What Do the Transcriptome and Proteome of Menstrual Blood-Derived Mesenchymal Stem Cells Tell Us about Endometriosis?
Abstract
:1. Introduction
2. Results
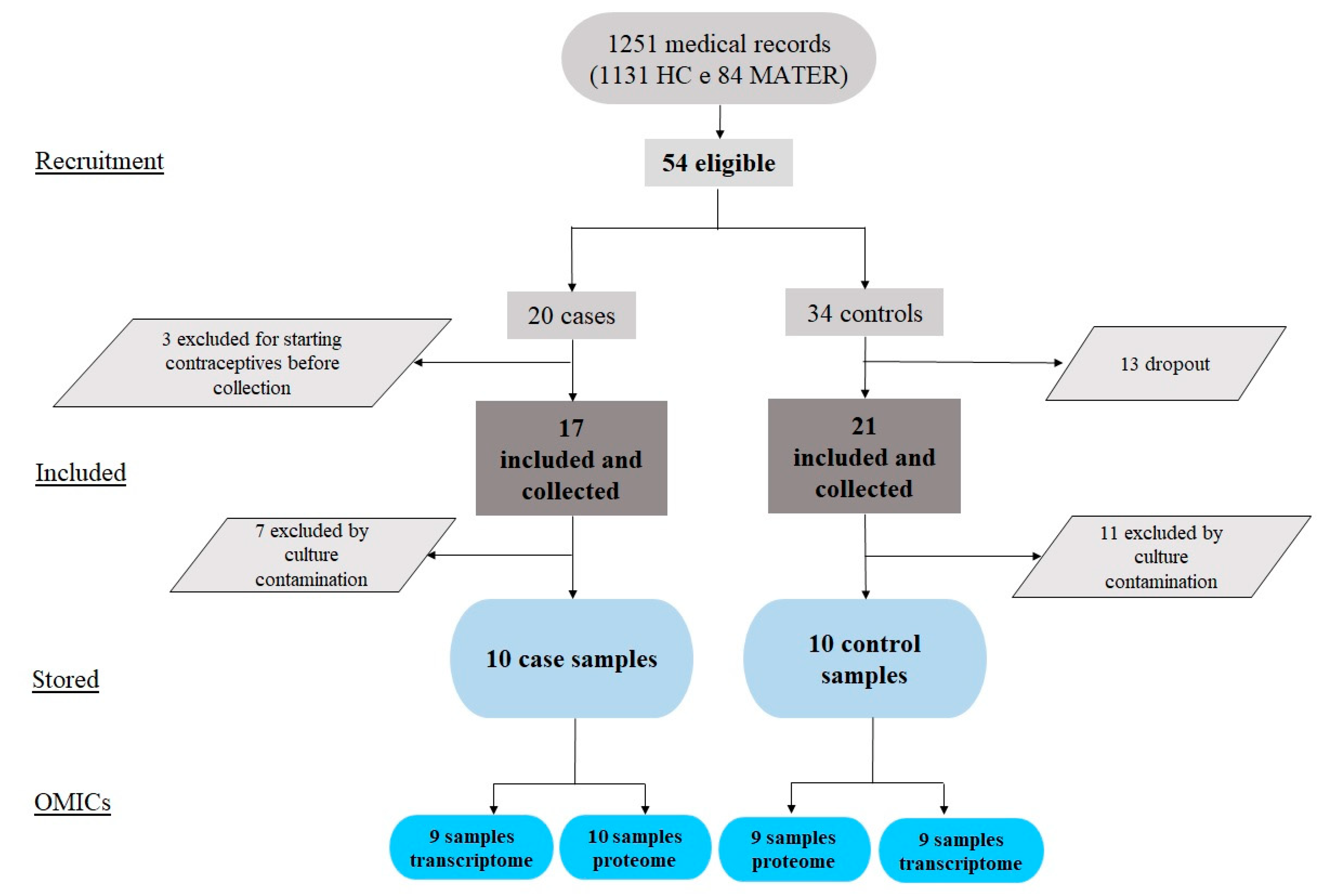
2.1. Study Flowchart, Clinical Variables, and the MenSCs In Vitro Model
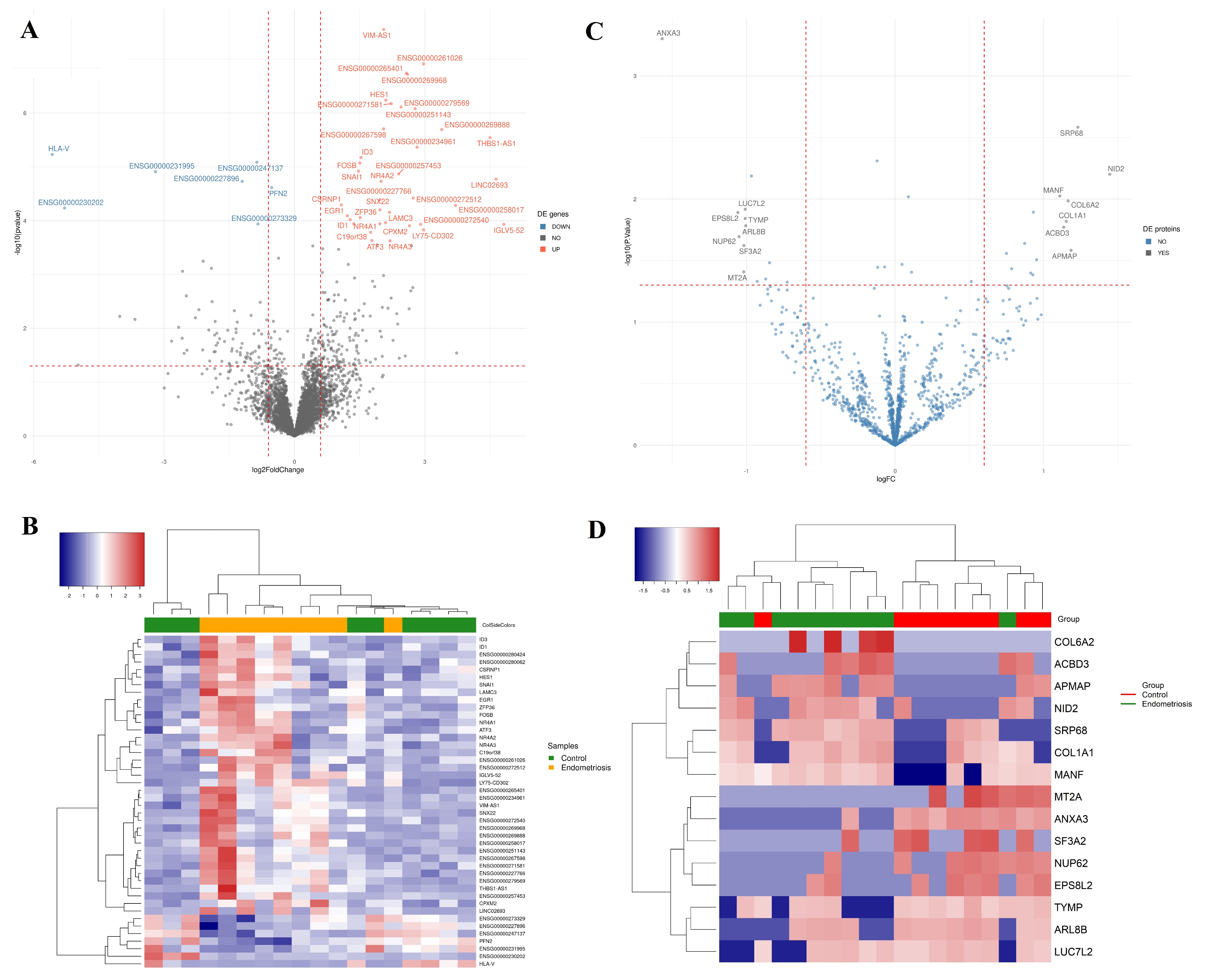
2.2. Differential Transcript Profile
2.3. Differential Protein Profile
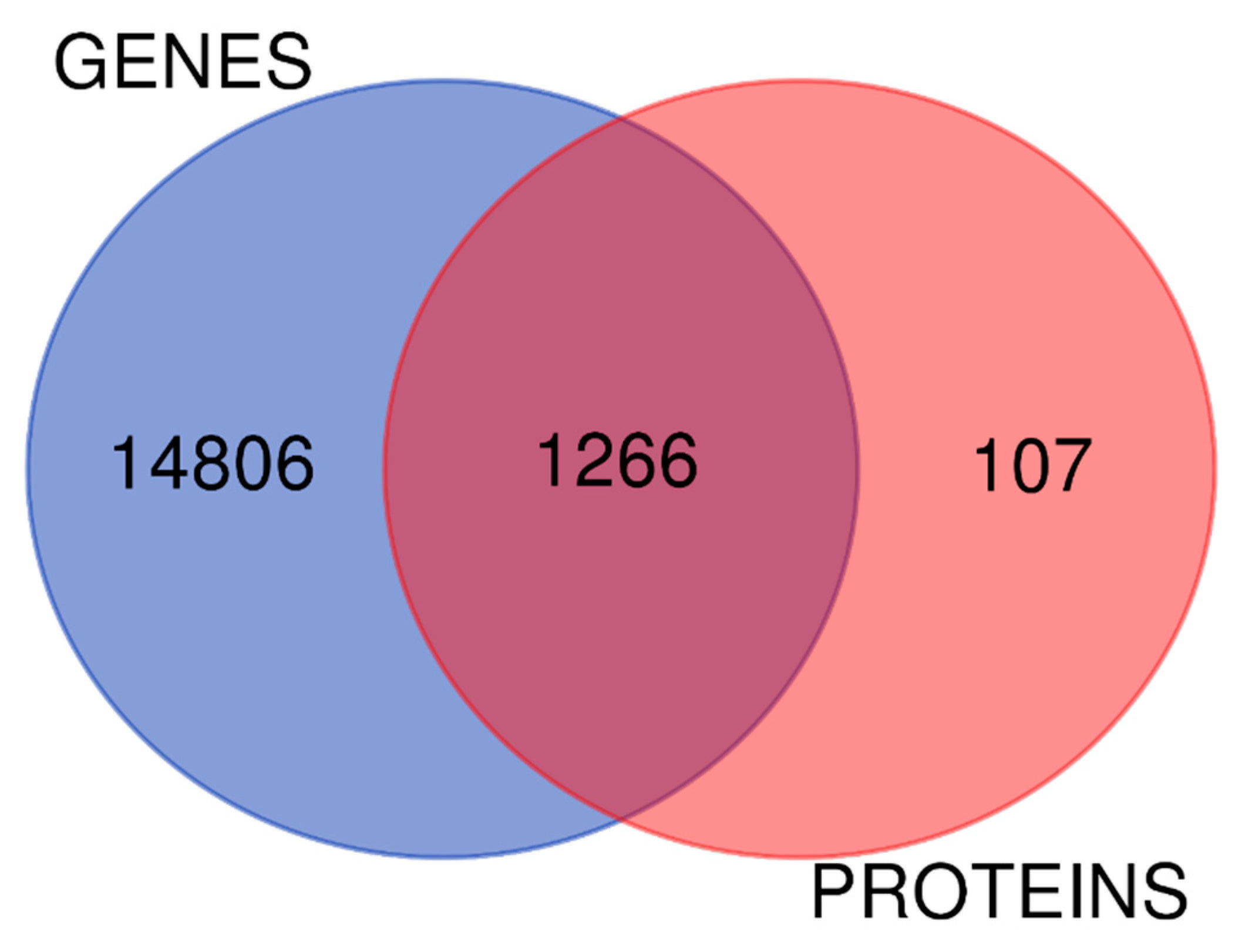
2.4. The Biology of Proteomic and Transcriptomic Systems in Endometriosis MenSCs Reveal Related Pathways
3. Discussion
4. Materials and Methods
4.1. Ethics Statement and Duration
4.2. Settings
4.3. Participants and Eligibility Criteria
4.4. Characterization and Establishment of the MenSCs In Vitro Model
4.5. OMICs Approaches
4.5.1. Transcriptome: Total RNA Extraction, RNA Integrity, and Quantification
4.5.2. Shotgun Proteomic: Extraction, Quantification, Trypsinization, and Desalination of Proteins
4.6. Enrichment Analysis
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darzi, S.; Werkmeister, J.A.; Deane, J.A.; Gargett, C.E. Identification and Characterization of Human Endometrial Mesenchymal Stem/Stromal Cells and Their Potential for Cellular Therapy. Stem Cells Transl. Med. 2016, 5, 1127–1132. [Google Scholar] [CrossRef]
- Chen, L.; Qu, J.; Xiang, C. The Multi-Functional Roles of Menstrual Blood-Derived Stem Cells in Regenerative Medicine. Stem Cell Res. Ther. 2019, 10, 1. [Google Scholar] [CrossRef]
- Bozorgmehr, M.; Gurung, S.; Darzi, S.; Nikoo, S.; Kazemnejad, S.; Zarnani, A.H.; Gargett, C.E. Endometrial and Menstrual Blood Mesenchymal Stem/Stromal Cells: Biological Properties and Clinical Application. Front. Cell Dev. Biol. 2020, 8, 497. [Google Scholar] [CrossRef]
- Meng, X.; Ichim, T.E.; Zhong, J.; Rogers, A.; Yin, Z.; Jackson, J.; Wang, H.; Ge, W.; Bogin, V.; Chan, K.W.; et al. Endometrial Regenerative Cells: A Novel Stem Cell Population. J. Transl. Med. 2007, 5, 57. [Google Scholar] [CrossRef]
- el Sabeh, M.; Afrin, S.; Singh, B.; Miyashita-Ishiwata, M.; Borahay, M. Uterine Stem Cells and Benign Gynecological Disorders: Role in Pathobiology and Therapeutic Implications. Stem Cell Rev. Rep. 2021, 17, 803. [Google Scholar] [CrossRef]
- Gargett, C.E.; Schwab, K.E.; Deane, J.A. Endometrial Stem/Progenitor Cells: The First 10 Years. Hum. Reprod. Update 2016, 22, 137–163. [Google Scholar] [CrossRef]
- Warren, L.A.; Shih, A.; Renteira, S.M.; Blau, B.; Simpfendorfer, K.; Lee, A.; Metz, C.N.; Gregersen, P.K. Analysis of Menstrual Effluent: Diagnostic Potential for Endometriosis. Mol. Med. 2018, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Shao, Y.; Ren, C.; Yang, G. Endometrial Stem/Progenitor Cells and Their Roles in Immunity, Clinical Application, and Endometriosis. Stem Cell Res. Ther. 2021, 12, 474. [Google Scholar] [CrossRef]
- Sahraei, S.S.; Davoodi Asl, F.; Kalhor, N.; Sheykhhasan, M.; Fazaeli, H.; Moud, S.S.; Sheikholeslami, A. A Comparative Study of Gene Expression in Menstrual Blood-Derived Stromal Cells between Endometriosis and Healthy Women. Biomed. Res. Int. 2022, 2022, 7053521. [Google Scholar] [CrossRef] [PubMed]
- Esfandyari, S.; Chugh, R.M.; Park, H.S.; Hobeika, E.; Ulin, M.; Al-Hendy, A. Mesenchymal Stem Cells as a Bio Organ for Treatment of Female Infertility. Cells 2020, 9, 2253. [Google Scholar] [CrossRef] [PubMed]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef]
- Missmer, S.A.; Cramer, D.W. The Epidemiology of Endometriosis. Obstet. Gynecol. Clin. N. Am. 2003, 30, 1–19. [Google Scholar] [CrossRef]
