Alterations in Hepatocellular Carcinoma-Specific Immune Responses Following Hepatitis C Virus Elimination by Direct-Acting Antivirals
Abstract
1. Introduction
2. Results
2.1. Patient Profile
2.2. The Treatment with DAAs Induced Different Immune Responses to TAA-Derived Peptides
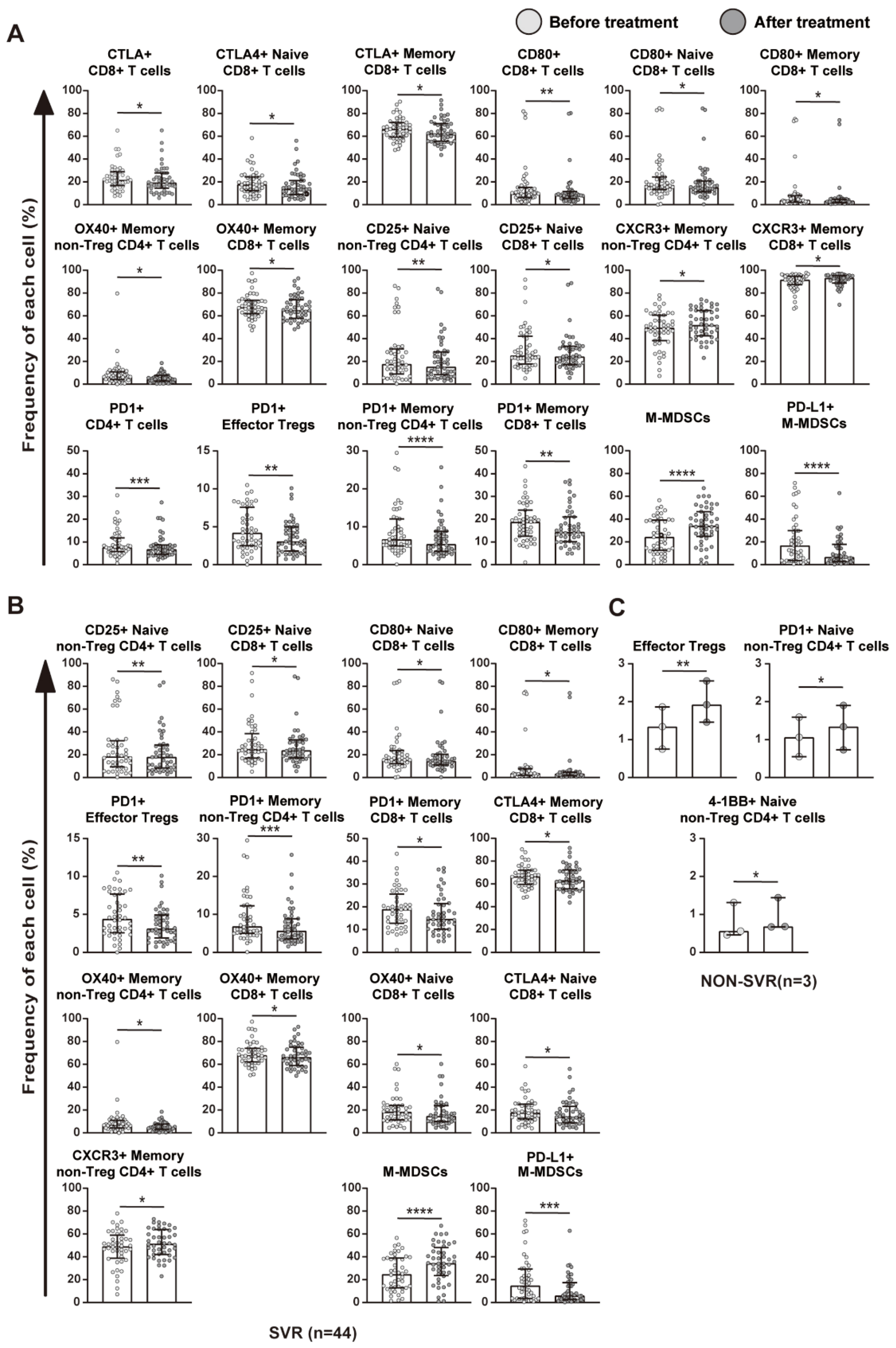
2.3. The Treatment with DAAs Significantly Altered Immune Cell Profiles
2.4. Relationship between Immune Cell Profiles and HCV Eradication
2.5. The Treatment with DAAs Enhanced Immune Responses by Decreasing the Frequency of PD-1-Expressing CD4+ and CD8+ T Cells
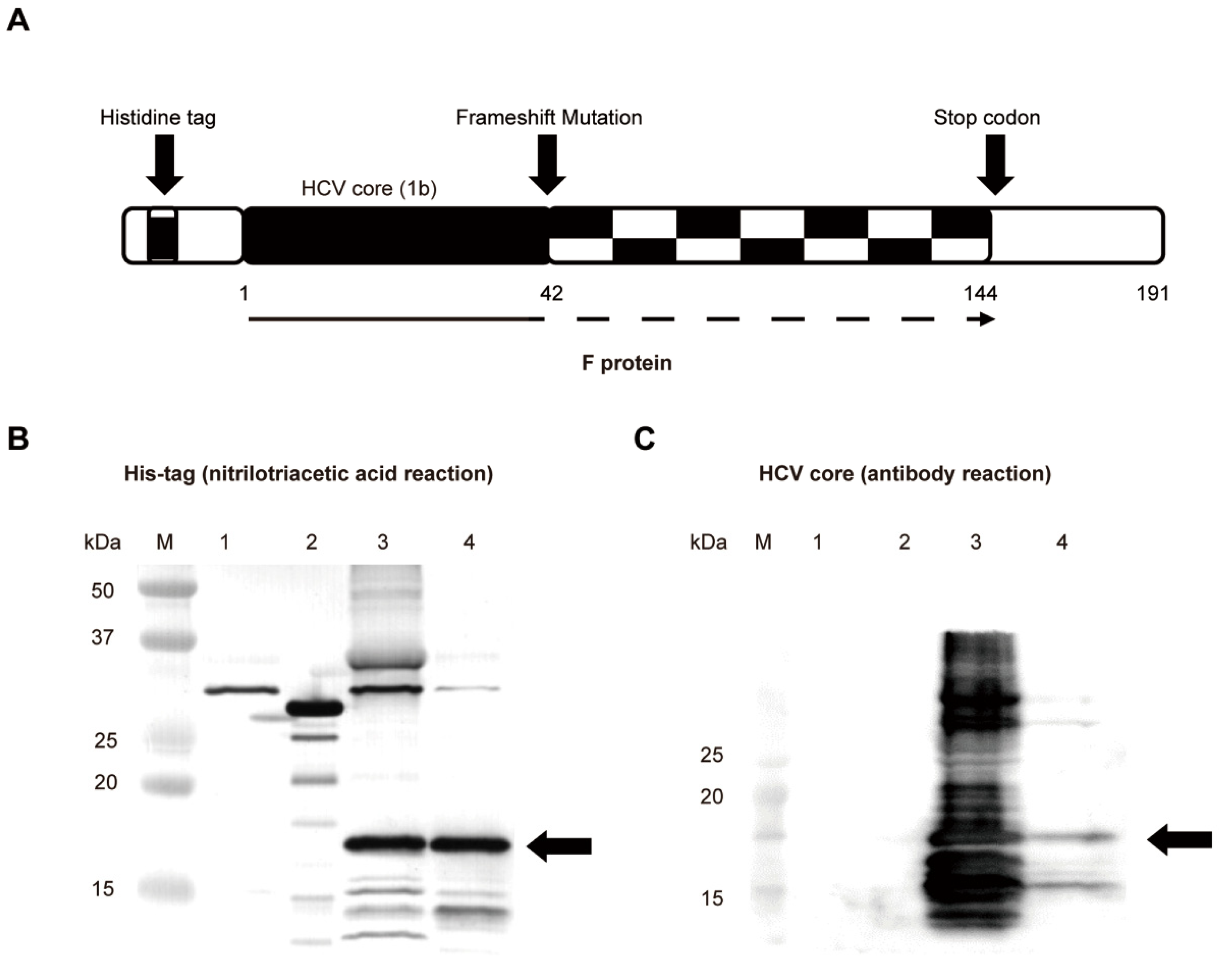
2.6. The F Protein Increased the Frequency of PD-1-Expressing CD4+ or CD8+ T Cells
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Preparation of Synthetic Peptides and PBMCs
4.3. IFN-γ ELISPOT Assay
4.4. Multicolor Fluorescence-Activated Cell Sorting Analysis
4.5. Expression and Purification of the HCV F Protein
4.6. Western Blotting
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S.; et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7. [Google Scholar] [CrossRef]
- Hajarizadeh, B.; Grebely, J.; Dore, G.J. Epidemiology and natural history of HCV infection. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 553–562. [Google Scholar] [CrossRef]
- Zhao, P.; Malik, S.; Xing, S. Epigenetic Mechanisms Involved in HCV-Induced Hepatocellular Carcinoma (HCC). Front. Oncol. 2021, 11, 677926. [Google Scholar] [CrossRef]
- Kumada, H.; Suzuki, Y.; Ikeda, K.; Toyota, J.; Karino, Y.; Chayama, K.; Kawakami, Y.; Ido, A.; Yamamoto, K.; Takaguchi, K. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology 2014, 59, 2083–2091. [Google Scholar] [CrossRef]
- Wang, H.L.; Lu, X.; Yang, X.; Xu, N. Effectiveness and safety of daclatasvir plus asunaprevir for hepatitis C virus genotype 1b: Systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2017, 32, 45–52. [Google Scholar] [CrossRef]
- Shrivastava, S.; Bhatta, M.; Ward, H.; Romani, S.; Lee, R.; Rosenthal, E.; Osinusi, A.; Kohli, A.; Masur, H.; Kottilil, S.; et al. Multitarget Direct-Acting Antiviral Therapy Is Associated With Superior Immunologic Recovery in Patients Coinfected With Human Immunodeficiency Virus and Hepatitis C Virus. Hepatol. Commun. 2018, 2, 1451–1466. [Google Scholar] [CrossRef]
