Mechanisms of Foreign Body Response Mitigation by Nitric Oxide Release
Abstract
:1. Introduction
2. Results and Discussion
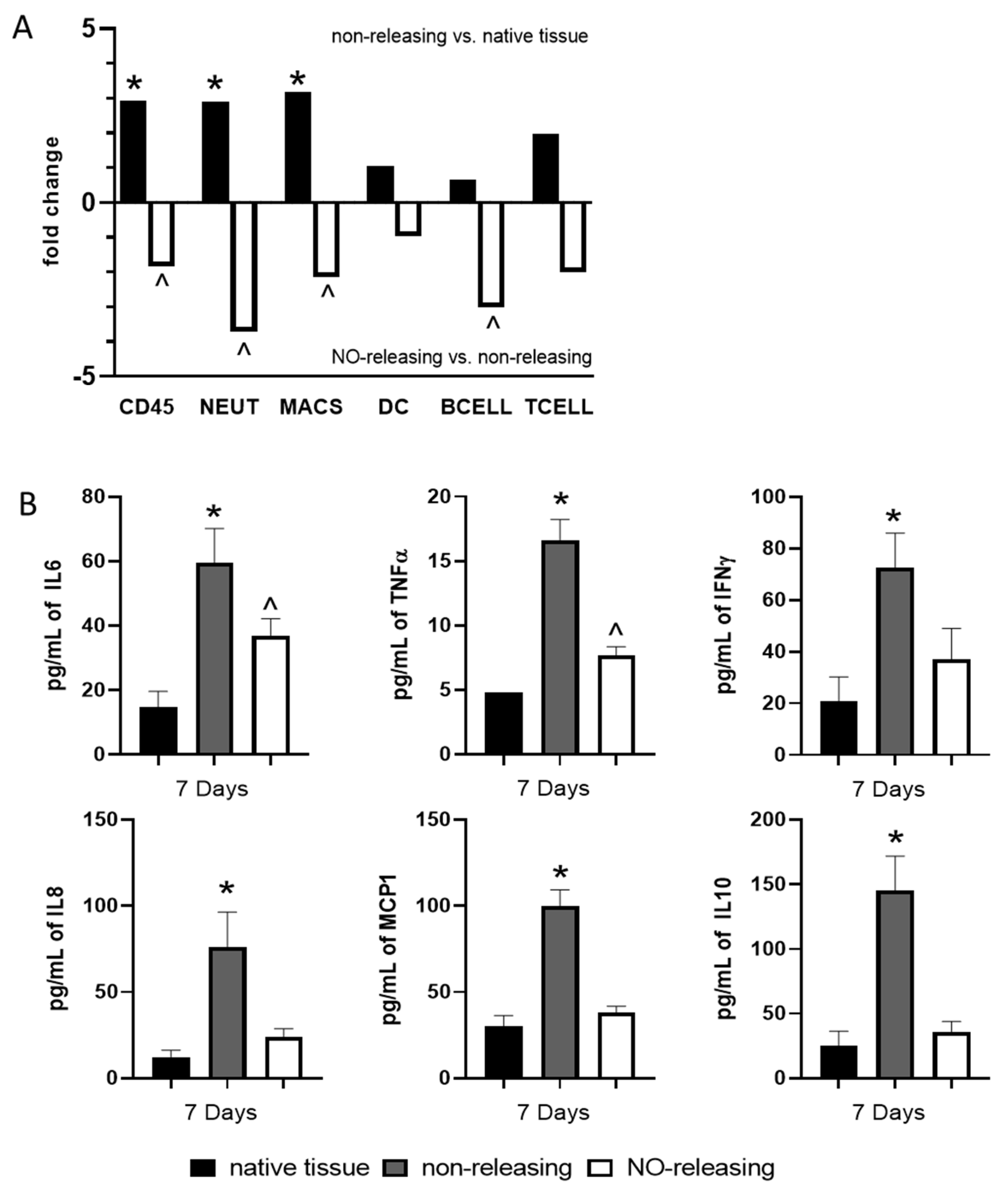
2.1. Subcutaneously Implanted Sensors with Sustained NO Release Abrogate Sensor-Induced Acute and Chronic Foreign Body Response through Modulation of the Tissue-Specific Immune Microenvironment
2.2. Subcutaneously Implanted Sensors with Sustained NO Release Abrogate the Sensor-Induced Acute and Chronic Foreign Body Response through Modulation of mRNA Encoding for Key Immunological Signaling Molecules and Pathways
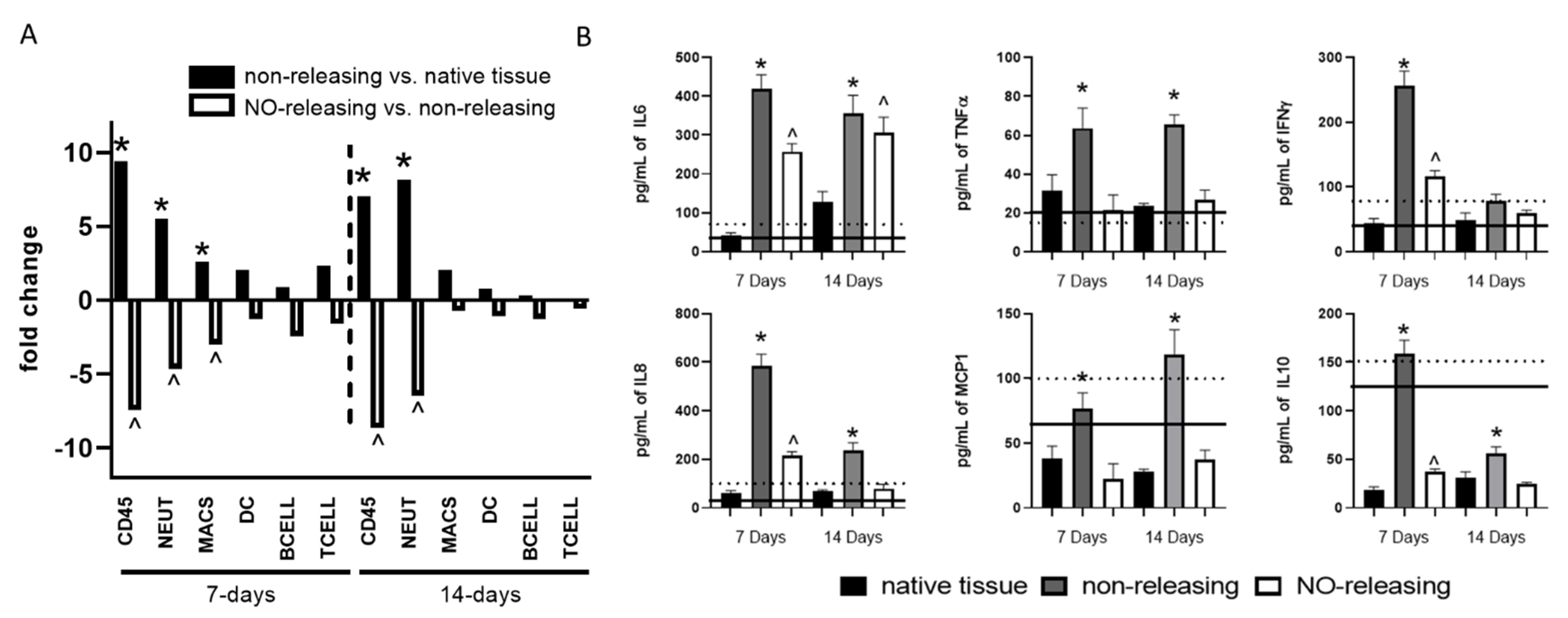
2.3. Sensor FBR in Diabetes Controlled by Sustained NO Release
3. Materials and Methods
3.1. Materials
3.2. Particle Synthesis and Preparation of Sensors
3.3. In Vivo Assessment of Foreign Body Response
3.4. Soluble Mediator Expression
3.5. mRNA Expression
3.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kieu, A.; King, J.; Govender, R.D.; Östlundh, L. The Benefits of Utilizing Continuous Glucose Monitoring of Diabetes Mellitus in Primary Care: A Systematic Review. J. Diabetes Sci. Technol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Benhamou, P.Y.; Catargi, B.; Delenne, B.; Guerci, B.; Hanaire, H.; Jeandidier, N.; Leroy, R.; Meyer, L.; Penfornis, A.; Radermecker, R.P.; et al. Real-Time Continuous Glucose Monitoring (CGM) Integrated into the Treatment of Type 1 Diabetes: Consensus of Experts from SFD, EVADIAC and SFE. Diabetes Metab. 2012, 38, S67–S83. [Google Scholar] [CrossRef] [Green Version]
- Ward, W.K. A Review of the Foreign-Body Response to Subcutaneously-Implanted Devices: The Role of Macrophages and Cytokines in Biofouling and Fibrosis. J. Diabetes Sci. Technol. 2008, 2, 768–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lotti, F.; Ranieri, F.; Vadala, G.; Zollo, L.; Di Pino, G. Invasive Intraneural Interfaces: Foreign Body Reaction Issues. Front. Neurosci. 2017, 11, 497. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M.; Rodriguez, A.; Chang, D.T. Foreign Body Reaction to Biomaterials. Semin. Immunol. 2008, 20, 86–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, N.N.; Rose, M.B.; Levinson, H.; Klitzman, B. Implant Healing in Experimental Animal Models of Diabetes. J. Diabetes Sci. Technol. 2011, 5, 605–618. [Google Scholar] [CrossRef] [Green Version]