- Eisenberg, V.H.; Weil, C.; Chodick, G.; Shalev, V. Epidemiology of Endometriosis: A Large Population-Based Database Study from a Healthcare Provider with 2 Million Members. BJOG 2018, 125, 55–62. [Google Scholar] [CrossRef]
- Agarwal, N.; Subramanian, A. Endometriosis—Morphology, Clinical Presentations and Molecular Pathology. J. Lab. Physicians 2010, 2, 001–009. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, D.; Mabrouk, M.; Zannoni, L.; Arena, A.; Zanello, M.; Benfenati, A.; Moro, E.; Paradisi, R.; Seracchioli, R. Severe Ureteral Endometriosis: Frequency and Risk Factors. J. Obstet. Gynaecol. 2017, 38, 257–260. [Google Scholar] [CrossRef]
- Hirata, T.; Koga, K.; Osuga, Y. Extra-pelvic Endometriosis: A Review. Reprod. Med. Biol. 2020, 19, 323. [Google Scholar] [CrossRef] [PubMed]
- Simoens, S.; Dunselman, G.; Dirksen, C.; Hummelshoj, L.; Bokor, A.; Brandes, I.; Brodszky, V.; Canis, M.; Colombo, G.L.; Deleire, T.; et al. The Burden of Endometriosis: Costs and Quality of Life of Women with Endometriosis and Treated in Referral Centres. Hum. Reprod. 2012, 27, 1292–1299. [Google Scholar] [CrossRef] [PubMed]
- Culley, L.; Law, C.; Hudson, N.; Denny, E.; Mitchell, H.; Baumgarten, M.; Raine-Fenning, N. The Social and Psychological Impact of Endometriosis on Women’s Lives: A Critical Narrative Review. Hum. Reprod. Update 2013, 19, 625–639. [Google Scholar] [CrossRef]
- Goulielmos, G.N.; Matalliotakis, M.; Matalliotaki, C.; Eliopoulos, E.; Matalliotakis, I.; Zervou, M.I. Endometriosis Research in the -Omics Era. Gene 2020, 741, 144545. [Google Scholar] [CrossRef] [PubMed]
- Sourial, S.; Tempest, N.; Hapangama, D.K. Theories on the Pathogenesis of Endometriosis. Int. J. Reprod Med. 2014, 2014, 179515. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, Z.; Yang, F.; Wang, H.; Liang, S.; Wang, H.; Yang, J.; Lin, J. The Role of Endometrial Stem Cells in the Pathogenesis of Endometriosis and Their Application to Its Early Diagnosis. Biol. Reprod. 2020, 102, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.A. Peritoneal Endometriosis Due to the Menstrual Dissemination of Endometrial Tissue into the Peritoneal Cavity. Am. J. Obstet. Gynecol. 1927, 14, 422–469. [Google Scholar] [CrossRef]
- Seeber, B.; Sammel, M.D.; Fan, X.; Gerton, G.L.; Shaunik, A.; Chittams, J.; Barnhart, K.T. Proteomic Analysis of Serum Yields Six Candidate Proteins That Are Differentially Regulated in a Subset of Women with Endometriosis. Fertil. Steril. 2010, 93, 2137–2144. [Google Scholar] [CrossRef]
- Nyholt, D.R.; Low, S.K.; Anderson, C.A.; Painter, J.N.; Uno, S.; Morris, A.P.; MacGregor, S.; Gordon, S.D.; Henders, A.K.; Martin, N.G.; et al. Genome-Wide Association Meta-Analysis Identifies New Endometriosis Risk Loci. Nat. Genet. 2012, 44, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Borghese, B.; Zondervan, K.T.; Abrao, M.S.; Chapron, C.; Vaiman, D. Recent Insights on the Genetics and Epigenetics of Endometriosis. Clin. Genet. 2017, 91, 254–264. [Google Scholar] [CrossRef]
- Poli-Neto, O.B.; Meola, J.; Rosa-e-Silva, J.C.; Tiezzi, D. Transcriptome Meta-Analysis Reveals Differences of Immune Profile between Eutopic Endometrium from Stage I-II and III-IV Endometriosis Independently of Hormonal Milieu. Sci. Rep. 2020, 10, 313. [Google Scholar] [CrossRef]
- Spitzer, T.L.B.; Rojas, A.; Zelenko, Z.; Aghajanova, L.; Erikson, D.W.; Meyer, F.B.; Tamaresis, J.S.; Hamilton, A.E.; Irwin, J.C.; Giudice, L.C. Perivascular Human Endometrial Mesenchymal Stem Cells Express Pathways Relevant to Self-Renewal, Lineage Specification, and Functional Phenotype. Biol. Reprod. 2012, 86, 58. [Google Scholar] [CrossRef]