- Tonnerre, P.; Wolski, D.; Subudhi, S.; Aljabban, J.; Hoogeveen, R.C.; Damasio, M.; Drescher, H.K.; Bartsch, L.M.; Tully, D.C.; Sen, D.R.; et al. Differentiation of exhausted CD8+ T cells after termination of chronic antigen stimulation stops short of achieving functional T cell memory. Nat. Immunol. 2021, 22, 1030–1041. [Google Scholar] [CrossRef]
- Hensel, N.; Gu, Z.; Sagar; Wieland, D.; Jechow, K.; Kemming, J.; Llewellyn-Lacey, S.; Gostick, E.; Sogukpinar, O.; Emmerich, F.; et al. Memory-like HCV-specific CD8+ T cells retain a molecular scar after cure of chronic HCV infection. Nat. Immunol. 2021, 22, 229–239. [Google Scholar] [CrossRef]
- Hsu, C.S.; Chao, Y.C.; Lin, H.H.; Chen, D.S.; Kao, J.H. Systematic Review: Impact of Interferon-based Therapy on HCV-related Hepatocellular Carcinoma. Sci. Rep. 2015, 5, 9954. [Google Scholar] [CrossRef]
- Conti, F.; Buonfiglioli, F.; Scuteri, A.; Crespi, C.; Bolondi, L.; Caraceni, P.; Foschi, F.G.; Lenzi, M.; Mazzella, G.; Verucchi, G.; et al. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J. Hepatol. 2016, 65, 727–733. [Google Scholar] [CrossRef]
- Kozbial, K.; Moser, S.; Schwarzer, R.; Laferl, H.; Al-Zoairy, R.; Stauber, R.; Stättermayer, A.F.; Beinhardt, S.; Graziadei, I.; Freissmuth, C.; et al. Unexpected high incidence of hepatocellular carcinoma in cirrhotic patients with sustained virologic response following interferon-free direct-acting antiviral treatment. J. Hepatol. 2016, 65, 856–858. [Google Scholar] [CrossRef] [PubMed]
- Bielen, R.; Moreno, C.; van Vlierberghe, H.; Bourgeois, S.; Mulkay, J.-P.; Vanwolleghem, T.; Verlinden, W.; Brixco, C.; Decaestecker, J.; de Galocsy, C.; et al. The risk of early occurrence and recurrence of hepatocellular carcinoma in hepatitis C-infected patients treated with direct-acting antivirals with and without pegylated interferon: A Belgian experience. J. Viral Hepat. 2017, 24, 976–981. [Google Scholar] [CrossRef]
- Calvaruso, V.; Cabibbo, G.; Cacciola, I.; Petta, S.; Madonia, S.; Bellia, A.; Tinè, F.; Distefano, M.; Licata, A.; Giannitrapani, L.; et al. Incidence of Hepatocellular Carcinoma in Patients With HCV-Associated Cirrhosis Treated With Direct-Acting Antiviral Agents. Gastroenterology 2018, 155, 411–421.e4. [Google Scholar] [CrossRef] [PubMed]
