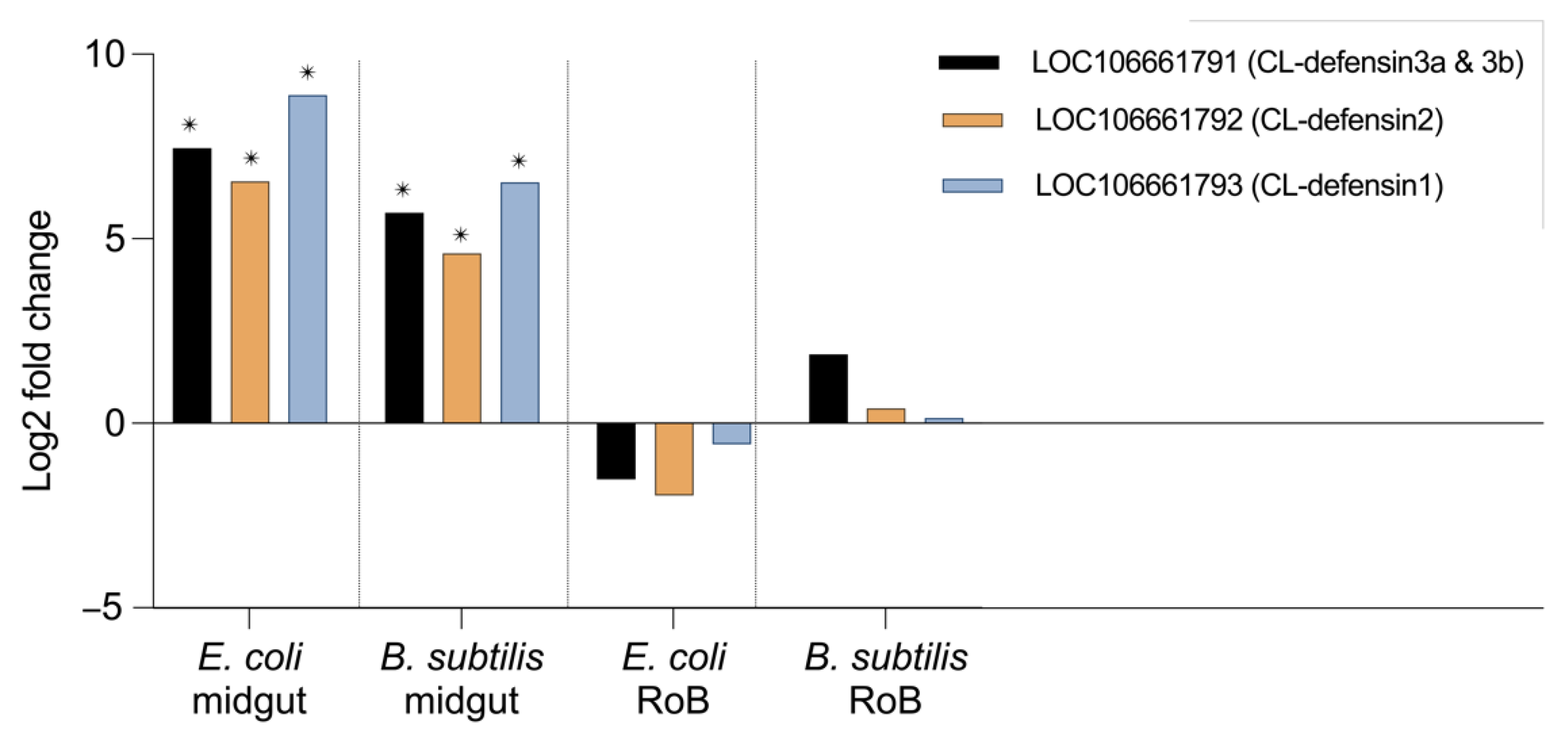
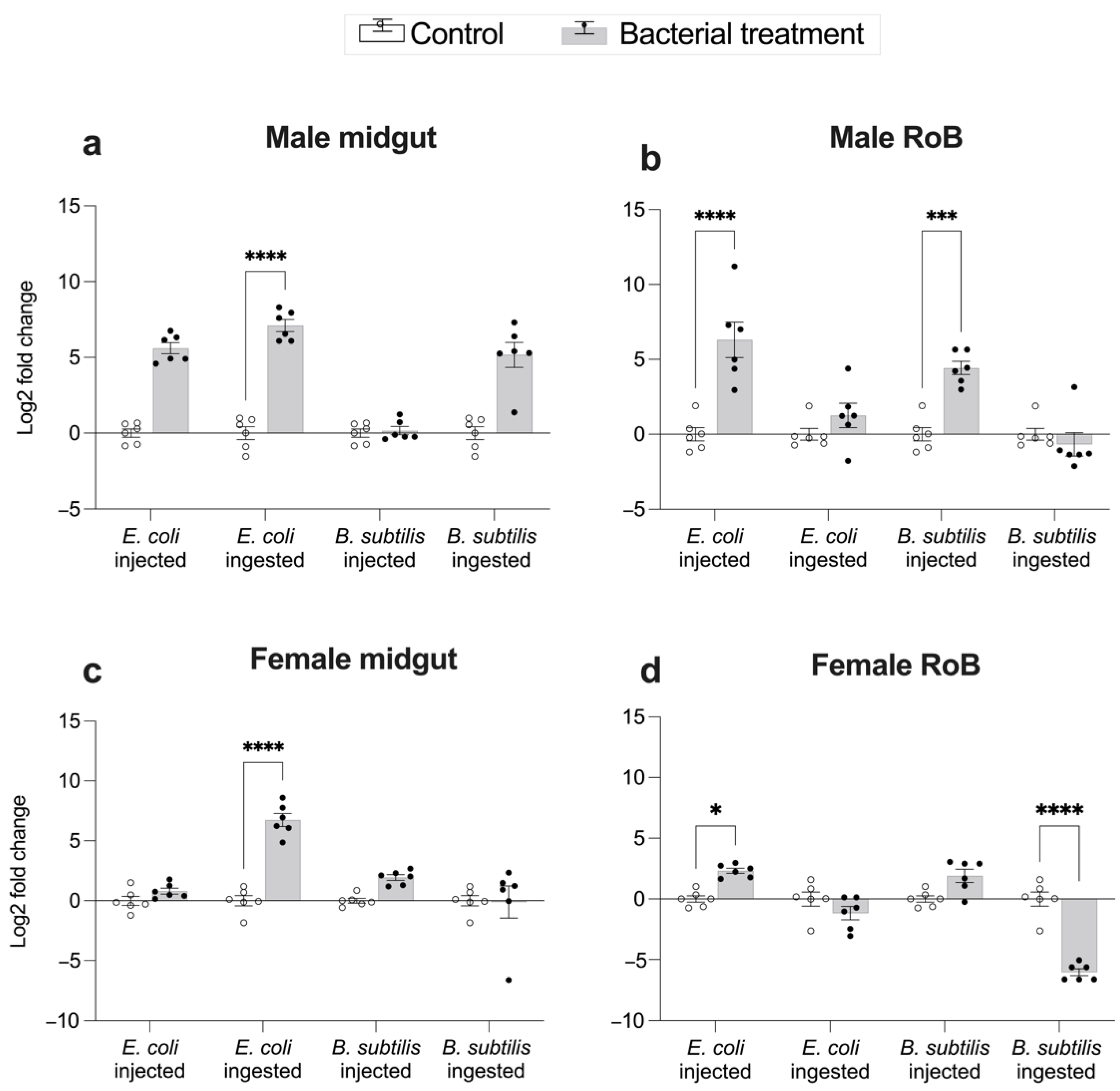
2.3. Midgut- and RoB-Distinct Gene Expression of CL-Defensins by Male and Female Bed Bugs Fed Bacteria-Infected Blood or Injected with Bacteria (Figure 4 and Figure 5)
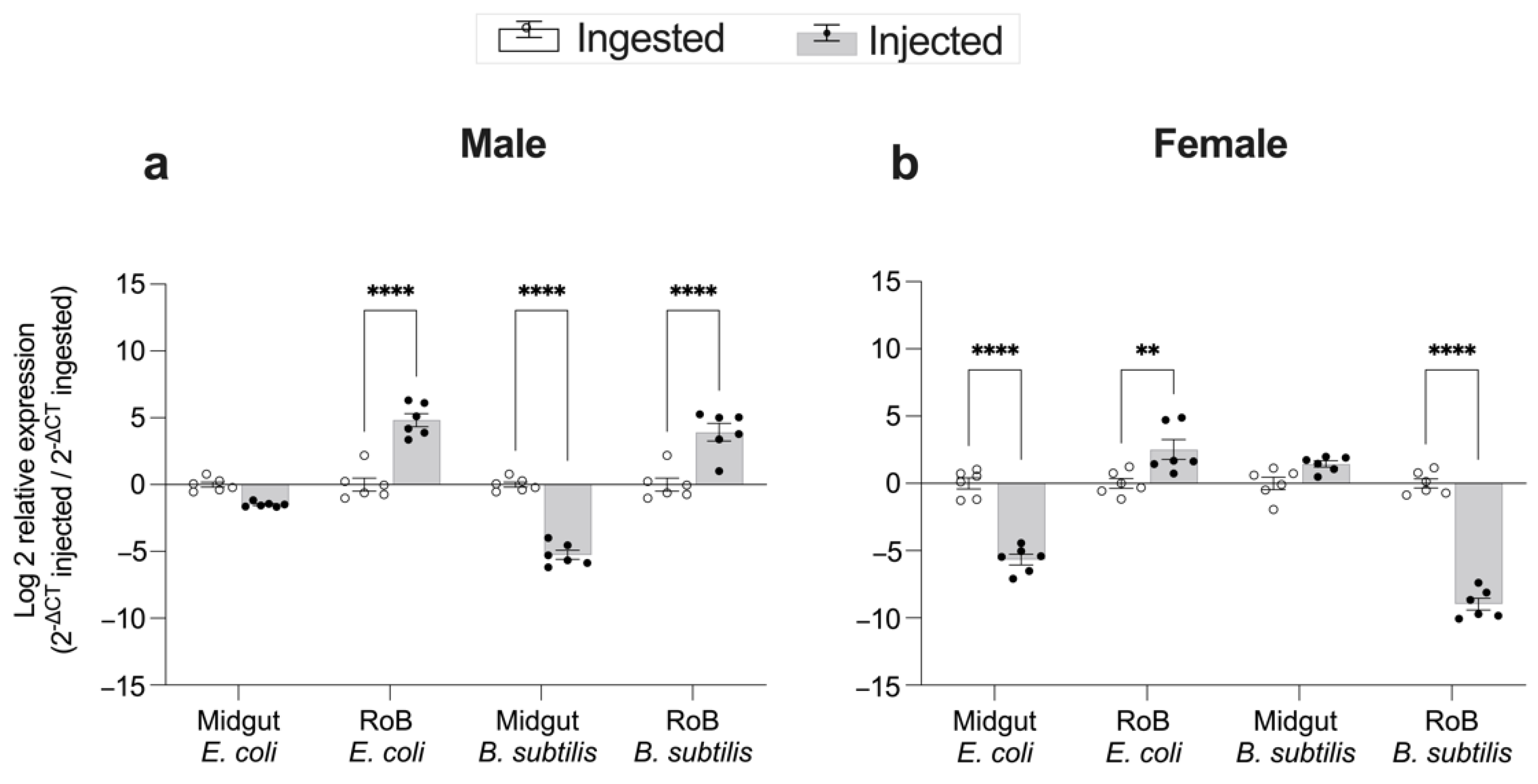
2.4. Effect of Bed Bug Sex and Mode of Immune Challenge on the Upregulation of CL-Defensins (Figure 6 and Figure 7)
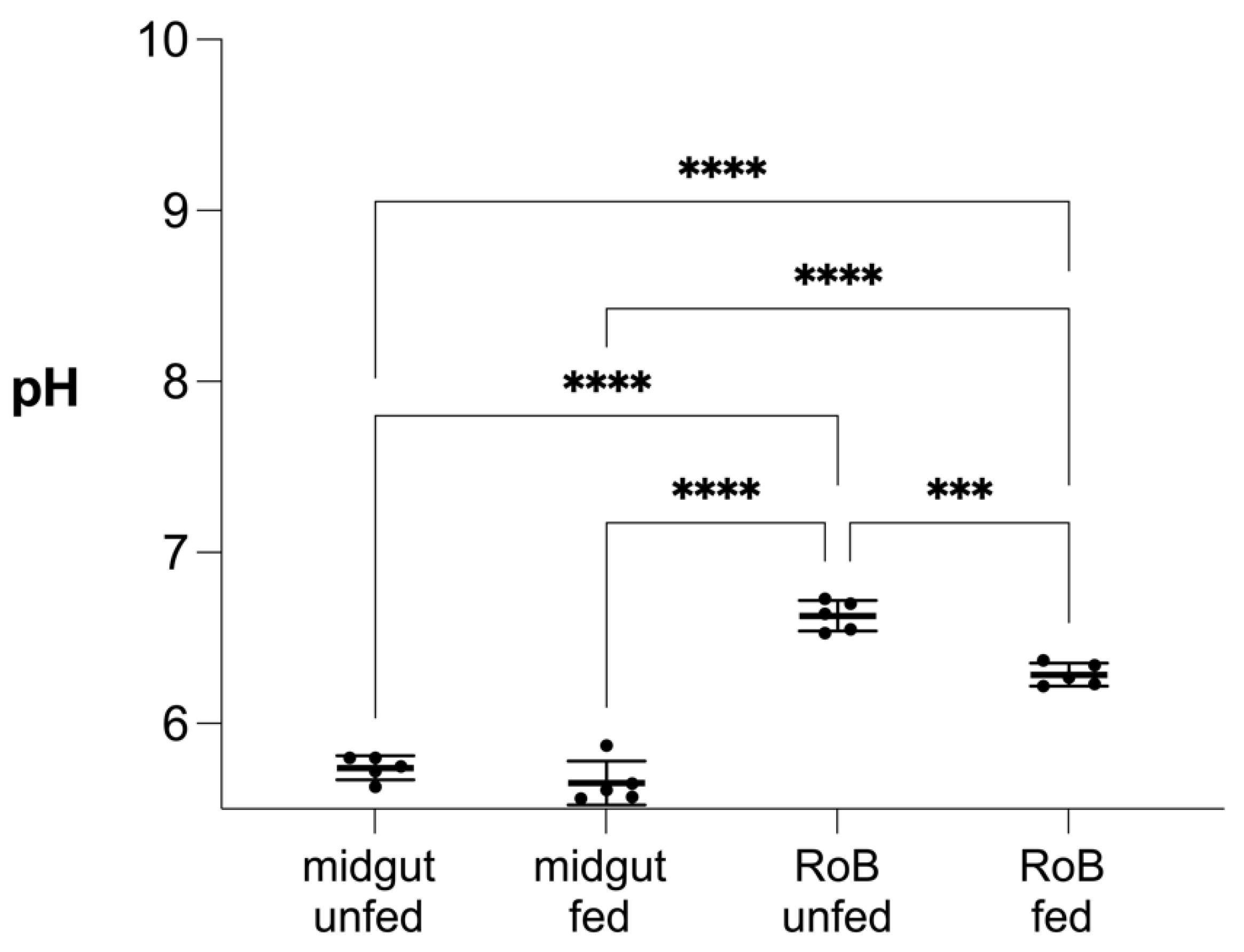
2.5. Effect of Blood Ingestion on the pH of Bed Bug Midgut and RoB (Figure 8)
3. Discussion
3.1. Induction of Immune Pathway Effector AMPs in Response to Gr– and Gr+ Bacteria in the Midgut and RoB
3.2. Effects of Bed bug Sex and Mode of Immune Challenge on Upregulation of AMPs
4. Materials and Methods
4.1. Laboratory Rearing of Bed Bugs
4.2. Growth and Preparation of Bacteria and Immune Challenges of Bed Bugs
4.3. Tissue Isolation and RNA Extraction
4.4. cDNA Synthesis and Quantitative Real-Time PCR (qPCR)
4.5. qPCR Analysis
4.6. Transcriptome Assembly: Library Preparation with PolyA Selection and HiSeq Sequencing and RNA-Seq Data Analysis
4.7. AMP Characteristic Identification
4.8. Phylogenetic Analysis
4.9. pH Measurements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doggett, S.L.; Dwyer, D.E.; Penas, P.F.; Russell, R.C. Bed bugs: Clinical relevance and control options. Clin. Microbiol. Rev. 2012, 25, 164–192. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Izri, A. Bedbugs. N. Engl. J. Med. 2020, 382, 2230–2237. [Google Scholar] [CrossRef] [PubMed]
- Balster, C.B. The Non-Infectious Disease Implications of Bed Bug Infestations. Master’s Thesis, Wright State University, Dayton, OH, USA, 2011. [Google Scholar]
- Burton, G.J. Bedbugs in relation to transmission of human diseases. Review of the literature. Public Health Rep. 1963, 78, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, P.; Blanc, V.; Del Giudice, P.; Levy-Bencheton, A.; Chosidow, O.; Marty, P.; Brouqui, P. Bedbugs and infectious diseases. Clin. Infect. Dis. 2011, 52, 200–210. [Google Scholar] [CrossRef]
- Lai, O.; Ho, D.; Glick, S.; Jagdeo, J. Bed bugs and possible transmission of human pathogens: A systematic review. Arch. Dermatol. Res. 2016, 308, 531–538. [Google Scholar] [CrossRef]
- El Hamzaoui, B.; Laroche, M.; Bechah, Y.; Berenger, J.M.; Parola, P. Testing the competence of Cimex lectularius bed bugs for the transmission of Borrelia recurrentis, the agent of relapsing fever. Am. J. Trop. Med. Hyg. 2019, 100, 1407–1412. [Google Scholar] [CrossRef]