- Barragan, F.; Irwin, J.C.; Balayan, S.; Erikson, D.W.; Chen, J.C.; Houshdaran, S.; Piltonen, T.T.; Spitzer, T.L.B.; George, A.; Rabban, J.T.; et al. Human Endometrial Fibroblasts Derived from Mesenchymal Progenitors Inherit Progesterone Resistance and Acquire an Inflammatory Phenotype in the Endometrial Niche in Endometriosis. Biol. Reprod. 2016, 94, 118. [Google Scholar] [CrossRef]
- Rekker, K.; Saare, M.; Eriste, E.; Tasa, T.; Kukuškina, V.; Mari Roost, A.; Anderson, K.; Samuel, K.; Karro, H.; Salumets, A.; et al. High-Throughput MRNA Sequencing of Stromal Cells from Endometriomas and Endometrium. Reproduction 2017, 154, 93–100. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, S.; Yang, F.; Sun, Y.; Niu, L.; Ren, Y.; Wang, H.; He, Y.; Du, J.; Yang, J.; et al. Biological Characteristics of Endometriotic Mesenchymal Stem Cells Isolated from Ectopic Lesions of Patients with Endometriosis. Stem Cell Res. Ther. 2020, 11, 346. [Google Scholar] [CrossRef]
- Hwang, J.H.; Oh, J.J.; Wang, T.; Jin, Y.C.; Lee, J.S.; Choi, J.R.; Lee, K.S.; Joo, J.K.; Lee, H.G. Identification of Biomarkers for Endometriosis in Eutopic Endometrial Cells from Patients with Endometriosis Using a Proteomics Approach. Mol. Med. Rep. 2013, 8, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Prašnikar, E.; Knez, J.; Kovačič, B.; Kunej, T. Molecular Signature of Eutopic Endometrium in Endometriosis Based on the Multi-Omics Integrative Synthesis. J. Assist. Reprod. Genet. 2020, 37, 1593–1611. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, R.Z.; de Oliveira Buono, F.; Cressoni, A.C.L.; Penariol, L.B.C.; Padovan, C.C.; Tozetti, P.A.; Poli-Neto, O.B.; Ferriani, R.A.; Orellana, M.D.; Rosa-e-Silva, J.C.; et al. Overexpression of MiR-200b-3p in Menstrual Blood-Derived Mesenchymal Stem Cells from Endometriosis Women. Reprod. Sci. 2022, 29, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells. The International Society for Cellular Therapy Position Statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Musina, R.A.; Belyavski, A.V.; Tarusova, O.V.; Solovyova, E.V.; Sukhikh, G.T. Endometrial Mesenchymal Stem Cells Isolated from the Menstrual Blood. Bull. Exp. Biol. Med. 2008, 145, 539–543. [Google Scholar] [CrossRef]
- Du, X.; Yuan, Q.; Qu, Y.; Zhou, Y.; Bei, J. Endometrial Mesenchymal Stem Cells Isolated from Menstrual Blood by Adherence. Stem Cells Int. 2016, 2016, 3573846. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Ding, S.; Yu, Q.; Wang, J.; Zhu, L.; Li, T.; Guo, X.; Zhang, X. Activation of ATF3/AP-1 Signaling Pathway Is Required for P2X3-Induced Endometriosis Pain. Hum. Reprod. 2020, 35, 1130–1144. [Google Scholar] [CrossRef]
- Mirza, Z.; Abdel-Dayem, U.A. Uncovering Potential Roles of Differentially Expressed Genes, Upstream Regulators, and Canonical Pathways in Endometriosis Using an In Silico Genomics Approach. Diagnostics 2020, 10, 416. [Google Scholar] [CrossRef]
- Bunis, D.G.; Wang, W.; Vallvé-Juanico, J.; Houshdaran, S.; Sen, S.; ben Soltane, I.; Kosti, I.; Vo, K.C.; Irwin, J.C.; Giudice, L.C.; et al. Whole-Tissue Deconvolution and ScRNAseq Analysis Identify Altered Endometrial Cellular Compositions and Functionality Associated with Endometriosis. Front. Immunol. 2022, 12, 788315. [Google Scholar] [CrossRef]
- Amirteimouri, S.; Ashini, M.; Ramazanali, F.; Aflatoonian, R.; Afsharian, P.; Shahhoseini, M. Epigenetic Role of the Nuclear Factor NF-Y on ID Gene Family in Endometrial Tissues of Women with Endometriosis: A Case Control Study. Reprod. Biol. Endocrinol. 2019, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Young, V.J.; Brown, J.K.; Saunders, P.T.K.; Duncan, W.C.; Horne, A.W. The Peritoneum Is Both a Source and Target of TGF-β in Women with Endometriosis. PLoS ONE 2014, 9, e106773. [Google Scholar] [CrossRef] [PubMed]