- Fahey, T.J.; Sadaty, A.; Jones, W.G.; Barber, A.; Smoller, B.; Shires, G.T. Diabetes Impairs the Late Inflammatory Response to Wound Healing. J. Surg. Res. 1991, 50, 308–313. [Google Scholar] [CrossRef]
- Koh, A.; Nichols, S.P.; Schoenfisch, M.H. Glucose Sensor Membranes for Mitigating the Foreign Body Response. J. Diabetes Sci. Technol. 2011, 5, 1052–1059. [Google Scholar] [CrossRef] [Green Version]
- Soto, R.J.; Merricks, E.P.; Bellinger, D.A.; Nichols, T.C.; Schoenfisch, M.H. Influence of Diabetes on the Foreign Body Response to Nitric Oxide-Releasing Implants. Biomaterials 2018, 157, 76–85. [Google Scholar] [CrossRef]
- Nichols, S.P.; Koh, A.; Storm, W.L.; Shin, J.H.; Schoenfisch, M.H. Biocompatible Materials for Continuous Glucose Monitoring Devices. Chem. Rev. 2013, 113, 2528–2549. [Google Scholar] [CrossRef] [Green Version]
- Soto, R.; Hall, J.R.; Brown, M.D.; Taylor, J.B.; Schoenfisch, M.H. In Vivo Chemical Sensors: Role of Biocompatibility on Performance and Utility. Anal. Chem. 2017, 89, 276–299. [Google Scholar] [CrossRef] [PubMed]
- Moilanen, E.; Vapaatalo, H. Nitric Oxide in Inflammation and Immune Response. Ann. Med. 1995, 27, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, R.; Lahti, A.; Kankaanranta, H.; Moilanen, E. Nitric Oxide Production and Signaling in Inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Guzik, T.J.; Korbut, R.; Adamek-Guzik, T. Nitric Oxide and Superoxide in Inflammation and Immune Regulation. J. Physiol. Pharmacol. 2003, 54, 469–487. [Google Scholar]
- Bogdan, C. Nitric Oxide and the Immune Response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef]
- Gifford, R.; Batchelor, M.M.; Lee, Y.; Gokulrangan, G.; Meyerhoff, M.E.; Wilson, G.S. Mediation Ofin Vivo Glucose Sensor Inflammatory Response via Nitric Oxide Release. J. Biomed. Mater. Res. Part A 2005, 75A, 755–766. [Google Scholar] [CrossRef]
- Nichols, S.P.; Le, N.N.; Klitzman, B.; Schoenfisch, M.H. Increased in Vivo Glucose Recovery via Nitric Oxide Release. Anal. Chem. 2011, 83, 1180–1184. [Google Scholar] [CrossRef] [Green Version]
- Soto, R.J.; Privett, B.J.; Schoenfisch, M.H. In Vivo Analytical Performance of Nitric Oxide-Releasing Glucose Biosensors. Anal. Chem. 2014, 86, 7141–7149. [Google Scholar] [CrossRef] [Green Version]