- Peterson, J.K.; Salazar, R.; Castillo-Neyra, R.; Borrini-Mayori, K.; Condori, C.; Bartow-McKenney, C.; Tracy, D.; Naquira, C.; Levy, M.Z. Trypanosoma cruzi infection does not decrease survival or reproduction of the common bed bug, Cimex lectularius. Am. J. Trop. Med. Hyg. 2018, 98, 724–734. [Google Scholar] [CrossRef]
- Salazar, R.; Castillo-Neyra, R.; Tustin, A.W.; Borrini-Mayori, K.; Naquira, C.; Levy, M.Z. Bed bugs (Cimex lectularius) as vectors of Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 2015, 92, 331–335. [Google Scholar] [CrossRef]
- Lowenberger, C. Innate immune response of Aedes aegypti. Insect. Biochem. Mol. Biol. 2001, 31, 219–229. [Google Scholar] [CrossRef]
- Meiser, C.K.; Pausch, J.K.; Schaub, G.A. Feeding-induced changes of bacteriolytic activity and the pattern of bacteriolytic compounds in the stomach and small intestine of the haematophagous bug Triatoma infestans (Klug, 1834) (Reduviidae, Triatominae). Parasitologia 2022, 2, 13–26. [Google Scholar] [CrossRef]
- Salcedo-Porras, N.; Lowenberger, C. The innate immune system of kissing bugs, vectors of chagas disease. Dev. Comp. Immunol. 2019, 98, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C.; Rollinghoff, M.; Diefenbach, A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 2000, 12, 64–76. [Google Scholar] [CrossRef]
- Nappi, A.J.; Vass, E. Cytotoxic reactions associated with insect immunity. Adv. Exp. Med. Biol. 2001, 484, 329–348. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef] [PubMed]
- Muta, T.; Iwanaga, S. The role of hemolymph coagulation in innate immunity. Curr. Opin. Immunol. 1996, 8, 41–47. [Google Scholar] [CrossRef]
- Aerts, A.M.; Francois, I.E.; Cammue, B.P.; Thevissen, K. The mode of antifungal action of plant, insect and human defensins. Cell. Mol. Life Sci. 2008, 65, 2069–2079. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef]
- Hoffmann, J.A.; Reichhart, J.M. Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 2002, 3, 121–126. [Google Scholar] [CrossRef]
- Morin-Poulard, I.; Vincent, A.; Crozatier, M. The Drosophila JAK-STAT pathway in blood cell formation and immunity. JAKSTAT 2013, 2, e25700. [Google Scholar] [CrossRef]
- Dong, Y.; Dong, S.; Dizaji, N.B.; Rutkowski, N.; Pohlenz, T.; Myles, K.; Dimopoulos, G. The Aedes aegypti siRNA pathway mediates broad-spectrum defense against human pathogenic viruses and modulates antibacterial and antifungal defenses. PLoS Biol. 2022, 20, e3001668. [Google Scholar] [CrossRef]