- Méar, L.; Com, E.; Fathallah, K.; Guillot, L.; Lavigne, R.; Guével, B.; Fauconnier, A.; Vialard, F.; Pineau, C. The Eutopic Endometrium Proteome in Endometriosis Reveals Candidate Markers and Molecular Mechanisms of Physiopathology. Diagnostics 2022, 12, 419. [Google Scholar] [CrossRef] [PubMed]
- Uimari, O.; Rahmioglu, N.; Nyholt, D.R.; Vincent, K.; Missmer, S.A.; Becker, C.; Morris, A.P.; Montgomery, G.W.; Zondervan, K.T. Genome-Wide Genetic Analyses Highlight Mitogen-Activated Protein Kinase (MAPK) Signaling in the Pathogenesis of Endometriosis. Hum. Reprod. 2017, 32, 780–793. [Google Scholar] [CrossRef]
- Tang, X.; Zheng, H.; Xu, H.; Wang, M.; Kang, X.; Wu, Z. NR4A1 Affects Endometrial Receptivity by Participating in Mesenchymal-Epithelial Transition of Endometrial Stromal Cells. Reprod. Sci. 2022, 29, 133–142. [Google Scholar] [CrossRef]
- Ma, L.; Andrieu, T.; McKinnon, B.; Duempelmann, L.; Peng, R.W.; Wotzkow, C.; Müller, C.; Mueller, M.D. Epithelial-to-Mesenchymal Transition Contributes to the Downregulation of Progesterone Receptor Expression in Endometriosis Lesions. J. Steroid Biochem. Mol. Biol 2021, 212, 105943. [Google Scholar] [CrossRef]
- Kazmi, I.; Alharbi, K.S.; Al-Abbasi, F.A.; Almalki, W.H.; Kumar, S.; Yasmeen, A.; Khan, A.; Gupta, G. Role of Epithelial-to-Mesenchymal Transition Markers in Different Stages of Endometriosis: Expression of the Snail, Slug, ZEB1, and Twist Genes. Crit. Rev. Eukaryot. Gene Expr. 2021, 31, 89–95. [Google Scholar] [CrossRef]
- Konrad, L.; Dietze, R.; Riaz, M.A.; Scheiner-Bobis, G.; Behnke, J.; Horné, F.; Hoerscher, A.; Reising, C.; Meinhold-Heerlein, I. Epithelial-Mesenchymal Transition in Endometriosis-When Does It Happen? J. Clin. Med. 2020, 9, 1915. [Google Scholar] [CrossRef]
- Xiong, W.; Zhang, L.; Liu, H.; Li, N.; Du, Y.; He, H.; Zhang, Z.; Liu, Y. E2-Mediated EMT by Activation of β-Catenin/Snail Signalling during the Development of Ovarian Endometriosis. J. Cell Mol. Med. 2019, 23, 8035–8045. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Sengupta, J.; Giudice, L.C.; Mittal, S.; Kumar, S.; Gupta, S.D.; Sharma, R.; Najwa, A.R.; Ghosh, D. CDNA-Based Transcript Analysis of Autologous Eutopic and Ectopic Endometrium of Women with Moderate and Severe Endometriosis. J. Endometr. 2011, 3, 8–33. [Google Scholar] [CrossRef]
- Laudanski, P.; Charkiewicz, R.; Kuzmicki, M.; Szamatowicz, J.; Świa̧tecka, J.; Mroczko, B.; Niklinski, J. Profiling of Selected Angiogenesis-Related Genes in Proliferative Eutopic Endometrium of Women with Endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014, 172, 85–92. [Google Scholar] [CrossRef]
- Ferguson, S.D.; Olson, M.; Fazleabas, A.T. Changes in Mesenchymal Stem Cell-Associated Gene Expression in Secretory and Menstrual Endometrium of Baboons after Experimental Induction of Endometriosis. Biol. Reprod. 2011, 85, 784. [Google Scholar] [CrossRef]
- Sha, G.; Wu, D.; Zhang, L.; Chen, X.; Lei, M.; Sun, H.; Lin, S.; Lang, J. Differentially Expressed Genes in Human Endometrial Endothelial Cells Derived from Eutopic Endometrium of Patients with Endometriosis Compared with Those from Patients without Endometriosis. Hum. Reprod. 2007, 22, 3159–3169. [Google Scholar] [CrossRef] [PubMed]
- Abu-Asab, M.; Zhang, M.; Amini, D.; Abu-Asab, N.; Amri, H. Endometriosis Gene Expression Heterogeneity and Biosignature: A Phylogenetic Analysis. Obstet. Gynecol. Int. 2011, 2011, 719059. [Google Scholar] [CrossRef]