- Malone-Povolny, M.J.; Merricks, E.P.; Wimsey, L.E.; Nichols, T.C.; Schoenfisch, M.H. Long-Term Accurate Continuous Glucose Biosensors via Extended Nitric Oxide Release. ACS Sens. 2019, 4, 3257–3264. [Google Scholar] [CrossRef]
- Malone-Povolny, M.J.; Bradshaw, T.M.; Merricks, E.P.; Long, C.T.; Nichols, T.C.; Schoenfisch, M.H. Combination of Nitric Oxide Release and Surface Texture for Mitigating the Foreign Body Response. ACS Biomater. Sci. Eng. 2021, 7, 2444–2452. [Google Scholar] [CrossRef]
- Shin, J.H.; Schoenfisch, M.H. Improving the Biocompatibility of In Vivo Sensors via Nitric Oxide Release. Analyst 2006, 131, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.R. Diffusion of Free Nitric Oxide. Methods Enzymol. 1996, 268, 31–50. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, M.W.; Kuo, L.; Liao, J.C. Effective Diffusion Distance of Nitric Oxide in the Microcirculation. Am. J. Physiol.-Heart Circ. Physiol. 1998, 274, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.R.; Maloney, S.E.; Jin, H.; Taylor, J.B.; Schoenfisch, M.H. Nitric Oxide Diffusion through Cystic Fibrosis-Relevant Media and Lung Tissue. RSC Adv. 2019, 9, 40176–40183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccio, D.A.; Schoenfisch, M.H. Nitric Oxide Release: Part I. Macromolecular Scaffolds. Chem. Soc. Rev. 2012, 41, 3731–3741. [Google Scholar] [CrossRef]
- Soto, R.J.; Yang, L.; Schoenfisch, M.H. Functionalized Mesoporous Silica via an Aminosilane Surfactant Ion Exchange Reaction: Controlled Scaffold Design and Nitric Oxide Release. ACS Appl. Mater. Interfaces 2016, 8, 2220–2231. [Google Scholar] [CrossRef]
- Mowery, K.A.; Schoenfisch, M.H.; Saavedra, J.E.; Keefer, L.K.; Meyerhoff, M.E. Preparation and Characterization of Hydrophobic Polymeric Films That Are Thromboresistant via Nitric Oxide Release. Biomaterials 2000, 21, 9–21. [Google Scholar] [CrossRef]
- Wu, Y.; Meyerhoff, M.E. Nitric Oxide-Releasing/Generating Polymers for the Development of Implantable Chemical Sensors with Enhanced Biocompatibility. Talanta 2008, 75, 642–650. [Google Scholar] [CrossRef] [Green Version]