- Mishra, A.K.; Sharma, V.; Mutsuddi, M.; Mukherjee, A. Signaling cross-talk during development: Context-specific networking of Notch, NF-kappaB and JNK signaling pathways in Drosophila. Cell. Signal. 2021, 82, 109937. [Google Scholar] [CrossRef] [PubMed]
- Nishide, Y.; Kageyama, D.; Yokoi, K.; Jouraku, A.; Tanaka, H.; Futahashi, R.; Fukatsu, T. Functional crosstalk across IMD and Toll pathways: Insight into the evolution of incomplete immune cascades. Proc. Biol. Sci. 2019, 286, 20182207. [Google Scholar] [CrossRef] [PubMed]
- Salcedo-Porras, N.; Noor, S.; Cai, C.; Oliveira, P.L.; Lowenberger, C. Rhodnius prolixus uses the peptidoglycan recognition receptor rpPGRP-LC/LA to detect Gram-negative bacteria and activate the IMD pathway. Curr. Res. Insect. Sci. 2021, 1, 100006. [Google Scholar] [CrossRef] [PubMed]
- Potts, R.; King, J.G.; Pietri, J.E. Ex. vivo. characterization of the circulating hemocytes of bed bugs and their responses to bacterial exposure. J. Invertebr. Pathol. 2020, 174, 107422. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999, 23, 329–344. [Google Scholar] [CrossRef]
- Lowenberger, C.; Bulet, P.; Charlet, M.; Hetru, C.; Hodgeman, B.; Christensen, B.M.; Hoffmann, J.A. Insect immunity: Isolation of three novel inducible antibacterial defensins from the vector mosquito, Aedes aegypti. Insect. Biochem. Mol. Biol. 1995, 25, 867–873. [Google Scholar] [CrossRef]
- Kaushal, A.; Gupta, K.; Shah, R.; van Hoek, M.L. Antimicrobial activity of mosquito cecropin peptides against Francisella. Dev. Comp. Immunol. 2016, 63, 171–180. [Google Scholar] [CrossRef]
- Lowenberger, C.A.; Smartt, C.T.; Bulet, P.; Ferdig, M.T.; Severson, D.W.; Hoffmann, J.A.; Christensen, B.M. Insect immunity: Molecular cloning, expression, and characterization of cDNAs and genomic DNA encoding three isoforms of insect defensin in Aedes aegypti. Insect Mol. Biol. 1999, 8, 107–118. [Google Scholar] [CrossRef]
- Charlet, M.; Lagueux, M.; Reichhart, J.M.; Hoffmann, D.; Braun, A.; Meister, M. Cloning of the gene encoding the antibacterial peptide drosocin involved in Drosophila immunity. Expression studies during the immune response. Eur. J. Biochem. 1996, 241, 699–706. [Google Scholar] [CrossRef]
- Welch, N.G.; Li, W.; Hossain, M.A.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. (Re)Defining the proline-rich antimicrobial peptide family and the identification of putative new members. Front. Chem. 2020, 8, 607769. [Google Scholar] [CrossRef]
