AI-Based Protein Interaction Screening and Identification (AISID)
Abstract
:1. Introduction
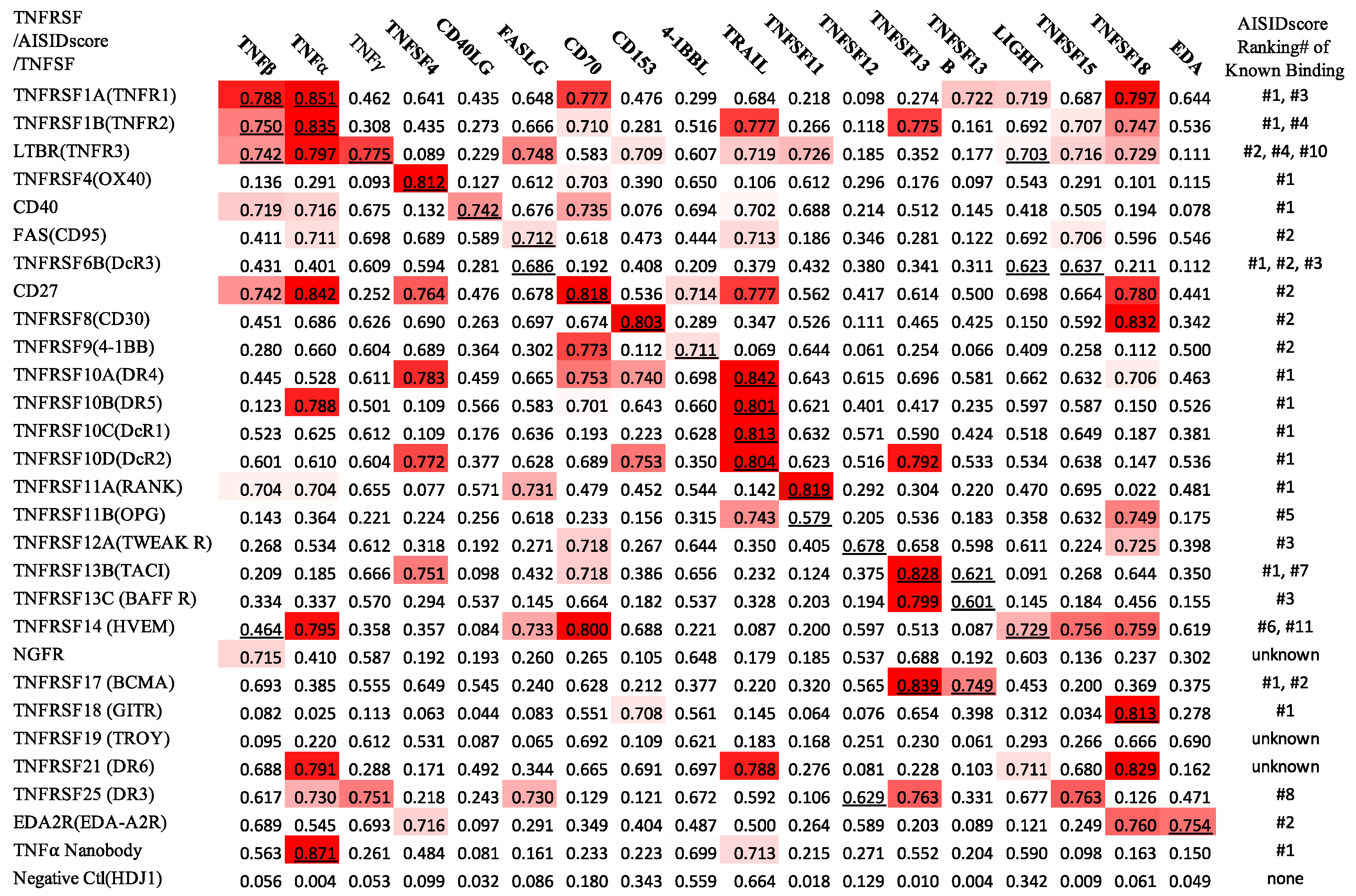
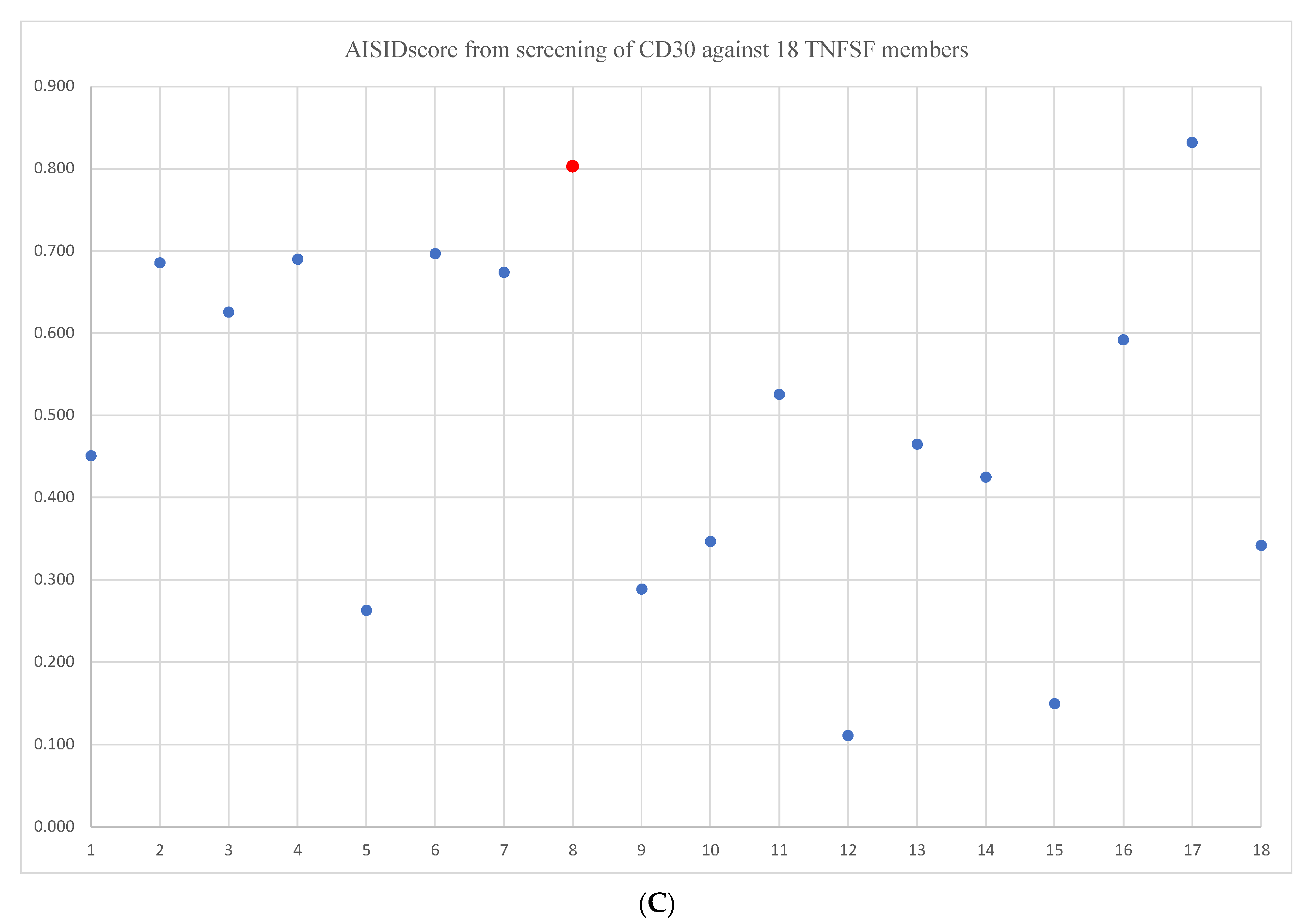
2. Results and Discussions
3. Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kryshtafovych, A.; Schwede, T.; Topf, M.; Fidelis, K.; Moult, J. Critical assessment of methods of protein structure prediction (CASP)-Round XIV. Proteins 2021, 89, 1607–1617. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13, 1265. [Google Scholar] [CrossRef]
- Humphreys, I.R.; Pei, J.; Baek, M.; Krishnakumar, A.; Anishchenko, I.; Ovchinnikov, S.; Zhang, J.; Ness, T.J.; Banjade, S.; Bagde, S.R.; et al. Computed structures of core eukaryotic protein complexes. Science 2021, 374, eabm4805. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Ho, Q.T.; Ou, Y.Y. Using two-dimensional convolutional neural networks for identifying GTP binding sites in Rab proteins. J. Bioinform. Comput. Biol. 2019, 17, 1950005. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Sandag, G.A.; Ou, Y.Y. Incorporating post translational modification information for enhancing the predictive performance of membrane transport proteins. Comput. Biol. Chem. 2018, 77, 251–260. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.W.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gao, M.; Nakajima An, D.; Parks, J.M.; Skolnick, J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 2022, 13, 1744. [Google Scholar] [CrossRef]
- Tordai, H.; Suhajda, E.; Sillitoe, I.; Nair, S.; Varadi, M.; Hegedus, T. Comprehensive Collection and Prediction of ABC Transmembrane Protein Structures in the AI Era of Structural Biology. Int. J. Mol. Sci. 2022, 23, 8877. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Feng, B.Y.; Varshney, A.; Pierce, B.G. Benchmarking AlphaFold for protein complex modeling reveals accuracy determinants. Protein Sci. 2022, 31, e4379. [Google Scholar] [CrossRef] [PubMed]