- Frost, M.C.; Reynolds, M.M.; Meyerhoff, M.E. Polymers Incorporating Nitric Oxide Releasing/Generating Substances for Improved Biocompatibility of Blood-Contacting Medical Devices. Biomaterials 2005, 26, 1685–1693. [Google Scholar] [CrossRef]
- Reynolds, M.M.; Frost, M.C.; Meyerhoff, M.E. Nitric Oxide-Releasing Hydrophobic Polymers: Preparation, Characterization, and Potential Biomedical Applications. Free. Radic. Biol. Med. 2004, 37, 926–936. [Google Scholar] [CrossRef]
- Koh, A.; Carpenter, A.W.; Slomberg, D.L.; Schoenfisch, M.H. Nitric Oxide-Releasing Silica Nanoparticle-Doped Polyurethane Electrospun Fibers. ACS Appl. Mater. Interfaces 2013, 5, 7956–7964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, R.J.; Schofield, J.B.; Walter, S.E.; Malone-Povolny, M.J.; Schoenfisch, M.H. Design Considerations for Silica-Particle-Doped Nitric-Oxide-Releasing Polyurethane Glucose Biosensor Membranes. ACS Sens. 2017, 2, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Hetrick, E.M.; Prichard, H.L.; Klitzman, B.; Schoenfisch, M.H. Reduced Foreign Body Response at Nitric Oxide-Releasing Subcutaneous Implants. Biomaterials 2007, 28, 4571–4580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichols, S.P.; Koh, A.; Brown, N.L.; Rose, M.B.; Sun, B.; Slomberg, D.L.; Riccio, D.A.; Klitzman, B.; Schoenfisch, M.H. The Effect of Nitric Oxide Surface Flux on the Foreign Body Response to Subcutaneous Implants. Biomaterials 2012, 33, 6305–6312. [Google Scholar] [CrossRef] [Green Version]
- Soto, R.J.; Schoenfisch, M.H. Preclinical Performance Evaluation of Percutaneous Glucose Biosensors: Experimental Considerations and Recommendations. J. Diabetes Sci. Technol. 2015, 9, 978–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danaher, P.; Warren, S.; Dennis, L.; D’Amico, L.; White, A.; Disis, M.L.; Geller, M.A.; Odunsi, K.; Beechem, J.; Fling, S.P. Gene Expression Markers of Tumor Infiltrating Leukocytes. J. ImmunoTherapy Cancer 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, K.; Mehta, H.; Drevets, D.A.; Coggeshall, K.M. IL-6 Increases B-Cell IgG Production in a Feed-Forward Proinflammatory Mechanism to Skew Hematopoiesis and Elevate Myeloid Production. Blood 2010, 115, 4699–4706. [Google Scholar] [CrossRef] [Green Version]