- Buonocore, F.; Fausto, A.M.; Della Pelle, G.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A promising class of insect antimicrobial peptides. Antibiotics 2021, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.L.; Bulet, P. Antimicrobial peptides in Drosophila: Structures, activities and gene regulation. Chem. Immunol. Allergy 2005, 86, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Stocklin, R. Insect antimicrobial peptides: Structures, properties and gene regulation. Protein Pept. Lett. 2005, 12, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Tzou, P.; De Gregorio, E.; Lemaitre, B. How Drosophila combats microbial infection: A model to study innate immunity and host-pathogen interaction. Curr. Opin. Microbiol. 2002, 5, 102–110. [Google Scholar] [CrossRef]
- Dassanayake, R.S.; Silva Gunawardene, Y.I.; Tobe, S.S. Evolutionary selective trends of insect/mosquito antimicrobial defensin peptides containing cysteine-stabilized alpha/beta motifs. Peptides 2007, 28, 62–75. [Google Scholar] [CrossRef]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; de Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldon, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21. [Google Scholar] [CrossRef]
- Boulanger, N.; Lowenberger, C.; Volf, P.; Ursic, R.; Sigutova, L.; Sabatier, L.; Svobodova, M.; Beverley, S.M.; Spath, G.; Brun, R.; et al. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania. major. Infect. Immun. 2004, 72, 7140–7146. [Google Scholar] [CrossRef]
- Cociancich, S.; Ghazi, A.; Hetru, C.; Hoffmann, J.A.; Letellier, L. Insect defensin, an inducible antibacterial peptide, forms voltage-dependent channels in Micrococcus luteus. J. Biol. Chem. 1993, 268, 19239–19245. [Google Scholar] [CrossRef]
- Eleftherianos, I.; Heryanto, C.; Bassal, T.; Zhang, W.; Tettamanti, G.; Mohamed, A. Haemocyte-mediated immunity in insects: Cells, processes and associated components in the fight against pathogens and parasites. Immunology 2021, 164, 401–432. [Google Scholar] [CrossRef]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef] [Green Version]
- Lopez, L.; Morales, G.; Ursic, R.; Wolff, M.; Lowenberger, C. Isolation and characterization of a novel insect defensin from Rhodnius prolixus, a vector of Chagas disease. Insect Biochem. Mol. Biol. 2003, 33, 439–447. [Google Scholar] [CrossRef]
- Ursic-Bedoya, R.J.; Lowenberger, C.A. Rhodnius prolixus: Identification of immune-related genes up-regulated in response to pathogens and parasites using suppressive subtractive hybridization. Dev. Comp. Immunol. 2007, 31, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Ursic-Bedoya, R.J.; Nazzari, H.; Cooper, D.; Triana, O.; Wolff, M.; Lowenberger, C. Identification and characterization of two novel lysozymes from Rhodnius prolixus, a vector of Chagas disease. J. Insect Physiol. 