Functional Mimicry of Eukaryotic Actin Assembly by Pathogen Effector Proteins
Abstract
:1. Introduction
2. Actin Assembly in Eukaryotes
2.1. Arp2/3
2.2. Formins
2.3. Ena/VASP
2.4. Tandem Actin Monomer-Binding Proteins
3. Actin Assembly by Pathogenic Effector Proteins
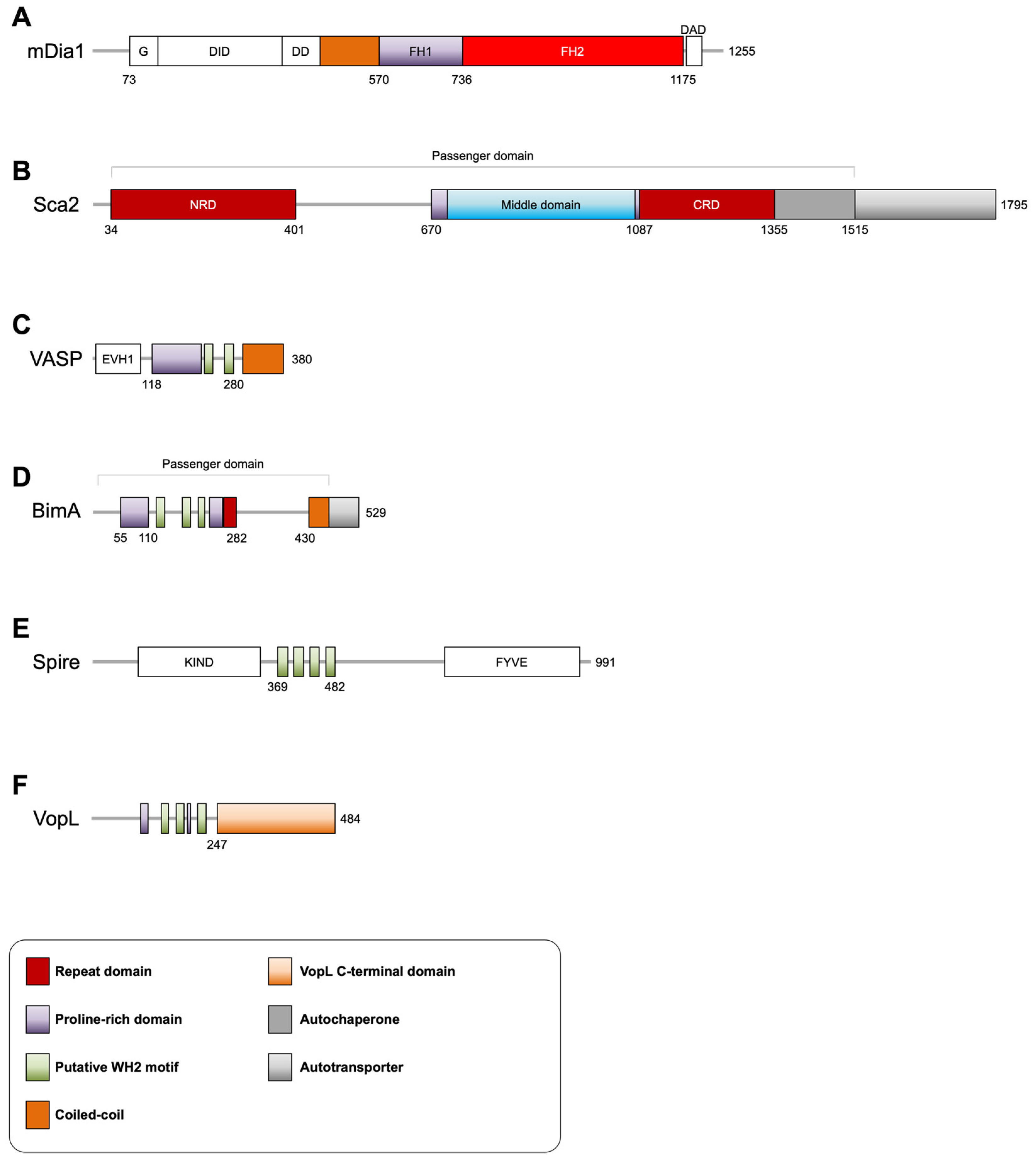
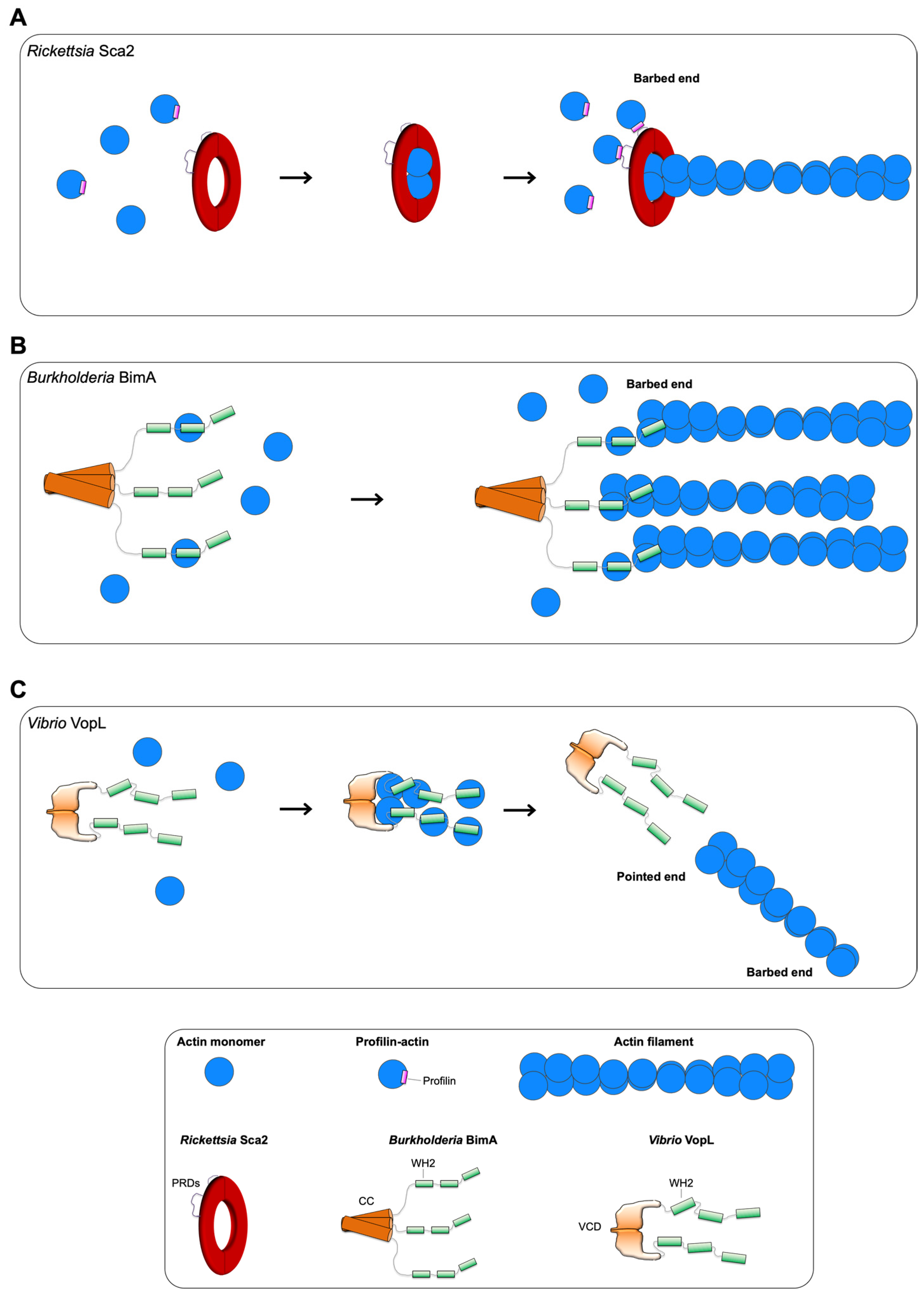
3.1. Rickettsia Sca2
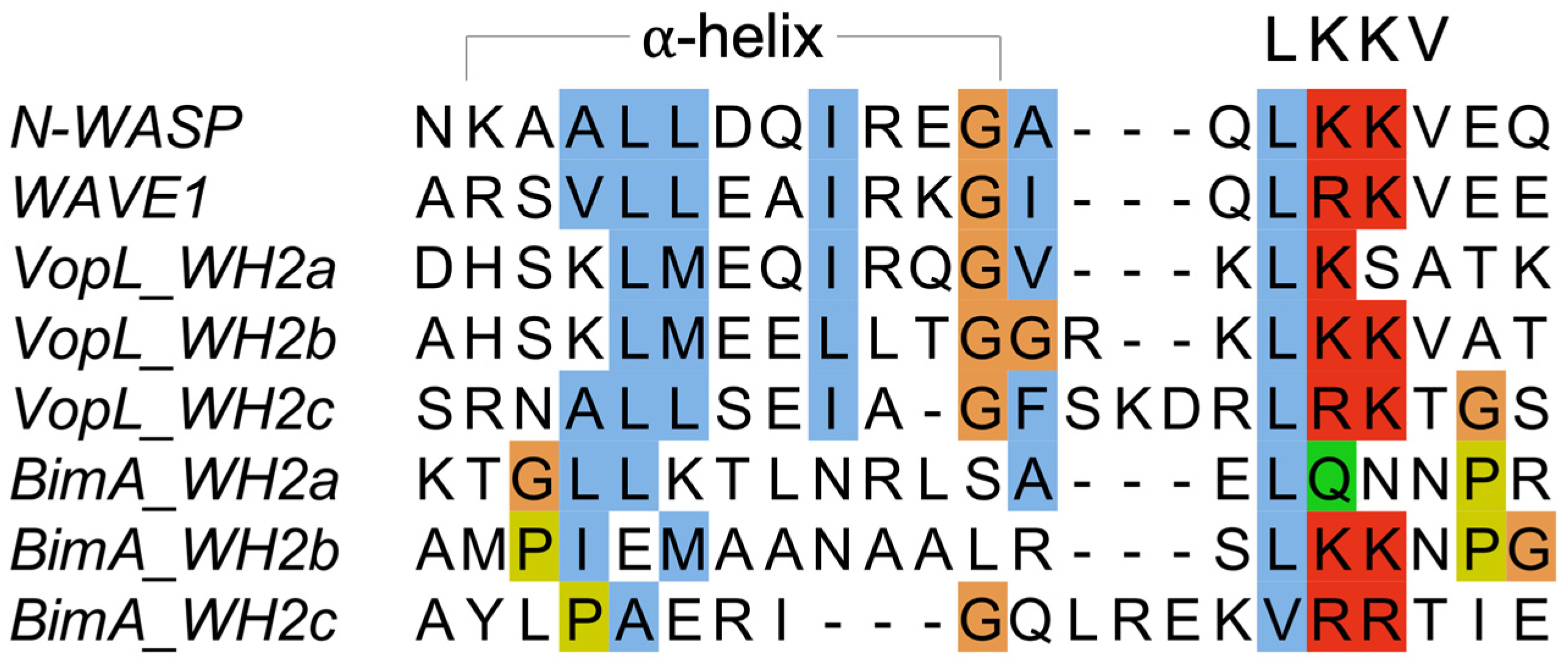
3.2. Burkholderia BimA
3.3. Vibrio VopL
3.4. Other Effectors
4. Lessons Learned
5. Outlook and Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Souza Santos, M.; Orth, K. Subversion of the cytoskeleton by intracellular bacteria: Lessons from Listeria, Salmonella and Vibrio. Cell. Microbiol. 2015, 17, 164–173. [Google Scholar] [CrossRef]
- Colonne, P.M.; Winchell, C.G.; Voth, D.E. Hijacking Host Cell Highways: Manipulation of the Host Actin Cytoskeleton by Obligate Intracellular Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 107. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, A.; Chen, D.; Alto, N.M. How Bacteria Subvert Animal Cell Structure and Function. Annu. Rev. Cell Dev. Biol. 2016, 32, 373–397. [Google Scholar] [CrossRef] [Green Version]
- Stradal, T.E.B.; Schelhaas, M. Actin Dynamics in Host-Pathogen Interaction. FEBS Lett. 2018, 592, 3658–3669. [Google Scholar] [CrossRef] [Green Version]
- Pollard, T.D.; Cooper, J.A. Actin, a Central Player in Cell Shape and Movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef] [Green Version]
- Cossart, P.; Helenius, A. Endocytosis of Viruses and Bacteria. Cold Spring Harb. Perspect. Biol. 2014, 6, a016972. [Google Scholar] [CrossRef] [Green Version]
- Choe, J.E.; Welch, M.D. Actin-based motility of bacterial pathogens: Mechanistic diversity and its impact on virulence. FEMS Pathog. Dis. 2016, 74, ftw099. [Google Scholar] [CrossRef]
- Lamason, R.L.; Welch, M.D. Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr. Opin. Microbiol. 2017, 35, 48–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popoff, M.R. Bacterial factors exploit eukaryotic Rho GTPase signaling cascades to promote invasion and proliferation within their host. Small GTPases 2014, 5, e983863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haglund, C.M.; Welch, M.D. Pathogens and polymers: Microbe–host interactions illuminate the cytoskeleton. J. Cell Biol. 2011, 195, 7–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Hayes, D.B.; Rebowski, G.; Tardieux, I.; Dominguez, R. Toxofilin from Toxoplasma gondii forms a ternary complex with an antiparallel actin dimer. Proc. Natl. Acad. Sci. USA 2007, 104, 16122–16127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aktories, K.; Lang, A.E.; Schwan, C.; Mannherz, H.G. Actin as target for modification by bacterial protein toxins. FEBS J. 2011, 278, 4526–4543. [Google Scholar] [CrossRef] [PubMed]