- Fernando, M.R.; Reyes, J.L.; Iannuzzi, J.; Leung, G.; McKay, D.M. The Pro-Inflammatory Cytokine, Interleukin-6, Enhances the Polarization of Alternatively Activated Macrophages. PLoS ONE 2014, 9, e94188. [Google Scholar] [CrossRef] [Green Version]
- Yabluchanskiy, A.; Ma, Y.; Iyer, R.P.; Hall, M.E.; Lindsey, M.L. Matrix Metalloproteinase-9: Many Shades of Function in Cardiovascular Disease. Physiology 2013, 28, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Jackson, S.E. Hsp90: Structure and Function. Top. Curr. Chem. 2012, 328, 155–240. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, S.; Lovisa, S.; Ambrose, C.G.; McAndrews, K.M.; Sugimoto, H.; Kalluri, R. Type-I Collagen Produced by Distinct Fibroblast Lineages Reveals Specific Function during Embryogenesis and Osteogenesis Imperfecta. Nat. Commun. 2021, 12, 7199. [Google Scholar] [CrossRef]
- Ramana, C.V.; Chatterjee-Kishore, M.; Nguyen, H.; Stark, G.R. Complex Roles of Stat1 in Regulating Gene Expression. Oncogene 2000, 19, 2619–2627. [Google Scholar] [CrossRef]
- Malone-Povolny, M.J.; Schoenfisch, M.H. Extended Nitric Oxide-Releasing Polyurethanes via S-Nitrosothiol-Modified Mesoporous Silica Nanoparticles. ACS Appl. Mater. Interfaces 2019, 11, 12216–12223. [Google Scholar] [CrossRef]
- Bellinger, D.A.; Merricks, E.P.; Nichols, T.C. Swine Models of Type 2 Diabetes Mellitus: Insulin Resistance, Glucose Tolerance, and Cardiovascular Complications. ILAR J. 2006, 47, 243–258. [Google Scholar] [CrossRef] [Green Version]

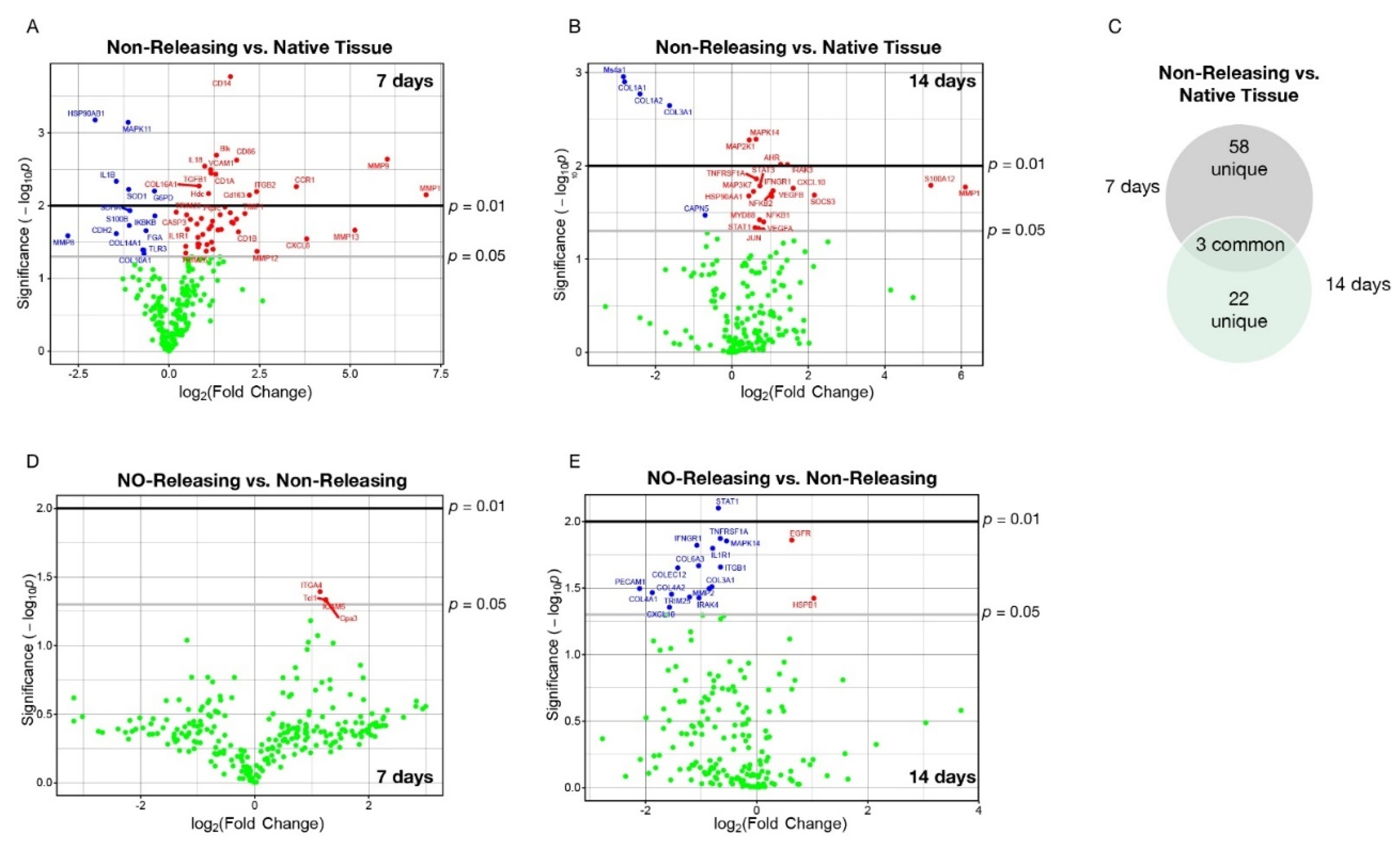

| 7 d Post Implantation | 14 d Post Implantation | |||||||
|---|---|---|---|---|---|---|---|---|
| mRNA | log2 Fold Change | p-Value | mRNA | log2 Fold Change | p-Value | mRNA | log2 Fold Change | p-Value |
| MMP1 | 7.093 | 0.0071 | CCL2 | 1.029 | 0.0421 | MMP1 | 6.110 | 0.0169 |
| MMP9 | 6.020 | 0.0023 | IL18 | 0.986 | 0.0029 | S100A12 | 5.210 | 0.0162 |
| MMP13 | 5.128 | 0.0217 | CASP1 | 0.934 | 0.0249 | SOCS3 | 2.160 | 0.0206 |
| CXCL8 | 3.799 | 0.0285 | Tpsab1 | 0.895 | 0.0149 | CXCL10 | 1.600 | 0.0174 |
| CCR1 | 3.513 | 0.0055 | COL16A1 | 0.832 | 0.0054 | IRAK3 | 1.450 | 0.0097 |
| MMP12 | 2.428 | 0.0424 | COL5A3 | 0.832 | 0.0338 | AHR | 1.280 | 0.0096 |
| ITGB2 | 2.417 | 0.0064 | PECAM1 | 0.807 | 0.0369 | IFNGR1 | 1.080 | 0.0185 |
| Cd163 | 2.217 | 0.0072 | SOCS1 | 0.799 | 0.0272 | VEGFB | 1.040 | 0.0213 |