- Kuan, K.K.W.; Gibson, D.A.; Whitaker, L.H.R.; Horne, A.W. Menstruation Dysregulation and Endometriosis Development. Front. Reprod. Health 2021, 3, 68. [Google Scholar] [CrossRef]
- Zheng, Y.; Khan, Z.; Zanfagnin, V.; Correa, L.F.; Delaney, A.A.; Daftary, G.S. Epigenetic Modulation of Collagen 1A1: Therapeutic Implications in Fibrosis and Endometriosis. Biol. Reprod. 2016, 94, 87. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Liu, J.; Kong, N.; Jiang, Y.; Jiang, R.; Zhen, X.; Zhou, J.; Li, C.; Sun, H.; et al. ATF3 Deficiency Impairs the Proliferative-Secretory Phase Transition and Decidualization in RIF Patients. Cell Death Dis. 2021, 12, 387. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Cibelli, G. Regulation of Life and Death by the Zinc Finger Transcription Factor Egr-1. J. Cell Physiol. 2002, 193, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Khalaj, K.; Ahn, S.H.; Bidarimath, M.; Nasirzadeh, Y.; Singh, S.S.; Fazleabas, A.T.; Young, S.L.; Lessey, B.A.; Koti, M.; Tayade, C. A Balancing Act: RNA Binding Protein HuR/TTP Axis in Endometriosis Patients. Sci. Rep. 2017, 7, 5883. [Google Scholar] [CrossRef]
- Hawkins, S.M.; Creighton, C.J.; Han, D.Y.; Zariff, A.; Anderson, M.L.; Gunaratne, P.H.; Matzuk, M.M. Functional MicroRNA Involved in Endometriosis. Mol. Endocrinol. 2011, 25, 821–832. [Google Scholar] [CrossRef] [Green Version]
- Saare, M.; Rekker, K.; Laisk-Podar, T.; Rahmioglu, N.; Zondervan, K.; Salumets, A.; Götte, M.; Peters, M. Challenges in Endometriosis MiRNA Studies—From Tissue Heterogeneity to Disease Specific MiRNAs. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2282–2292. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Hu, Y.; Cui, Z.; Jia, H. Human Menstrual Blood: A Renewable and Sustainable Source of Stem Cells for Regenerative Medicine. Stem Cell Res. Ther. 2018, 9, 325. [Google Scholar] [CrossRef] [PubMed]
- Barra, F.; Ferro Desideri, L.; Ferrero, S. Inhibition of PI3K/AKT/MTOR Pathway for the Treatment of Endometriosis. Br. J. Pharmacol. 2018, 175, 3626–3627. [Google Scholar] [CrossRef] [PubMed]
- Makker, A.; Goel, M.M.; Das, V.; Agarwal, A. PI3K-Akt-MTOR and MAPK Signaling Pathways in Polycystic Ovarian Syndrome, Uterine Leiomyomas and Endometriosis: An Update. Gynecol. Endocrinol. 2012, 28, 175–181. [Google Scholar] [CrossRef]
- Liu, Y.; Qin, X.; Lu, X.; Jiang, J. Effects of Inhibiting the PI3K/Akt/MTOR Signaling Pathway on the Pain of Sciatic Endometriosis in a Rat Model. Can. J. Physiol. Pharmacol. 2019, 97, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Samartzis, E.P.; Noske, A.; Dedes, K.J.; Fink, D.; Imesch, P. ARID1A Mutations and PI3K/AKT Pathway Alterations in Endometriosis and Endometriosis-Associated Ovarian Carcinomas. Int. J. Mol. Sci 2013, 14, 18824–18849. [Google Scholar] [CrossRef]
- Porta, C.; Paglino, C.; Mosca, A. Targeting PI3K/Akt/MTOR Signaling in Cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef]
- Kvaskoff, M.; Mu, F.; Terry, K.L.; Harris, H.R.; Poole, E.M.; Farland, L.; Missmer, S.A. Endometriosis: A High-Risk Population for Major Chronic Diseases? Hum. Reprod. Update 2015, 21, 500–516. [Google Scholar] [CrossRef]
- Dong, P.; Konno, Y.; Watari, H.; Hosaka, M.; Noguchi, M.; Sakuragi, N. The Impact of MicroRNA-Mediated PI3K/AKT Signaling on Epithelial-Mesenchymal Transition and Cancer Stemness in Endometrial Cancer. J. Transl. Med. 2014, 12, 231. [Google Scholar] [CrossRef]
- Kim, L.C.; Cook, R.S.; Chen, J. MTORC1 and MTORC2 in Cancer and the Tumor Microenvironment. Oncogene 2017, 36, 2191. [Google Scholar] [CrossRef] [Green Version]