- Heisler, D.B.; Kudryashova, E.; Grinevich, D.O.; Suarez, C.; Winkelman, J.D.; Birukov, K.G.; Kotha, S.R.; Parinandi, N.L.; Vavylonis, D.; Kovar, D.R.; et al. ACD toxin–produced actin oligomers poison formin-controlled actin polymerization. Science 2015, 349, 535–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domínguez, R. Subversive bacteria reveal new tricks in their cytoskeleton-hijacking arsenal. Nat. Struct. Mol. Biol. 2015, 22, 178–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhu, W.; Tan, Y.; Nakayasu, E.S.; Staiger, C.J.; Luo, Z.-Q. A Legionella Effector Disrupts Host Cytoskeletal Structure by Cleaving Actin. PLoS Pathog. 2017, 13, e1006186. [Google Scholar] [CrossRef] [Green Version]
- Bugalhão, J.N.; Mota, L.J.; Franco, I.S. Bacterial nucleators: Actin’ on actin. Pathog. Dis. 2015, 73, ftv078. [Google Scholar] [CrossRef] [Green Version]
- Elde, N.C.; Malik, H.S. The evolutionary conundrum of pathogen mimicry. Nat. Rev. Genet. 2009, 7, 787–797. [Google Scholar] [CrossRef]
- Stebbins, C.E.; Galan, J.E. Structural mimicry in bacterial virulence. Nature 2001, 412, 701–705. [Google Scholar] [CrossRef]
- Rosenbloom, A.D.; Kovar, E.W.; Kovar, D.R.; Loew, L.M.; Pollard, T.D. Mechanism of actin filament nucleation. Biophys. J. 2021, 120, 4399–4417. [Google Scholar] [CrossRef]
- Funk, J.; Merino, F.; Schaks, M.; Rottner, K.; Raunser, S.; Bieling, P. A barbed end interference mechanism reveals how capping protein promotes nucleation in branched actin networks. Nat. Commun. 2021, 12, 5329. [Google Scholar] [CrossRef]
- Zweifel, M.E.; Sherer, L.A.; Mahanta, B.; Courtemanche, N. Nucleation limits the lengths of actin filaments assembled by formin. Biophys. J. 2021, 120, 4442–4456. [Google Scholar] [CrossRef] [PubMed]
- Gautreau, A.M.; Fregoso, F.E.; Simanov, G.; Dominguez, R. Nucleation, stabilization, and disassembly of branched actin networks. Trends Cell Biol. 2021, 32, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, R. Actin filament nucleation and elongation factors—Structure-function relationships. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 351–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domínguez, R. Structural insights into de novo actin polymerization. Curr. Opin. Struct. Biol. 2010, 20, 217–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merino, F.; Pospich, S.; Raunser, S. Towards a structural understanding of the remodeling of the actin cytoskeleton. Semin. Cell Dev. Biol. 2020, 102, 51–64. [Google Scholar] [CrossRef]
- Lappalainen, P.; Kotila, T.; Jégou, A.; Romet-Lemonne, G. Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol. 2022, 1–17. [Google Scholar] [CrossRef]
- Sept, D.; McCammon, J.A. Thermodynamics and Kinetics of Actin Filament Nucleation. Biophys. J. 2001, 81, 667–674. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, R. The beta-Thymosin/WH2 Fold: Multifunctionality and Structure. Ann. N. Y. Acad. Sci. 2007, 1112, 86–94. [Google Scholar] [CrossRef]