| TIMP1 | 2.094 | 0.0128 | TLR2 | 0.799 | 0.0335 | NFKB2 | 1.030 | 0.0200 |
| CD1B | 1.918 | 0.0229 | IL10RB | 0.774 | 0.0179 | NFKB1 | 0.831 | 0.0399 |
| CD86 | 1.868 | 0.0024 | MYD88 | 0.595 | 0.0155 | VEGFA | 0.819 | 0.0485 |
| Cd68 | 1.864 | 0.0153 | IL1R1 | 0.506 | 0.0211 | STAT3 | 0.730 | 0.0164 |
| Csf3r | 1.774 | 0.0174 | CASP3 | 0.485 | 0.0134 | MYD88 | 0.727 | 0.0379 |
| ICAM1 | 1.731 | 0.0168 | CD40LG | 0.465 | 0.0362 | JUN | 0.697 | 0.0470 |
| CD14 | 1.696 | 0.0002 | TRIM26 | 0.465 | 0.0446 | TNFRSF1A | 0.642 | 0.0138 |
| Nkg7 | 1.692 | 0.0125 | TRIM25 | 0.202 | 0.0123 | MAPK14 | 0.631 | 0.0052 |
| Ptprc | 1.546 | 0.0104 | IKBKB | −0.390 | 0.0138 | STAT1 | 0.605 | 0.0460 |
| Fcgr4 | 1.444 | 0.0212 | G6PD | −0.401 | 0.0063 | MAP3K7 | 0.566 | 0.0188 |
| RUNX1 | 1.411 | 0.0133 | FGA | −0.632 | 0.0220 | MAP2K1 | 0.449 | 0.0053 |
| CCR5 | 1.350 | 0.0216 | COL10A1 | −0.687 | 0.0456 | HSP90AA1 | 0.440 | 0.0210 |
| Blk | 1.310 | 0.0020 | TLR3 | −0.696 | 0.0407 | CAPN5 | −0.698 | 0.0339 |
| CD1A | 1.293 | 0.0037 | COL14A1 | −0.722 | 0.0404 | COL3A1 | −1.630 | 0.0023 |
| CCR2 | 1.214 | 0.0396 | SDHA | −1.077 | 0.0117 | COL1A2 | −2.400 | 0.0017 |
| IL33 | 1.202 | 0.0163 | S100B | −1.098 | 0.0187 | COL1A1 | −2.810 | 0.0013 |
| MMP14 | 1.176 | 0.0317 | SOD1 | −1.111 | 0.0060 | Ms4a1 | −2.840 | 0.0011 |
| COL7A1 | 1.170 | 0.0190 | MAPK11 | −1.124 | 0.0007 | |||
| TGFB1 | 1.163 | 0.0036 | IL1B | −1.454 | 0.0046 | |||
| VCAM1 | 1.151 | 0.0032 | CDH2 | −1.454 | 0.0242 | |||
| CCL19 | 1.104 | 0.0208 | HSP90AB1 | −2.036 | 0.0007 | |||
| Hdc | 1.091 | 0.0068 | MMP8 | −2.789 | 0.0258 | |||
| Cd3d | 1.050 | 0.0342 | ||||||
| 7 d Post Implantation | 14 d Post Implantation | ||||
|---|---|---|---|---|---|
| mRNA | log2 Fold Change | p-Value | mRNA | log2 Fold Change | p-Value |
| Cpa3 | 2.370 | 0.0464 | HSPB1 | 1.030 | 0.0377 |
| ICAM5 | 2.370 | 0.0464 | EGFR | 0.632 | 0.0138 |
| Tcl1 | 2.370 | 0.0464 | MAPK14 | −0.542 | 0.0140 |
| ITGA4 | 2.210 | 0.0404 | ITGB1 | −0.650 | 0.0220 |
| TNFRSF1A | −0.655 | 0.0134 | |||
| STAT1 | −0.689 | 0.0079 | |||
| IL1R1 | −0.791 | 0.0159 | |||
| COL3A1 | −0.806 | 0.0309 | |||
| MMP2 | −0.856 | 0.0319 | |||
| COL6A3 | −1.040 | 0.0215 | |||
| IRAK4 | −1.040 | 0.0375 | |||
| IFNGR1 | −1.080 | 0.0151 | |||