- Young, V.J.; Ahmad, S.F.; Duncan, W.C.; Horne, A.W. The Role of TGF-β in the Pathophysiology of Peritoneal Endometriosis. Hum. Reprod. Update 2017, 23, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.L.; Stoikos, C.; Findlay, J.K.; Salamonsen, L.A. TGF-Beta Superfamily Expression and Actions in the Endometrium and Placenta. Reproduction 2006, 132, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Omwandho, C.O.A.; Konrad, L.; Halis, G.; Oehmke, F.; Tinneberg, H.R. Role of TGF-Betas in Normal Human Endometrium and Endometriosis. Hum. Reprod. 2010, 25, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Li, W.N.; Wu, M.H.; Tsai, S.J. Hypoxia and Reproductive Health: The Role of Hypoxia in the Development and Progression of Endometriosis. Reproduction 2021, 161, F19–F31. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, K.Y.; Chang, N.; Tsai, J.L.; Lin, S.C.; Tsai, S.J.; Wu, M.H. Hypoxia-Inhibited DUSP2 Expression Promotes IL-6/STAT3 Signaling in Endometriosis. Am. J. Reprod. Immunol. 2017, 78, e12690. [Google Scholar] [CrossRef] [PubMed]
- Birt, J.A.; Nabli, H.; Stilley, J.A.; Windham, E.A.; Frazier, S.R.; Sharpe-Timms, K.L. Elevated Peritoneal Fluid TNF-α Incites Ovarian Early Growth Response Factor 1 Expression and Downstream Protease Mediators: A Correlation with Ovulatory Dysfunction in Endometriosis. Reprod. Sci. 2013, 20, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Situmorang, H.; Hestiantoro, A.; Purbadi, S.; Flamandita, D.; Sahlan, M. IN-SILICO Dynamic Analysis of Sulawesi Propolis as Anti-Endometriosis Drug: Interaction Study with TNF Alpha Receptor, NF-KB, Estrogen Receptor, Progesterone Receptor and Prostaglandin Receptor. Ann. Med. Surg. 2021, 67, 102459. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, S.; Darcha, C. Epithelial to Mesenchymal Transition-like and Mesenchymal to Epithelial Transition-like Processes Might Be Involved in the Pathogenesis of Pelvic Endometriosis. Hum. Reprod. 2012, 27, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.M.; Yang, W.X. Epithelial-to-Mesenchymal Transition in the Development of Endometriosis. Oncotarget 2017, 8, 41679–41689. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhou, Y.; Xu, H.; Hill, C.; Ewing, R.M.; He, D.; Zhang, X.; Wang, Y. Bioinformatic Analysis Reveals the Importance of Epithelial-Mesenchymal Transition in the Development of Endometriosis. Sci. Rep. 2020, 10, 8442. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, C.; Fan, L.; Wang, J.; Li, T.; Liu, Z.; Sheng, J.; Qian, R.Y.; Duan, A.; Lu, D. MiR-199a-5p Targets ZEB1 to Inhibit the Epithelial-Mesenchymal Transition of Ovarian Ectopic Endometrial Stromal Cells Via PI3K/Akt/MTOR Signal Pathway In Vitro and In Vivo. Reprod. Sci. 2020, 27, 110–118. [Google Scholar] [CrossRef]
- Jing, X.; Peng, J.; Dou, Y.; Sun, J.; Ma, C.; Wang, Q.; Zhang, L.; Luo, X.; Kong, B.; Zhang, Y.; et al. Macrophage ERα Promoted Invasion of Endometrial Cancer Cell by MTOR/KIF5B-Mediated Epithelial to Mesenchymal Transition. Immunol. Cell Biol. 2019, 97, 563–576. [Google Scholar] [CrossRef]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 Polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef]
- Bonatz, G.; Hansmann, M.L.; Buchholz, F.; Mettler, L.; Radzun, H.J.; Semm, K. Macrophage- and Lymphocyte-Subtypes in the Endometrium during Different Phases of the Ovarian Cycle. Int. J. Gynaecol. Obstet. 1992, 37, 29–36. [Google Scholar] [CrossRef]
- Maybin, J.A.; Critchley, H.O.D. Menstrual Physiology: Implications for Endometrial Pathology and Beyond. Hum. Reprod. Update 2015, 21, 748–761. [Google Scholar] [CrossRef]