- Pollard, T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 451–477. [Google Scholar] [CrossRef]
- Welch, M.D.; Way, M. Arp2/3-mediated actin-based motility: A tail of pathogen abuse. Cell Host Microbe 2013, 14, 242–255. [Google Scholar] [CrossRef] [Green Version]
- Newsome, T.P.; Marzook, N.B. Viruses that ride on the coat-tails of actin nucleation. Semin. Cell Dev. Biol. 2015, 46, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.L.; Eck, M.J. Mechanism and Function of Formins in the Control of Actin Assembly. Annu. Rev. Biochem. 2007, 76, 593–627. [Google Scholar] [CrossRef] [PubMed]
- Courtemanche, N. Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 2018, 10, 1553–1569. [Google Scholar] [CrossRef] [PubMed]
- Funk, J.; Merino, F.; Venkova, L.; Heydenreich, L.; Kierfeld, J.; Vargas, P.; Raunser, S.; Piel, M.; Bieling, P. Profilin and formin constitute a pacemaker system for robust actin filament growth. eLife 2019, 8, e50963. [Google Scholar] [CrossRef] [PubMed]
- Homa, K.E.; Zsolnay, V.; Anderson, C.A.; O’Connell, M.E.; Neidt, E.M.; Voth, G.A.; Bidone, T.C.; Kovar, D.R. Formin Cdc12′s specific actin assembly properties are tailored for cytokinesis in fission yeast. Biophys. J. 2021, 120, 2984–2997. [Google Scholar] [CrossRef]
- Breitsprecher, D.; Kiesewetter, A.K.; Linkner, J.; Vinzenz, M.; Stradal, T.E.B.; Small, J.V.; Curth, U.; Dickinson, R.B.; Faix, J. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 2011, 30, 456–467. [Google Scholar] [CrossRef]
- Ferron, F.; Rebowski, G.; Lee, S.H.; Dominguez, R. Structural basis for the recruitment of profilin–actin complexes during filament elongation by Ena/VASP. EMBO J. 2007, 26, 4597–4606. [Google Scholar] [CrossRef] [Green Version]
- Krause, M.; Dent, E.W.; Bear, J.E.; Loureiro, J.J.; Gertler, F.B. Ena/VASP Proteins: Regulators of the Actin Cytoskeleton and Cell Migration. Annu. Rev. Cell Dev. Biol. 2003, 19, 541–564. [Google Scholar] [CrossRef] [Green Version]
- Hansen, S.D.; Mullins, R.D. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 2010, 191, 571–584. [Google Scholar] [CrossRef] [Green Version]
- Winkelman, J.D.; Bilancia, C.G.; Peifer, M.; Kovar, D.R. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc. Natl. Acad. Sci. USA 2014, 111, 4121–4126. [Google Scholar] [CrossRef] [Green Version]
- Brühmann, S.; Ushakov, D.S.; Winterhoff, M.; Dickinson, R.B.; Curth, U.; Faix, J. Distinct VASP tetramers synergize in the processive elongation of individual actin filaments from clustered arrays. Proc. Natl. Acad. Sci. USA 2017, 114, E5815–E5824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harker, A.J.; Katkar, H.; Bidone, T.C.; Aydin, F.; Voth, G.A.; Applewhite, D.A.; Kovar, D.R. Ena/VASP processive elongation is modulated by avidity on actin filaments bundled by the