- Kamp, W.M.; Sellers, C.M.; Stein, S.; Lim, J.K.; Kim, H.S. Impact of Direct Acting Antivirals on Survival in Patients with Chronic Hepatitis C and Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Rich, N.E.; Mehta, N.; Branch, A.D.; Pillai, A.; Hoteit, M.; Volk, M.; Odewole, M.; Scaglione, S.; Guy, J.; et al. Direct-Acting Antiviral Therapy for Hepatitis C Virus Infection Is Associated With Increased Survival in Patients With a History of Hepatocellular Carcinoma. Gastroenterology 2019, 157, 1253–1263.e2. [Google Scholar] [CrossRef]
- Smits, M.; Zoldan, K.; Ishaque, N.; Gu, Z.; Jechow, K.; Wieland, D.; Conrad, C.; Eils, R.; Fauvelle, C.; Baumert, F.T. Follicular T helper cells shape the HCV-specific CD4+ T cell repertoire after virus elimination. J. Clin. Investig. 2020, 130, 998–1009. [Google Scholar] [CrossRef]
- Inada, Y.; Mizukoshi, E.; Seike, T.; Tamai, T.; Iida, N.; Kitahara, M.; Yamashita, T.; Arai, K.; Terashima, T.; Fushimi, K.; et al. Characteristics of Immune Response to Tumor-Associated Antigens and Immune Cell Profile in Patients With Hepatocellular Carcinoma. Hepatology 2019, 69, 653–665. [Google Scholar] [CrossRef]
- Mizukoshi, E.; Yamashita, T.; Arai, K.; Sunagozaka, H.; Ueda, T.; Arihara, F.; Kagaya, T.; Yamashita, T.; Fushimi, K.; Kaneko, S. Enhancement of tumor-associated antigen-specific T cell responses by radiofrequency ablation of hepatocellular carcinoma. Hepatology 2013, 57, 1448–1457. [Google Scholar] [CrossRef]
- Kao, C.; Oestreich, K.J.; Paley, M.A.; Crawford, A.; Angelosanto, J.M.; Ali, M.A.; Intlekofer, A.M.; Boss, J.M.; Reiner, S.L.; Weinmann, A.S.; et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat. Immunol. 2011, 12, 663–671. [Google Scholar] [CrossRef]
- Zhu, D.Y.; Deng, X.Z.; Jiang, L.F.; Xiao, W.; Pei, J.P.; Li, B.J.; Wang, C.J.; Zhang, J.H.; Zhang, Q.; Zhou, Z.X.; et al. Potential Role of Hepatitis C Virus Alternate Reading Frame Protein in Negative Regulation of T-Bet Gene Expression. Inflammation 2015, 38, 1823–1834. [Google Scholar] [CrossRef]
- Mohamadi, M.; Azarbayjani, K.; Mozhgani, S.H.; Bamdad, T.; Alamdary, A.; Nikoo, H.R.; Hashempour, T.; Yaghoobi, M.H.; Ajorloo, M. Hepatitis C virus alternative reading frame protein (ARFP): Production, features, and pathogenesis. J. Med. Virol. 2020, 92, 2930–2937. [Google Scholar] [CrossRef] [PubMed]