| TRIM25 | −1.210 | 0.0369 | |||
| COLEC12 | −1.420 | 0.0223 | |||
| COL4A2 | −1.530 | 0.0352 | |||
| CXCL10 | −1.570 | 0.0441 | |||
| COL4A1 | −1.880 | 0.0342 | |||
| PECAM1 | −2.110 | 0.0319 | |||
| 7 d Post Implantation | 14 d Post Implantation | |||||||
|---|---|---|---|---|---|---|---|---|
| mRNA | log2 Fold Change | p-Value | mRNA | log2 Fold Change | p-Value | mRNA | log2 Fold Change | p-Value |
| MMP13 | 6.730 | 0.0019 | Cd209e | 1.510 | 0.0223 | S100A12 | 4.960 | 0.0120 |
| MMP1 | 6.520 | 0.0012 | Fcgr4 | 1.460 | 0.0102 | IL1RN | 1.350 | 0.0438 |
| IL6 | 4.570 | 0.0170 | ITGAL | 1.350 | 0.0273 | MAPK11 | 1.290 | 0.0306 |
| S100A12 | 4.330 | 0.0078 | TGFB1 | 1.290 | 0.0093 | Tpsab1 | 1.210 | 0.0304 |
| MMP3 | 4.200 | 0.0301 | Cd244a | 1.240 | 0.0416 | IL23R | 1.140 | 0.0250 |
| MMP9 | 3.760 | 0.0171 | CD1C | 1.240 | 0.0443 | ICAM3 | 1.090 | 0.0419 |
| TIMP1 | 3.080 | 0.0030 | CXCL10 | 1.220 | 0.0229 | IGF1 | 1.080 | 0.0226 |
| CD1B | 3.040 | 0.0092 | MMP2 | 1.160 | 0.0298 | CCL19 | 0.994 | 0.0408 |
| COL6A5 | 2.870 | 0.0134 | IL1R1 | 1.130 | 0.0216 | ICAM1 | 0.872 | 0.0113 |
| CCL19 | 2.870 | 0.0139 | Ms4a2 | 1.060 | 0.0263 | IL17RC | 0.832 | 0.0322 |
| SOCS3 | 2.660 | 0.0096 | TLR2 | 1.010 | 0.0147 | PECAM1 | 0.736 | 0.0214 |
| Cd163 | 2.630 | 0.0052 | AHR | 0.982 | 0.0059 | MMP7 | 0.719 | 0.0133 |
| ITGB2 | 2.570 | 0.0051 | IL1RN | 0.952 | 0.0034 | AHR | 0.573 | 0.0143 |
| Il21r | 2.460 | 0.0445 | NFKB2 | 0.939 | 0.0092 | STAT3 | 0.360 | 0.0213 |
| CCL2 | 2.250 | 0.0220 | STAT1 | 0.906 | 0.0150 | MYD88 | 0.292 | 0.0328 |
| IL27 | 2.130 | 0.0370 | MMP7 | 0.903 | 0.0330 | TRIM25 | 0.289 | 0.0090 |
| ICAM1 | 2.070 | 0.0191 | TNFRSF1A | 0.874 | 0.0099 | STAT1 | 0.199 | 0.0470 |
| RUNX1 | 1.980 | 0.0072 | MYD88 | 0.856 | 0.0006 | CASP3 | −0.280 | 0.0421 |
| Klrb1 | 1.970 | 0.0339 | TRIM8 | 0.849 | 0.0242 | SMAD2 | −0.332 | 0.0079 |
| Tpsab1 | 1.950 | 0.0064 | CSF1 | 0.794 | 0.0050 | TOLLIP | −0.396 | 0.0382 |
| IL33 | 1.920 | 0.0044 | IL17RA | 0.752 | 0.0271 | GATA3 | −0.425 | 0.0013 |
| CD1A | 1.850 | 0.0060 | STAT3 | 0.728 | 0.0113 | Hsd11b1 | −0.544 | 0.0068 |
| CXCL2 | 1.820 | 0.0165 | MMP14 | 0.678 | 0.0278 | CD34 | −0.625 | 0.0272 |
| CCR5 | 1.760 | 0.0212 | IL10RB | 0.619 | 0.0000 | TGFB2 | −0.941 | 0.0129 |