filopodia cross-linker fascin. Mol. Biol. Cell 2019, 30, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, M.E.; Heuser, J.E.; Kerkhoff, E.; Mullins, R.D. Drosophila Spire is an actin nucleation factor. Nature 2005, 433, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, R.; Pinyol, R.; Reichenbach, N.; Custer, L.; Klingensmith, J.; Kessels, M.M.; Qualmann, B. Cordon-Bleu Is an Actin Nucleation Factor and Controls Neuronal Morphology. Cell 2007, 131, 337–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, V.M.; Dominguez, R. Tropomodulins and Leiomodins: Actin Pointed End Caps and Nucleators in Muscles. Biophys. J. 2017, 112, 1742–1760. [Google Scholar] [CrossRef] [Green Version]
- Rebowski, G.; Boczkowska, M.; Hayes, D.B.; Guo, L.; Irving, T.C.; Dominguez, R. X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc. Natl. Acad. Sci. USA 2008, 105, 10785–10790. [Google Scholar] [CrossRef] [Green Version]
- Qualmann, B.; Kessels, M.M. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 2009, 19, 276–285. [Google Scholar] [CrossRef]
- Rebowski, G.; Namgoong, S.; Boczkowska, M.; Leavis, P.C.; Navaza, J.; Dominguez, R. Structure of a Longitudinal Actin Dimer Assembled by Tandem W Domains: Implications for Actin Filament Nucleation. J. Mol. Biol. 2010, 403, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, R. The WH2 Domain and Actin Nucleation: Necessary but Insufficient. Trends Biochem. Sci. 2016, 41, 478–490. [Google Scholar] [CrossRef] [Green Version]
- McGinn, J.; Lamason, R.L. The enigmatic biology of rickettsiae: Recent advances, open questions and outlook. Pathog. Dis. 2021, 79, ftab019. [Google Scholar] [CrossRef]
- Voss, O.H.; Rahman, M.S. Rickettsia-Host interaction: Strategies of intracytosolic host colonization. Pathog. Dis. 2021, 79, ftab015. [Google Scholar] [CrossRef] [PubMed]
- Narra, H.P.; Sahni, A.; Walker, D.H.; Sahni, S.K. Recent research milestones in the pathogenesis of human rickettsioses and opportunities ahead. Futur. Microbiol. 2020, 15, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Salje, J. Cells within cells: Rickettsiales and the obligate intracellular bacterial lifestyle. Nat. Rev. Genet. 2021, 19, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Haglund, C.M.; Choe, J.E.; Skau, C.T.; Kovar, D.R.; Welch, M.D. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 2010, 12, 1057–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleba, B.; Clark, T.R.; Lutter, E.I.; Ellison, D.W.; Hackstadt, T. Disruption of the Rickettsia rickettsii Sca2 Autotransporter Inhibits Actin-Based Motility. Infect. Immun. 2010, 78, 2240–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, S.C.; Lamason, R.L.; Risca, V.I.; Abernathy, E.; Welch, M.D. Rickettsia Actin-Based Motility Occurs in Distinct Phases Mediated by Different Actin Nucleators. Curr. Biol. 2014, 24, 98–103. [Google Scholar] [CrossRef] [Green Version]
