Genes, Structural, and Biochemical Characterization of Four Chlorophyllases from Solanum lycopersicum
Abstract
:1. Introduction
2. Results
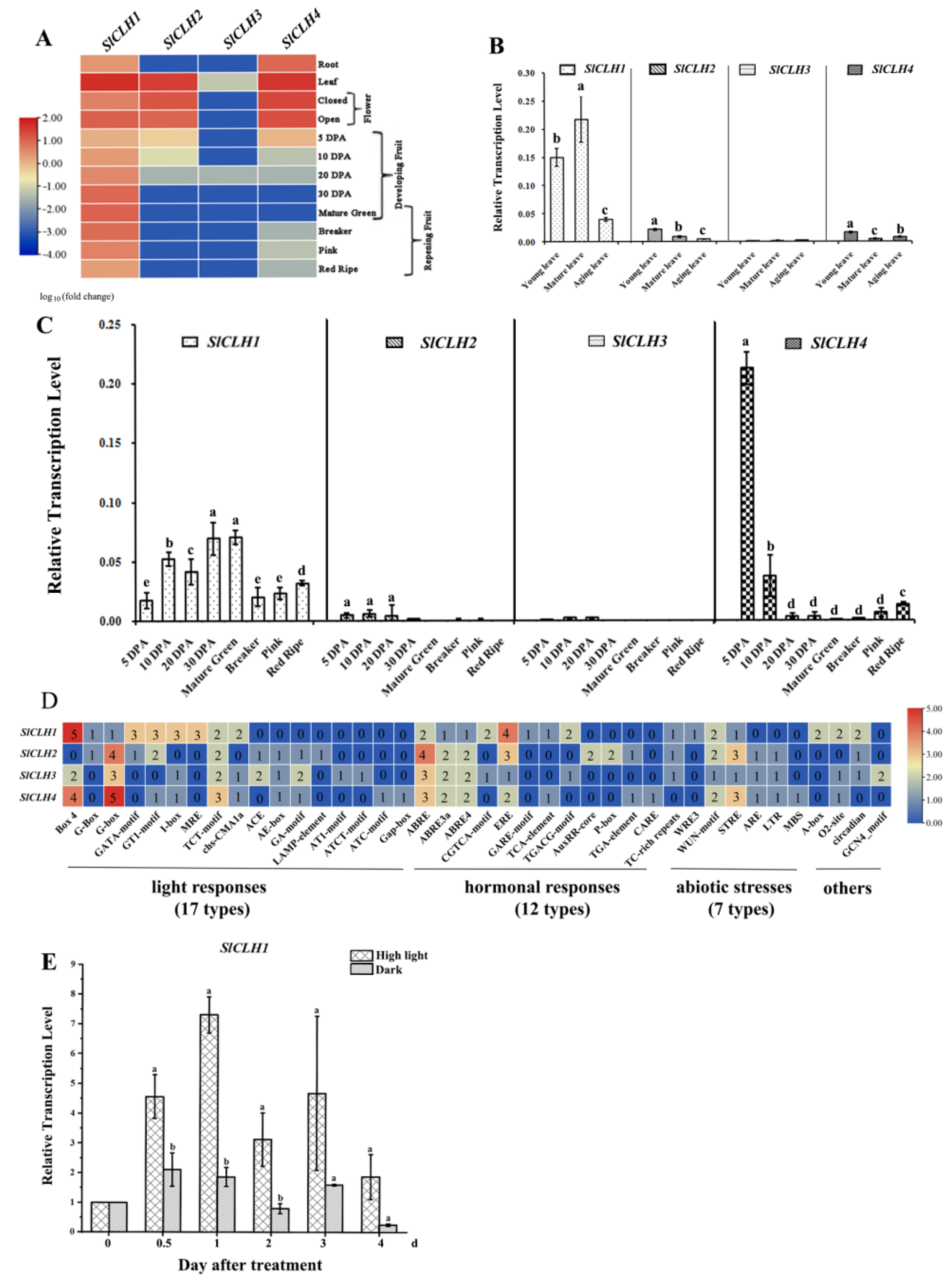
2.1. Differential Expression Pattern