- Seike, T.; Mizukoshi, E.; Yamada, K.; Okada, H.; Kitahara, M.; Yamashita, T.; Arai, K.; Terashima, T.; Iida, N.; Fushimi, K.; et al. Fatty acid-driven modifications in T-cell profiles in non-alcoholic fatty liver disease patients. J. Gastroenterol. 2020, 55, 701–711. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.L.; Fouser, L.A.; Jussif, J.; Fitz, L.; Deng, B.; Wood, C.R.; Collins, M.; Honjo, T.; Freeman, G.J.; Carreno, B.M. PD-1:PD-L inhibitory pathway affects both CD4+ and CD8+ T cells and is overcome by IL-2. Eur. J. Immunol. 2002, 32, 634–643. [Google Scholar] [CrossRef]
- Huang, H.W.; Chang, C.C.; Wang, C.S.; Lin, K.H. Association between Inflammation and Function of Cell Adhesion Molecules Influence on Gastrointestinal Cancer Development. Cells 2021, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Kuroda, H.; Matsunaga, T.; Osaki, T.; Ikeguchi, M. Increased PD-1 expression on CD4+ and CD8+ T cells is involved in immune evasion in gastric cancer. J. Surg. Oncol. 2013, 107, 517–522. [Google Scholar] [CrossRef]
- Kim, H.D.; Song, G.W.; Park, S.; Jung, M.K.; Kim, M.H.; Kang, H.J.; Yoo, C.; Yi, K.; Kim, K.H.; Eo, S.; et al. Association Between Expression Level of PD1 by Tumor-Infiltrating CD8+ T cells and Features of Hepatocellular Carcinoma. Gastroenterology 2018, 155, 1936–1950.e17. [Google Scholar] [CrossRef]
- Dalagiorgou, G.; Vassilaki, N.; Foka, P.; Boumlic, A.; Kakkanas, A.; Kochlios, E.; Khalili, S.; Aslanoglou, E.; Veletza, S.; Orfanoudakis, G.; et al. High levels of HCV core+1 antibodies in HCV patients with hepatocellular carcinoma. J. Gen. Virol. 2011, 92, 1343–1351. [Google Scholar] [CrossRef]
- Komurian-Pradel, F.; Rajoharison, A.; Berland, J.-L.; Khouri, V.; Perret, M.; van Roosmalen, M.; Pol, S.; Negro, F.; Paranhos-Baccalà, G. Antigenic relevance of F protein in chronic hepatitis C virus infection. Hepatology 2004, 40, 900–909. [Google Scholar] [CrossRef]
- Xiao, W.; Jiang, L.F.; Deng, X.Z.; Zhu, D.Y.; Pei, J.P.; Xu, M.L.; Li, B.J.; Wang, C.J.; Zhang, J.H.; Zhang, Q.; et al. PD-1/PD-L1 signal pathway participates in HCV F protein-induced T cell dysfunction in chronic HCV infection. Immunol. Res. 2016, 64, 412–423. [Google Scholar] [CrossRef]
- Gomi, S.; Nakao, M.; Niiya, F.; Imamura, Y.; Kawano, K.; Nishizaka, S.; Hayashi, A.; Sobao, Y.; Oizumi, K.; Itoh, K. A cyclophilin B gene encodes antigenic epitopes recognized by HLA-A24-restricted and tumor-specific CTLs. J. Immunol. 1999, 163, 4994–5004. [Google Scholar] [PubMed]