- Abomaray, F.; Gidlöf, S.; Götherström, C. Mesenchymal Stromal Cells Are More Immunosuppressive In Vitro If They Are Derived from Endometriotic Lesions than from Eutopic Endometrium. Stem Cells Int. 2017, 2017, 3215962. [Google Scholar] [CrossRef]
- Revised American Society for Reproductive Medicine Classification of Endometriosis: 1996. Fertil. Steril. 1997, 67, 817–821. [CrossRef]
- Cheng, Y.; Li, L.; Wang, D.; Guo, Q.; He, Y.; Liang, T.; Sun, L.; Wang, X.; Cheng, Y.; Zhang, G. Characteristics of Human Endometrium-Derived Mesenchymal Stem Cells and Their Tropism to Endometriosis. Stem Cells Int. 2017, 2017, 4794827. [Google Scholar] [CrossRef] [PubMed]
- Zucherato, V.S.; Penariol, L.B.C.; Silva, L.E.C.M.; Padovan, C.C.; Poli-Neto, O.B.; Rosa-E-Silva, J.C.; Ferriani, R.A.; Meola, J. Identification of Suitable Reference Genes for Mesenchymal Stem Cells from Menstrual Blood of Women with Endometriosis. Sci. Rep. 2021, 11, 5422. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhao, S.; Yin, L.; Guo, Y.; Sheng, Q.; Shyr, Y. Package “heatmap3” Type Package Title An Improved Heatmap Package. 2021. Available online: https://cran.r-project.org/web/packages/heatmap3/index.html (accessed on 1 June 2021).
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Rogers-Broadway, K.R.; Kumar, J.; Sisu, C.; Wander, G.; Mazey, E.; Jeyaneethi, J.; Pados, G.; Tsolakidis, D.; Klonos, E.; Grunt, T.; et al. Differential Expression of MTOR Components in Endometriosis and Ovarian Cancer: Effects of Rapalogues and Dual Kinase Inhibitors on MTORC1 and MTORC2 Stoichiometry. Int. J. Mol. Med. 2019, 43, 47. [Google Scholar] [CrossRef]

| Ensemble ID. | Official Gene Symbol | Gene Type | Chromosome Location | Gene Name | Log2fc | p-Value | P-Adj | Endometriosis-Related Gene/Protein in the Literature |
|---|---|---|---|---|---|---|---|---|
| ENSG00000162772 | ATF3 | Protein coding | 1q32.3 | Activating transcription factor 3 | 1.782043 | 0.000234 | 0.099158 | [38] |
| ENSG00000214212 | C19orf38 | Protein coding | 19p13.2 | Chromosome 19 open reading frame 38 | 1.756059 | 0.000165 | 0.073847 | |
| ENSG00000121898 | CPXM2 | Protein coding | 10q26.13 | Carboxypeptidase X, M14 family member 2 | 2.647503 | 0.000124 | 0.058576 | |
| ENSG00000144655 | CSRNP1 | Protein coding | 3p22.2 | Cysteine and serine-rich nuclear protein 1 | 1.076952 | 5.07 × 10−5 | 0.036267 | |
| ENSG00000120738 | EGR1 | Protein coding | 5q31.2 | Early growth response 1 | 1.217478 | 8.09 × 10−5 | 0.049207 | [39] |
| ENSG00000125740 | FOSB | Protein coding | 19q13.32 | FosB proto-oncogene, AP-1 transcription factor subunit | 1.50465 | 8.48 × 10−6 | 0.009987 | [26,40] |
| ENSG00000114315 | HES1 | Protein coding | 3q29 | Hes family bHLH transcription factor 1 | 2.103473 | 5.75 × 10−7 | 0.001951 | |
| ENSG00000125968 | ID1 | Protein coding | 20q11.21 | Inhibitor of DNA binding 1, HLH protein | 1.282674 | 9.56 × 10−5 | 0.054622 | [41,42] |
| ENSG00000117318 | ID3 | Protein coding | 1p36.12 | Inhibitor of DNA binding 3, HLH protein | 1.530359 | 6.71 × 10−6 | 0.009036 | [41,42] |
| ENSG00000211643 | IGLV5–52 | Protein coding | 22q11.22 | Immunoglobulin Lambda Variable 5–52 | 4.818943 | 0.000117 | 0.056648 | |
| ENSG00000050555 | LAMC3 | Protein coding | 9q34.12 | Laminin subunit gamma 3 | 2.187422 | 6.99 × 10−5 | 0.043905 | [43,44] |
| ENSG00000248672 | LY75-CD302 | Protein coding | 2q24.2 | LY75-CD302 readthrough | 2.969177 | 0.000148 | 0.068122 | |
| ENSG00000123358 | NR4A1 | Protein coding | 12q13.13 | Nuclear receptor subfamily 4 group A member 1 | 1.965574 | 0.000114 | 0.056648 | [45] |
| ENSG00000153234 | NR4A2 | Protein coding | 2q24.1 | Nuclear receptor subfamily 4 group A member 2 | 1.993967 | 1.86 × 10−5 | 0.015948 | |