- Verburgt, J.; Zhang, Z.; Kihara, D. Multi-level analysis of intrinsically disordered protein docking methods. Methods 2022, 204, 55–63. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Taldaev, A.; Lisitsa, A.V.; Ponomarenko, E.A.; Archakov, A.I. Prediction of Monomeric and Dimeric Structures of CYP102A1 Using AlphaFold2 and AlphaFold Multimer and Assessment of Point Mutation Effect on the Efficiency of Intra- and Interprotein Electron Transfer. Molecules 2022, 27, 1386. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Dostert, C.; Grusdat, M.; Letellier, E.; Brenner, D. The TNF Family of Ligands and Receptors: Communication Modules in the Immune System and Beyond. Physiol. Rev. 2019, 99, 115–160. [Google Scholar] [CrossRef]
- Kucka, K.; Wajant, H. Receptor Oligomerization and Its Relevance for Signaling by Receptors of the Tumor Necrosis Factor Receptor Superfamily. Front. Cell Dev. Biol. 2020, 8, 615141. [Google Scholar] [CrossRef]
- Degli-Esposti, M.A.; Dougall, W.C.; Smolak, P.J.; Waugh, J.Y.; Smith, C.A.; Goodwin, R.G. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity 1997, 7, 813–820. [Google Scholar] [CrossRef] [Green Version]
- Emery, J.G.; McDonnell, P.; Burke, M.B.; Deen, K.C.; Lyn, S.; Silverman, C.; Dul, E.; Appelbaum, E.R.; Eichman, C.; DiPrinzio, R.; et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J. Biol. Chem. 1998, 273, 14363–14367. [Google Scholar] [CrossRef] [Green Version]
- Hargreaves, P.G.; Al-Shamkhani, A. Soluble CD30 binds to CD153 with high affinity and blocks transmembrane signaling by CD30. Eur. J. Immunol. 2002, 32, 163–173. [Google Scholar] [CrossRef]