- Meuskens, I.; Saragliadis, A.; Leo, J.C.; Linke, D. Type V Secretion Systems: An Overview of Passenger Domain Functions. Front. Microbiol. 2019, 10, 1163. [Google Scholar] [CrossRef] [Green Version]
- Madasu, Y.; Suarez, C.; Kast, D.J.; Kovar, D.R.; Dominguez, R. Rickettsia Sca2 has evolved formin-like activity through a different molecular mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, E2677–E2686. [Google Scholar] [CrossRef] [Green Version]
- Alqassim, S.S.; Lee, I.-G.; Dominguez, R. Rickettsia Sca2 Recruits Two Actin Subunits for Nucleation but Lacks WH2 Domains. Biophys. J. 2019, 116, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, H.D. Looks can be deceiving: Recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol. Microbiol. 2015, 97, 205–215. [Google Scholar] [CrossRef]
- Xu, Y.; Moseley, J.B.; Sagot, I.; Poy, F.; Pellman, D.; Goode, B.L.; Eck, M.J. Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture. Cell 2004, 116, 711–723. [Google Scholar] [CrossRef] [Green Version]
- Otomo, T.; Tomchick, D.R.; Otomo, C.; Panchal, S.C.; Machius, M.; Rosen, M.K. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 2005, 433, 488–494. [Google Scholar] [CrossRef]
- Sherer, L.A.; Zweifel, M.E.; Courtemanche, N. Dissection of two parallel pathways for formin-mediated actin filament elongation. J. Biol. Chem. 2018, 293, 17917–17928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zweifel, M.E.; Courtemanche, N. Competition for delivery of profilin–actin to barbed ends limits the rate of formin-mediated actin filament elongation. J. Biol. Chem. 2020, 295, 4513–4525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gassiep, I.; Armstrong, M.; Norton, R. Human Melioidosis. Clin. Microbiol. Rev. 2020, 33, e00006-19. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.P.; Stevens, J.M.; Jeng, R.L.; Taylor, L.A.; Wood, M.W.; Hawes, P.; Monaghan, P.; Welch, M.D.; Galyov, E.E. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 2005, 56, 40–53. [Google Scholar] [CrossRef]
- Sitthidet, C.; Korbsrisate, S.; Layton, A.N.; Field, T.R.; Stevens, M.P.; Stevens, J.M. Identification of motifs of Burkholderia pseudomallei BimA required for intracellular motility, actin binding, and actin polymerization. J. Bacteriol. 2011, 193, 1901–1910. [Google Scholar] [CrossRef] [Green Version]
- French, C.T.; Toesca, I.J.; Wu, T.-H.; Teslaa, T.; Beaty, S.M.; Wong, W.; Liu, M.; Schröder, I.; Chiou, P.-Y.; Teitell, M.A.; et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc. Natl. Acad. Sci. USA 2011, 108, 12095–12100. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Singh, P.; Robertson, J.D.; LeRoux, M.; Skerrett, S.J.; Goodlett, D.R.; West, T.E.; Mougous, J.D. VgrG-5 Is a Burkholderia Type VI Secretion System-Exported Protein Required for Multinucleated Giant Cell Formation and Virulence. Infect. Immun. 2014, 82, 1445–1452. [Google Scholar] [CrossRef] [Green Version]