| Ptprc | 1.720 | 0.0097 | MYC | 0.593 | 0.0265 | Cd209e | −1.000 | 0.0128 |
| CD86 | 1.720 | 0.0242 | TLR3 | 0.572 | 0.0404 | Ms4a1 | −1.700 | 0.0469 |
| VCAM1 | 1.630 | 0.0135 | IFNGR1 | 0.534 | 0.0036 | |||
| Ms4a4a | 1.580 | 0.0316 | TRIM25 | 0.526 | 0.0342 | |||
| Cd68 | 1.560 | 0.0101 | IKBKB | 0.278 | 0.0113 | |||
| 7 d Post Implantation | 14 d Post Implantation | ||||
|---|---|---|---|---|---|
| mRNA | log2 Fold Change | p-Value | mRNA | log2 Fold Change | p-Value |
| MTOR | 0.139 | 0.0074 | Cd163 | 1.810 | 0.0464 |
| MMP7 | −0.652 | 0.0246 | MMP3 | 1.700 | 0.0139 |
| IL33 | −0.856 | 0.0301 | TLR4 | 0.660 | 0.0092 |
| SOCS3 | −1.490 | 0.0410 | ICAM1 | 0.594 | 0.0257 |
| TIMP1 | −1.570 | 0.0234 | VCAM1 | 0.544 | 0.0329 |
| CD1B | −1.930 | 0.0383 | MAPKAPK2 | 0.469 | 0.0287 |
| IL6 | −2.780 | 0.0339 | MAPKAPK3 | −0.528 | 0.0279 |
| PIK3CB | −0.628 | 0.0074 | |||
| GATA3 | −0.633 | 0.0152 | |||
| IL1RL2 | −0.831 | 0.0350 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, J.B.; Malone-Povolny, M.J.; Merricks, E.P.; Wimsey, L.E.; Soliman, D.; Nichols, T.C.; Wallet, S.M.; Maile, R.; Schoenfisch, M.H. Mechanisms of Foreign Body Response Mitigation by Nitric Oxide Release. Int. J. Mol. Sci. 2022, 23, 11635. https://doi.org/10.3390/ijms231911635
Taylor JB, Malone-Povolny MJ, Merricks EP, Wimsey LE, Soliman D, Nichols TC, Wallet SM, Maile R, Schoenfisch MH. Mechanisms of Foreign Body Response Mitigation by Nitric Oxide Release. International Journal of Molecular Sciences. 2022; 23(19):11635. https://doi.org/10.3390/ijms231911635
Chicago/Turabian StyleTaylor, James B., Maggie J. Malone-Povolny, Elizabeth P. Merricks, Lauren E. Wimsey, Daniel Soliman, Timothy C. Nichols, Shannon M. Wallet, Robert Maile, and Mark H. Schoenfisch. 2022. "Mechanisms of Foreign Body Response Mitigation by Nitric Oxide Release" International Journal of Molecular Sciences 23, no. 19: 11635. https://doi.org/10.3390/ijms231911635
APA StyleTaylor, J. B., Malone-Povolny, M. J., Merricks, E. P., Wimsey, L. E., Soliman, D., Nichols, T. C., Wallet, S. M., Maile, R., & Schoenfisch, M. H. (2022). Mechanisms of Foreign Body Response Mitigation by Nitric Oxide Release. International Journal of Molecular Sciences, 23(19), 11635. https://doi.org/10.3390/ijms231911635