- Mizukoshi, E.; Fushimi, K.; Arai, K.; Yamashita, T.; Honda, M.; Kaneko, S. Expression of chondroitin-glucuronate C5-epimerase and cellular immune responses in patients with hepatocellular carcinoma. Liver Int. 2012, 32, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Kaji, K.; Mizukoshi, E.; Yamashita, T.; Arai, K.; Sunagozaka, H.; Fushimi, K.; Nakagawa, H.; Yamada, K.; Terashima, T.; Kitahara, M.; et al. Cellular immune responses for squamous cell carcinoma antigen recognized by T cells 3 in patients with hepatocellular carcinoma. PLoS ONE 2017, 12, e0170291. [Google Scholar] [CrossRef]
- Umano, Y.; Tsunoda, T.; Tanaka, H.; Matsuda, K.; Yamaue, H.; Tanimura, H. Generation of cytotoxic T cell responses to an HLA-A24 restricted epitope peptide derived from wild-type p53. Br. J. Cancer 2001, 84, 1052–1057. [Google Scholar] [CrossRef] [PubMed]
- Mizukoshi, E.; Honda, M.; Arai, K.; Yamashita, T.; Nakamoto, Y.; Kaneko, S. Expression of multidrug resistance-associated protein 3 and cytotoxic T cell responses in patients with hepatocellular carcinoma. J. Hepatol. 2008, 49, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Mizukoshi, E.; Nakamoto, Y.; Tsuji, H.; Yamashita, T.; Kaneko, S. Identification of α-fetoprotein-derived peptides recognized by cytotoxic T lymphocytes in HLA-A24+ patients with hepatocellular carcinoma. Int. J. Cancer 2006, 118, 1194–1204. [Google Scholar] [CrossRef]
- Mizukoshi, E.; Nakamoto, Y.; Marukawa, Y.; Arai, K.; Yamashita, T.; Tsuji, H.; Kuzushima, K.; Takiguchi, M.; Kaneko, S. Cytotoxic T cell responses to human telomerase reverse transcriptase in patients with hepatocellular carcinoma. Hepatology 2006, 43, 1284–1294. [Google Scholar] [CrossRef]
- Tsuboi, A.; Oka, Y.; Udaka, K.; Murakami, M.; Masuda, T.; Nakano, A.; Nakajima, H.; Yasukawa, M.; Hiraki, A.; Oji, Y.; et al. Enhanced induction of human WT1-specific cytotoxic T lymphocytes with a 9-mer WT1 peptide modified at HLA-A*2402-binding residues. Cancer Immunol. Immunother. 2002, 51, 614–620. [Google Scholar] [CrossRef]
- Ogata, R.; Matsueda, S.; Yao, A.; Noguchi, M.; Itoh, K.; Harada, M. Identification of polycomb group protein enhancer of zeste homolog 2 (EZH2)-derived peptides immunogenic in HLA-A24+ prostate cancer patients. Prostate 2004, 60, 273–281. [Google Scholar] [CrossRef]
- Komori, H.; Nakatsura, T.; Senju, S.; Yoshitake, Y.; Motomura, Y.; Ikuta, Y.; Fukuma, D.; Yokomine, K.; Harao, M.; Beppu, T.; et al. Identification of HLA-A2- or HLA-A24-restricted CTL epitopes possibly useful for glypican-3-specific immunotherapy of hepatocellular carcinoma. Clin. Cancer Res. 2006, 12, 2689–2697. [Google Scholar] [CrossRef]