- Benanti, E.L.; Nguyen, C.M.; Welch, M.D. Virulent Burkholderia Species Mimic Host Actin Polymerases to Drive Actin-Based Motility. Cell 2015, 161, 348–360. [Google Scholar] [CrossRef] [Green Version]
- Dautin, N.; Bernstein, H.D. Protein Secretion in Gram-Negative Bacteria via the Autotransporter Pathway. Annu. Rev. Microbiol. 2007, 61, 89–112. [Google Scholar] [CrossRef]
- Cotter, S.E.; Surana, N.K.; St Geme, J.W., 3rd. Trimeric autotransporters: A distinct subfamily of autotransporter proteins. Trends Microbiol. 2005, 13, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.D.; Mullins, R.D. Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. eLife 2015, 4, e06585. [Google Scholar] [CrossRef] [PubMed]
- Reidl, J.; Klose, K.E. Vibrio cholerae and cholera: Out of the water and into the host. FEMS Microbiol. Rev. 2002, 26, 125–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, J.G., Jr. Cholera and other types of vibriosis: A story of human pandemics and oysters on the half shell. Clin. Infect. Dis. 2003, 37, 272–280. [Google Scholar] [CrossRef]
- Yeung, P.S.; Boor, K.J. Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog. Dis. 2004, 1, 74–88. [Google Scholar] [CrossRef]
- Janda, J.M.; Newton, A.E.; Bopp, C.A. Vibriosis. Clin. Lab. Med. 2015, 35, 273–288. [Google Scholar] [CrossRef]
- Dutta, D.; Kaushik, A.; Kumar, D.; Bag, S. Foodborne Pathogenic Vibrios: Antimicrobial Resistance. Front. Microbiol. 2021, 12, 638331. [Google Scholar] [CrossRef]
- Tam, V.C.; Serruto, D.; Dziejman, M.; Brieher, W.; Mekalanos, J.J. A Type III Secretion System in Vibrio cholerae Translocates a Formin/Spire Hybrid-like Actin Nucleator to Promote Intestinal Colonization. Cell Host Microbe 2007, 1, 95–107. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.D.S.; Salomon, D.; Orth, K. T3SS effector VopL inhibits the host ROS response, promoting the intracellular survival of Vibrio parahaemolyticus. PLoS Pathog. 2017, 13, e1006438. [Google Scholar] [CrossRef] [Green Version]
- Liverman, A.D.; Cheng, H.C.; Trosky, J.E.; Leung, D.W.; Yarbrough, M.L.; Burdette, D.L.; Rosen, M.K.; Orth, K. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. USA 2007, 104, 17117–17122. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Cheng, H.-C.; Brautigam, C.A.; Tomchick, D.R.; Rosen, M.K. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat. Struct. Mol. Biol. 2011, 18, 1068–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namgoong, S.; Boczkowska, M.; Glista, M.J.; Winkelman, J.D.; Rebowski, G.; Kovar, D.R.; Dominguez, R. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat. Struct. Mol. Biol. 2011, 18, 1060–1067. [Google Scholar] [CrossRef]