2008, 54, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.S.; Waniek, P.J.; Mattos, D.P.; Castro, D.P.; Mello, C.B.; Ratcliffe, N.A.; Garcia, E.S.; Azambuja, P. Humoral responses in Rhodnius prolixus: Bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasites Vectors 2014, 7, 232. [Google Scholar] [CrossRef]
- Ursic-Bedoya, R.; Buchhop, J.; Joy, J.B.; Durvasula, R.; Lowenberger, C. Prolixicin: A novel antimicrobial peptide isolated from Rhodnius prolixus with differential activity against bacteria and Trypanosoma cruzi. Insect Mol. Biol. 2011, 20, 775–786. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Cornet, B.; Bonmatin, J.M.; Hetru, C.; Hoffmann, J.A.; Ptak, M.; Vovelle, F. Refined three-dimensional solution structure of insect defensin A. Structure 1995, 3, 435–448. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Stokes, B.A.; Yadav, S.; Shokal, U.; Smith, L.C.; Eleftherianos, I. Bacterial and fungal pattern recognition receptors in homologous innate signaling pathways of insects and mammals. Front. Microbiol. 2015, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Ren, M.; Liu, X.; Xia, H.; Chen, K. Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 2019, 106, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Benoit, J.B.; Adelman, Z.N.; Reinhardt, K.; Dolan, A.; Poelchau, M.; Jennings, E.C.; Szuter, E.M.; Hagan, R.W.; Gujar, H.; Shukla, J.N.; et al. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat. Commun. 2016, 7, 10165. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.L.; Cury, J.C.; Gurgel-Goncalves, R.; Bahia, A.C.; Monteiro, F.A. Field-collected Triatoma sordida from central Brazil display high microbiota diversity that varies with regard to developmental stage and intestinal segmentation. PLoS Negl. Trop. Dis. 2018, 12, e0006709. [Google Scholar] [CrossRef] [PubMed]
- Salcedo-Porras, N.; Guarneri, A.; Oliveira, P.L.; Lowenberger, C. Rhodnius prolixus: Identification of missing components of the IMD immune signaling pathway and functional characterization of its role in eliminating bacteria. PLoS ONE 2019, 14, e0214794. [Google Scholar] [CrossRef] [PubMed]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef]
- Xiao, X.; Liu, Y.; Zhang, X.; Wang, J.; Li, Z.; Pang, X.; Wang, P.; Cheng, G. Complement-related proteins control the flavivirus infection of Aedes aegypti by inducing antimicrobial peptides. PLoS Pathog. 2014, 10, e1004027. [Google Scholar] [CrossRef]