- Korangy, F.; Ormandy, L.A.; Bleck, J.S.; Klempnauer, J.; Wilkens, L.; Manns, M.P.; Greten, T.F. Spontaneous tumor-specific humoral and cellular immune responses to NY-ESO-1 in hepatocellular carcinoma. Clin. Cancer Res. 2004, 10, 4332–4341. [Google Scholar] [CrossRef]
- Homma, S.; Harada, M.; Yano, H.; Ogasawara, S.; Shichijo, S.; Matsueda, S.; Komatsu, N.; Shomura, H.; Maeda, Y.; Sato, Y.; et al. Identification of squamous cell carcinoma antigen-derived peptides having the capacity of inducing cancer-reactive CTLs in HLA-A24+ cancer patients. Int. J. Oncol. 2006, 29, 577–587. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okochi, M.; Hayashi, H.; Ito, A.; Kato, R.; Tamura, Y.; Sato, N.; Honda, H. Identification of HLA-A24-restricted epitopes with high affinities to Hsp70 using peptide arrays. J. Biosci. Bioeng. 2008, 105, 198–203. [Google Scholar] [CrossRef]
- Kuzushima, K.; Hayashi, N.; Kimura, H.; Tsurumi, T. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8+ T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 2001, 98, 1872–1881. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Deng, X.; Wang, Z.; Yang, J.; Zhang, Y.; Yu, J. Hepatitis C virus F protein: A double-edged sword in the potential contribution of chronic inflammation to carcinogenesis. Mol. Med. Rep. 2009, 2, 461–469. [Google Scholar] [PubMed]




| Participants (N = 47) | |||
|---|---|---|---|
| Characteristic | DAAs for 24 Weeks | ||
| SVR (n = 44) | Non-SVR (n = 3) | p Value * | |
| Age, years median ± SD | 65 ± 9.66 | 65 ± 6.25 | NS |
| Sex, M/F | 15/29 | 0/3 | NS |
| BMI (kg/m2), median ± SD | 22.48 ± 3.48 | 23.15 ± 0.51 | NS |
| HCV RNA, median ± SD | 6.0 ± 0.64 | 6.2 ± 0.15 | NS |
| IL28B, N (%) | NS | ||
| Major | 22(50) | 2(67) | |
| hetero | 21(48) | 1(33) | |
| minor | 1(2) | 0(0) | |
| With cirrhosis, N (%) | 19(43) | 2(67) | NS |
| With HCC, N (%) | 6(14) | 0(0) | NS |
| L31 (NS5A) (+), N (%) | 1(2) | 0(0) | NS |
| Y93 (NS5A) (+), N (%) | 0(0) | 2(67) | 0.003 |
| ALT(IU/L) | 48.9 ± 34.58 | 48.67 ± 24.01 | NS |
| AST(IU/L) | 53 ± 34.60 | 38.7 ± 15.04 | NS |
| HCV treatment naive, N (%) | 14(32) | 1(33) | NS |
| Peptide No. | Peptide Name | Source | Reference | Amino Acid Sequence |
|---|---|---|---|---|
| 1 | Cyp-B109 | Cyp-B | [31] | KFHRVIKDF |
| 2 | SART2899 | SART2 | [32] | SYTRLFLIL |