| ENSG00000119508 | NR4A3 | Protein coding | 9q31.1 | Nuclear receptor subfamily 4 group A member 3 | 2.200431 | 0.000237 | 0.099158 | |
| ENSG00000124216 | SNAI1 | Protein coding | 20q13.13 | Snail family transcriptional repressor 1 | 1.473873 | 1.20 × 10−5 | 0.012836 | [46,47,48,49] |
| ENSG00000157734 | SNX22 | Protein coding | 15q22.31 | Sorting nexin 22 | 1.968101 | 6.28 × 10−5 | 0.040804 | |
| ENSG00000128016 | ZFP36 | Protein coding | 19q13.2 | ZFP36 ring finger protein | 1.512248 | 8.75 × 10−5 | 0.05156 | [39] |
| ENSG00000070087 | PFN2 | Protein coding | 3q25.1 | Profilin 2 | −0.52405 | 2.42 × 10−5 | 0.019852 |
| Official Protein Symbol. | Chromosome Location | Protein Name | Log2fc | p-Value | Endometriosis-Related Gene/Protein in the Literature |
|---|---|---|---|---|---|
| ANXA3 | 4q21.21 | Annexin A3 | −1.57 | 0.00050 | |
| EPS8L2 | 11p15.5 | EPS8 like 2 | −1.06 | 0.01287 | |
| NUP62 | 19q13.33 | Nucleoporin 62 | −1.05 | 0.02018 | |
| MT2A | 16q13 | Metallothionein 2A | −1.02 | 0.03902 | [50] |
| SF3A2 | 19p13.3 | Splicing factor 3a subunit 2 | −1.02 | 0.02382 | |
| TYMP | 22q13.33 | Thymidine phosphorylase | −1.01 | 0.01438 | [51] |
| LUC7L2 | 7q34 | LUC7 like 2, pre-mRNA splicing factor | −1.01 | 0.01209 | |
| ARL8B | 3p26.1 | ADP ribosylation factor-like GTPase 8B | −1.01 | 0.01648 | |
| MANF | 3p21.2 | Mesencephalic astrocyte-derived neurotrophic factor | 1.11 | 0.00944 | |
| ACBD3 | 1q42.12 | Acyl-CoA binding domain-containing 3 | 1.14 | 0.01693 | |
| COL1A1 | 17q21.33 | Collagen type I alpha 1 chain | 1.15 | 0.01517 | [52] |
| COL6A2 | 21q22.3 | Collagen type VI alpha 2 chain | 1.16 | 0.01035 | [53] |
| APMAP | 20p11.21 | Adipocyte plasma membrane-associated protein | 1.18 | 0.02613 | |
| SRP68 | 17q25.1 | Signal recognition particle 68 | 1.23 | 0.00261 | |
| NID2 | 14q22.1 | Nidogen 2 | 1.45 | 0.00629 | [54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penariol, L.B.C.; Thomé, C.H.; Tozetti, P.A.; Paier, C.R.K.; Buono, F.O.; Peronni, K.C.; Orellana, M.D.; Covas, D.T.; Moraes, M.E.A.; Silva, W.A., Jr.; et al. What Do the Transcriptome and Proteome of Menstrual Blood-Derived Mesenchymal Stem Cells Tell Us about Endometriosis? Int. J. Mol. Sci. 2022, 23, 11515. https://doi.org/10.3390/ijms231911515
Penariol LBC, Thomé CH, Tozetti PA, Paier CRK, Buono FO, Peronni KC, Orellana MD, Covas DT, Moraes MEA, Silva WA Jr., et al. What Do the Transcriptome and Proteome of Menstrual Blood-Derived Mesenchymal Stem Cells Tell Us about Endometriosis? International Journal of Molecular Sciences. 2022; 23(19):11515. https://doi.org/10.3390/ijms231911515
Chicago/Turabian StylePenariol, Letícia B. C., Carolina H. Thomé, Patrícia A. Tozetti, Carlos R. K. Paier, Fabiana O. Buono, Kamila C. Peronni, Maristela D. Orellana, Dimas T. Covas, Maria E. A. Moraes, Wilson A. Silva, Jr., and et al. 2022. "What Do the Transcriptome and Proteome of Menstrual Blood-Derived Mesenchymal Stem Cells Tell Us about Endometriosis?" International Journal of Molecular Sciences 23, no. 19: 11515. https://doi.org/10.3390/ijms231911515
APA StylePenariol, L. B. C., Thomé, C. H., Tozetti, P. A., Paier, C. R. K., Buono, F. O., Peronni, K. C., Orellana, M. D., Covas, D. T., Moraes, M. E. A., Silva, W. A., Jr., Rosa-e-Silva, J. C., Ferriani, R. A., Faça, V. M., Poli-Neto, O. B., Tiezzi, D. G., & Meola, J. (2022). What Do the Transcriptome and Proteome of Menstrual Blood-Derived Mesenchymal Stem Cells Tell Us about Endometriosis? International Journal of Molecular Sciences, 23(19), 11515. https://doi.org/10.3390/ijms231911515