- Reinhardt, K.; Naylor, R.; Siva-Jothy, M.T. Reducing a cost of traumatic insemination: Female bedbugs evolve a unique organ. Proc. Biol. Sci. 2003, 270, 2371–2375. [Google Scholar] [CrossRef]
- Cole, P.A.; Shen, K.; Qiao, Y.; Wang, D. Protein tyrosine kinases Src and Csk: A tail’s tale. Curr. Opin. Chem. Biol. 2003, 7, 580–585. [Google Scholar] [CrossRef]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Yeung, A.T.; Gellatly, S.L.; Hancock, R.E. Multifunctional cationic host defence peptides and their clinical applications. Cell. Mol. Life Sci. 2011, 68, 2161–2176. [Google Scholar] [CrossRef]
- Fujii, G.; Selsted, M.E.; Eisenberg, D. Defensins promote fusion and lysis of negatively charged membranes. Protein Sci. 1993, 2, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Del Pilar Corena, M.; VanEkeris, L.; Salazar, M.I.; Bowers, D.; Fiedler, M.M.; Silverman, D.; Tu, C.; Linser, P.J. Carbonic anhydrase in the adult mosquito midgut. J. Exp. Biol. 2005, 208, 3263–3273. [Google Scholar] [CrossRef]
- Bartholomay, L.C.; Fuchs, J.F.; Cheng, L.L.; Beck, E.T.; Vizioli, J.; Lowenberger, C.; Christensen, B.M. Reassessing the role of defensin in the innate immune response of the mosquito, Aedes aegypti. Insect Mol. Biol. 2004, 13, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, J.G.; Jurat-Fuentes, J.L. Physiology and ecology of host defense against microbial invaders. In Insect Pathology, 2nd ed.; Vega, F.E., Kaya, H.K., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2012; pp. 461–480. [Google Scholar]
- Oliveira, J.H.; Goncalves, R.L.; Lara, F.A.; Dias, F.A.; Gandara, A.C.; Menna-Barreto, R.F.; Edwards, M.C.; Laurindo, F.R.; Silva-Neto, M.A.; Sorgine, M.H.; et al. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 2011, 7, e1001320. [Google Scholar] [CrossRef] [PubMed]
- Meraj, S.; Mohr, E.; Ketabchi, N.; Bogdanovic, A.; Lowenberger, C.; Gries, G. Time- and tissue-specific antimicrobial activity of the common bed bug in response to blood feeding and immune activation by bacterial injection. J. Insect Physiol. 2021, 135, 104322. [Google Scholar] [CrossRef] [PubMed]
- Boutros, M.; Agaisse, H.; Perrimon, N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 2002, 3, 711–722. [Google Scholar] [CrossRef]
- Lemaitre, B.; Reichhart, J.M.; Hoffmann, J.A. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 1997, 94, 14614–14619. [Google Scholar] [CrossRef]
- Rutschmann, S.; Jung, A.C.; Hetru, C.; Reichhart, J.M.; Hoffmann, J.A.; Ferrandon, D. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 2000, 12, 569–580. [Google Scholar] [CrossRef]