- Zahm, J.A.; Padrick, S.B.; Chen, Z.; Pak, C.W.; Yunus, A.A.; Henry, L.; Tomchick, D.R.; Chen, Z.; Rosen, M.K. The Bacterial Effector VopL Organizes Actin into Filament-like Structures. Cell 2013, 155, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Burke, T.A.; Harker, A.J.; Dominguez, R.; Kovar, D.R. The bacterial virulence factors VopL and VopF nucleate actin from the pointed end. J. Cell Biol. 2017, 216, 1267–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, M.E.; Hilgert, S.; Bedrossian, A.; Mullins, R.D.; Kerkhoff, E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 2007, 179, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, S.; Weiss, S.; Pleiser, S.; Kerkhoff, E. Structural and functional insights into the Spir/formin actin nucleator complex. Biol. Chem. 2013, 394, 1649–1660. [Google Scholar] [CrossRef]
- Schwintzer, L.; Koch, N.; Ahuja, R.; Grimm, J.; Kessels, M.M.; Qualmann, B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011, 30, 3147–3159. [Google Scholar] [CrossRef]
- Wayt, J.; Bretscher, A. Cordon Bleu serves as a platform at the basal region of microvilli, where it regulates microvillar length through its WH2 domains. Mol. Biol. Cell 2014, 25, 2817–2827. [Google Scholar] [CrossRef] [Green Version]
- Grega-Larson, N.E.; Crawley, S.W.; Erwin, A.L.; Tyska, M.J. Cordon bleu promotes the assembly of brush border microvilli. Mol. Biol. Cell 2015, 26, 3803–3815. [Google Scholar] [CrossRef]
- Pernier, J.; Orban, J.; Avvaru, B.S.; Jégou, A.; Romet-Lemonne, G.; Guichard, B.; Carlier, M.-F. Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat. Struct. Mol. Biol. 2013, 20, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Avvaru, B.S.; Pernier, J.; Carlier, M.-F. Dimeric WH2 repeats of VopF sequester actin monomers into non-nucleating linear string conformations: An X-ray scattering study. J. Struct. Biol. 2015, 190, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Jewett, T.J.; Fischer, E.R.; Mead, D.J.; Hackstadt, T. Chlamydial TARP is a bacterial nucleator of actin. Proc. Natl. Acad. Sci. USA 2006, 103, 15599–15604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caven, L.; Carabeo, R.A. Pathogenic Puppetry: Manipulation of the Host Actin Cytoskeleton by Chlamydia trachomatis. Int. J. Mol. Sci. 2019, 21, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiwani, S.; Ohr, R.J.; Fischer, E.R.; Hackstadt, T.; Alvarado, S.; Romero, A.; Jewett, T.J. Chlamydia trachomatis Tarp cooperates with the Arp2/3 complex to increase the rate of actin polymerization. Biochem. Biophys. Res. Commun. 2012, 420, 816–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jewett, T.J.; Miller, N.J.; Dooley, C.A.; Hackstadt, T. The Conserved Tarp Actin Binding Domain Is Important for Chlamydial Invasion. PLoS Pathog. 2010, 6, e1000997. [Google Scholar] [CrossRef] [Green Version]
- Tolchard, J.; Walpole, S.J.; Miles, A.J.; Maytum, R.; Eaglen, L.A.; Hackstadt, T.; Wallace, B.A.; Blumenschein, T.M.A. The intrinsically disordered Tarp protein from chlamydia binds actin with a partially preformed helix. Sci. Rep. 2018, 8, 1960. [Google Scholar] [CrossRef] [Green Version]