| 3 | SART3109 | SART3 | [33] | VYDYNCHVDL |
| 4 | p53161 | p53 | [34] | AIYKQSQHM |
| 5 | MRP3765 | MRP3 | [35] | VYSDADIFL |
| 6 | MRP3692 | MRP3 | [35] | AYVPQQAWI |
| 7 | AFP403 | AFP | [36] | KYIQESQAL |
| 8 | AFP434 | AFP | [36] | AYTKKAPQL |
| 9 | AFP357 | AFP | [36] | EYSRRHPQL |
| 10 | hTERT167 | hTERT | [37] | AYQVCGPPL |
| 11 | hTERT461 | hTERT | [37] | VYGFVRACL |
| 12 | hTERT324 | hTERT | [37] | VYAETKHFL |
| 13 | WT-1235 | WT-1 | [38] | CYTWNQMNL |
| 14 | EZH2291 | EZH2 | [39] | KYDCFLHPF |
| 15 | GPC3298 | GPC3 | [40] | EYILSLEEL |
| 16 | NY-ESO-1158 | NY-ESO-1 | [41] | LLMWITQCF |
| 17 | SCCA112 | SCCA | [42] | TYLFLQEYL |
| 18 | IMP-3508 | IMP-3 | [17] | KTVNELQNL |
| 19 | Hsp70136 | Hsp70 | [43] | GYPVTNAVI |
| 20 | CMV pp65328 | CMV pp65 | [44] | QYDPVAALF |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Mizukoshi, E.; Kawaguchi, K.; Miura, M.; Nishino, M.; Shimakami, T.; Arai, K.; Yamashita, T.; Sakai, Y.; Yamashita, T.; et al. Alterations in Hepatocellular Carcinoma-Specific Immune Responses Following Hepatitis C Virus Elimination by Direct-Acting Antivirals. Int. J. Mol. Sci. 2022, 23, 11623. https://doi.org/10.3390/ijms231911623
Li S, Mizukoshi E, Kawaguchi K, Miura M, Nishino M, Shimakami T, Arai K, Yamashita T, Sakai Y, Yamashita T, et al. Alterations in Hepatocellular Carcinoma-Specific Immune Responses Following Hepatitis C Virus Elimination by Direct-Acting Antivirals. International Journal of Molecular Sciences. 2022; 23(19):11623. https://doi.org/10.3390/ijms231911623
Chicago/Turabian StyleLi, Shihui, Eishiro Mizukoshi, Kazunori Kawaguchi, Miyabi Miura, Michiko Nishino, Tetsuro Shimakami, Kuniaki Arai, Taro Yamashita, Yoshio Sakai, Tatsuya Yamashita, and et al. 2022. "Alterations in Hepatocellular Carcinoma-Specific Immune Responses Following Hepatitis C Virus Elimination by Direct-Acting Antivirals" International Journal of Molecular Sciences 23, no. 19: 11623. https://doi.org/10.3390/ijms231911623
APA StyleLi, S., Mizukoshi, E., Kawaguchi, K., Miura, M., Nishino, M., Shimakami, T., Arai, K., Yamashita, T., Sakai, Y., Yamashita, T., Honda, M., & Kaneko, S. (2022). Alterations in Hepatocellular Carcinoma-Specific Immune Responses Following Hepatitis C Virus Elimination by Direct-Acting Antivirals. International Journal of Molecular Sciences, 23(19), 11623. https://doi.org/10.3390/ijms231911623