- Siva-Jothy, M.T.; Zhong, W.; Naylor, R.; Heaton, L.; Hentley, W.; Harney, E. Female bed bugs (Cimex lectularius L.) anticipate the immunological consequences of traumatic insemination via feeding cues. Proc. Natl. Acad. Sci. USA 2019, 116, 14682–14687. [Google Scholar] [CrossRef]
- Reinhardt, K.; Anthes, N.; Lange, R. Copulatory wounding and traumatic insemination. Cold Spring Harb. Perspect. Biol. 2015, 7, a017582. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, K.; Naylor, R.A.; Siva-Jothy, M.T. Potential sexual transmission of environmental microbes in a traumatically inseminating insect. Ecol. Entomol. 2005, 30, 607–611. [Google Scholar] [CrossRef]
- Fisher, M.L.; Levine, J.F.; Guy, J.S.; Mochizuki, H.; Breen, M.; Schal, C.; Watson, D.W. Lack of influence by endosymbiont Wolbachia on virus titer in the common bed bug, Cimex lectularius. Parasites Vectors 2019, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Meriweather, M.; Matthews, S.; Rio, R.; Baucom, R.S. A 454 survey reveals the community composition and core microbiome of the common bed bug (Cimex lectularius) across an urban landscape. PLoS ONE 2013, 8, e61465. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, J.M.; Rasgon, J.L. Endosymbiotic bacteria of bed bugs: Evolution, ecology and genetics. Am. Entomol. 2006, 52, 119–122. [Google Scholar] [CrossRef]
- Hickin, M.L.; Kakumanu, M.L.; Schal, C. Effects of Wolbachia elimination and B-vitamin supplementation on bed bug development and reproduction. Sci. Rep. 2022, 12, 10270. [Google Scholar] [CrossRef]
- Salcedo-Porras, N.; Umana-Diaz, C.; Bitencourt, R.O.B.; Lowenberger, C. The role of bacterial symbionts in triatomines: An evolutionary perspective. Microorganisms 2020, 8, 1438. [Google Scholar] [CrossRef]
- Bennett, G.M.; Moran, N.A. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 2015, 112, 10169–10176. [Google Scholar] [CrossRef]
- The International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8, e1000313. [Google Scholar] [CrossRef]
- Zaidman-Remy, A.; Vigneron, A.; Weiss, B.L.; Heddi, A. What can a weevil teach a fly, and reciprocally? Interaction of host immune systems with endosymbionts in Glossina and Sitophilus. BMC Microbiol. 2018, 18, 150. [Google Scholar] [CrossRef] [Green Version]
- Castro, D.P.; Moraes, C.S.; Gonzalez, M.S.; Ratcliffe, N.A.; Azambuja, P.; Garcia, E.S. Trypanosoma cruzi immune response modulation decreases microbiota in Rhodnius prolixus gut and is crucial for parasite survival and development. PLoS ONE 2012, 7, e36591. [Google Scholar] [CrossRef] [PubMed]