- Hayward, R.D.; Koronakis, V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999, 18, 4926–4934. [Google Scholar] [CrossRef]
- Chang, J.; Myeni, S.K.; Lin, T.L.; Wu, C.C.; Staiger, C.J.; Zhou, D. SipC multimerization promotes actin nucleation and contributes to Salmonella-induced inflammation. Mol. Microbiol. 2007, 66, 1548–1556. [Google Scholar] [CrossRef]
- Myeni, S.K.; Zhou, D. The C Terminus of SipC Binds and Bundles F-actin to Promote Salmonella Invasion. J. Biol. Chem. 2010, 285, 13357–13363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, I.S.; Shohdy, N.; Shuman, H.A. The Legionella pneumophila Effector VipA Is an Actin Nucleator That Alters Host Cell Organelle Trafficking. PLoS Pathog. 2012, 8, e1002546. [Google Scholar] [CrossRef] [Green Version]
- Bugalhão, J.N.; Mota, L.J.; Franco, I.S. Identification of regions within the Legionella pneumophila VipA effector protein involved in actin binding and polymerization and in interference with eukaryotic organelle trafficking. Microbiologyopen 2016, 5, 118–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Zhu, X.; Li, C.; Ma, Z.; Han, X.; Luo, Y.; Yang, L.; Yu, J.; Miao, Y. Xanthomonas effector XopR hijacks host actin cytoskeleton via complex coacervation. Nat. Commun. 2021, 12, 4064. [Google Scholar] [CrossRef]
- Dominguez, R. Actin-binding proteins—A unifying hypothesis. Trends Biochem. Sci. 2004, 29, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Holt, M.R.; Koffer, A. Cell motility: Proline-rich proteins promote protrusions. Trends Cell Biol. 2001, 11, 38–46. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Bhattacharyya, R.P.; Lim, W.A. The structure and function of proline recognition domains. Sci. STKE 2003, 2003, RE8. [Google Scholar] [CrossRef]
- Welch, M.D. Why should cell biologists study microbial pathogens? Mol. Biol. Cell 2015, 26, 4295–4301. [Google Scholar] [CrossRef]
- Kuhn, S.; Enninga, J. The actin comet guides the way: How Listeria actin subversion has impacted cell biology, infection biology and structural biology. Cell. Microbiol. 2020, 22, e13190. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alqassim, S.S. Functional Mimicry of Eukaryotic Actin Assembly by Pathogen Effector Proteins. Int. J. Mol. Sci. 2022, 23, 11606. https://doi.org/10.3390/ijms231911606
Alqassim SS. Functional Mimicry of Eukaryotic Actin Assembly by Pathogen Effector Proteins. International Journal of Molecular Sciences. 2022; 23(19):11606. https://doi.org/10.3390/ijms231911606
Chicago/Turabian StyleAlqassim, Saif S. 2022. "Functional Mimicry of Eukaryotic Actin Assembly by Pathogen Effector Proteins" International Journal of Molecular Sciences 23, no. 19: 11606. https://doi.org/10.3390/ijms231911606