- Beirnaert, E.; Desmyter, A.; Spinelli, S.; Lauwereys, M.; Aarden, L.; Dreier, T.; Loris, R.; Silence, K.; Pollet, C.; Cambillau, C.; et al. Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions. Front. Immunol. 2017, 8, 867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toto, A.; Malagrino, F.; Visconti, L.; Troilo, F.; Pagano, L.; Brunori, M.; Jemth, P.; Gianni, S. Templated folding of intrinsically disordered proteins. J. Biol. Chem. 2020, 295, 6586–6593. [Google Scholar] [CrossRef] [PubMed]


| TNFSF/AISIDscore | Default | Cutoff Date 1 January 2016 |
|---|---|---|
| TNFα | 0.871 | 0.869 |
| TRAIL | 0.713 | 0.705 |
| 4-1BBL | 0.699 | 0.392 |
| LIGHT | 0.590 | 0.709 |
| TNFβ | 0.563 | 0.515 |
| TNFSF13 | 0.552 | 0.454 |
| TNFSF4 | 0.484 | 0.118 |
| TNFSF12 | 0.271 | 0.101 |
| TNFγ | 0.261 | 0.311 |
| CD70 | 0.233 | 0.353 |
| CD153 | 0.223 | 0.096 |
| TNFSF11 | 0.215 | 0.265 |
| TNFSF13B | 0.204 | 0.253 |
| TNFSF18 | 0.163 | 0.276 |
| FASLG | 0.161 | 0.155 |
| EDA | 0.150 | 0.158 |
| TNFSF15 | 0.098 | 0.089 |
| CD40LG | 0.081 | 0.082 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Z.-Q.; Sha, H.L.; Sha, B. AI-Based Protein Interaction Screening and Identification (AISID). Int. J. Mol. Sci. 2022, 23, 11685. https://doi.org/10.3390/ijms231911685
Fu Z-Q, Sha HL, Sha B. AI-Based Protein Interaction Screening and Identification (AISID). International Journal of Molecular Sciences. 2022; 23(19):11685. https://doi.org/10.3390/ijms231911685
Chicago/Turabian StyleFu, Zheng-Qing, Hansen L. Sha, and Bingdong Sha. 2022. "AI-Based Protein Interaction Screening and Identification (AISID)" International Journal of Molecular Sciences 23, no. 19: 11685. https://doi.org/10.3390/ijms231911685