- Azambuja, P.; Feder, D.; Garcia, E.S. Isolation of Serratia marcescens in the midgut of Rhodnius prolixus: Impact on the establishment of the parasite Trypanosoma cruzi in the vector. Exp. Parasitol. 2004, 107, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Gries, R.; Britton, R.; Holmes, M.; Zhai, H.; Draper, J.; Gries, G. Bed bug aggregation pheromone finally identified. Angew. Chem. Int. Ed. Engl. 2015, 54, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. Lysogeny at mid-twentieth century: P1, P2, and other experimental, systems. J. Bacteriol. 2004, 186, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Mamidala, P.; Rajarapu, S.P.; Jones, S.C.; Mittapalli, O. Identification and validation of reference genes for quantitative real-time polymerase chain reaction in Cimex lectularius. J. Med. Entomol. 2011, 48, 947–951. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Z.; Zhu, R.; Wang, F.; Cheng, Y.; Liu, Y. Three differential expression analysis methods for RNA sequencing: Limma, EdgeR, DESeq2. J. Vis. Exp. 2021, 175, e62528. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Clamp, M.; Cuff, J.; Searle, S.M.; Barton, G.J. The Jalview Java alignment editor. Bioinformatics 2004, 20, 426–427. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]

| Gene | Primer Sequence (5′–3′) |
|---|---|
| RPL18 | FW: AAAGGCACGGTTACATCAAAGGTG RV: TAGTCTTGAACCTATAGGGGTCCC |
| CL-defensin1 (LOC106661793) | FW: AAG AGC GAC CTG CGA TTT GT RV: TGT GCC TTT ACA ATA CCC TCC TC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meraj, S.; Dhari, A.S.; Mohr, E.; Lowenberger, C.; Gries, G. Characterization of New Defensin Antimicrobial Peptides and Their Expression in Bed Bugs in Response to Bacterial Ingestion and Injection. Int. J. Mol. Sci. 2022, 23, 11505. https://doi.org/10.3390/ijms231911505
Meraj S, Dhari AS, Mohr E, Lowenberger C, Gries G. Characterization of New Defensin Antimicrobial Peptides and Their Expression in Bed Bugs in Response to Bacterial Ingestion and Injection. International Journal of Molecular Sciences. 2022; 23(19):11505. https://doi.org/10.3390/ijms231911505
Chicago/Turabian StyleMeraj, Sanam, Arshvir Singh Dhari, Emerson Mohr, Carl Lowenberger, and Gerhard Gries. 2022. "Characterization of New Defensin Antimicrobial Peptides and Their Expression in Bed Bugs in Response to Bacterial Ingestion and Injection" International Journal of Molecular Sciences 23, no. 19: 11505. https://doi.org/10.3390/ijms231911505