of Four SlCLH Genes in Various Tissues
2.2. Various cis-Acting Elements Related to Abiotic Stresses and Phytohormone Response Are Present in the Promoters of Four SlCLH Genes
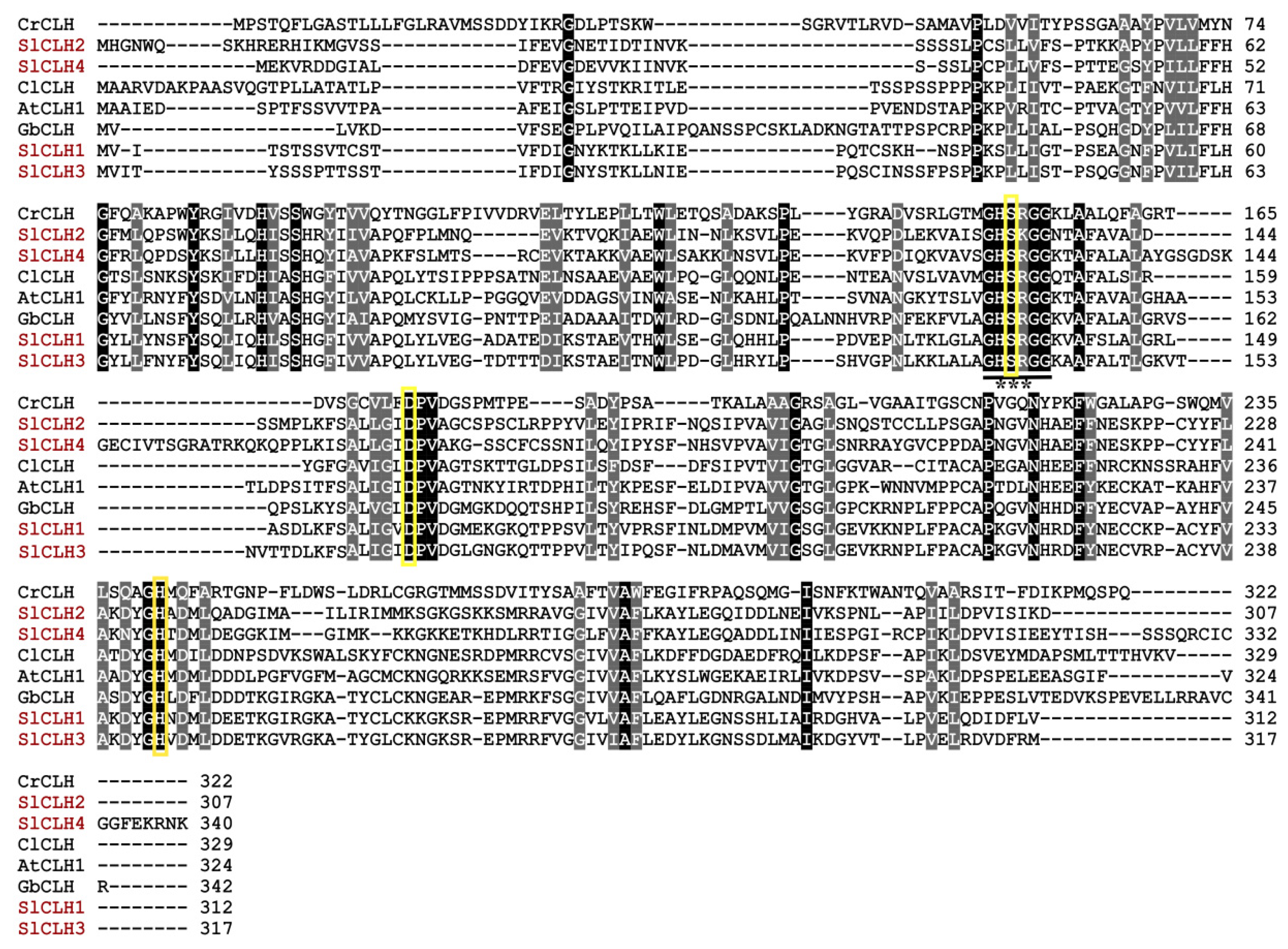
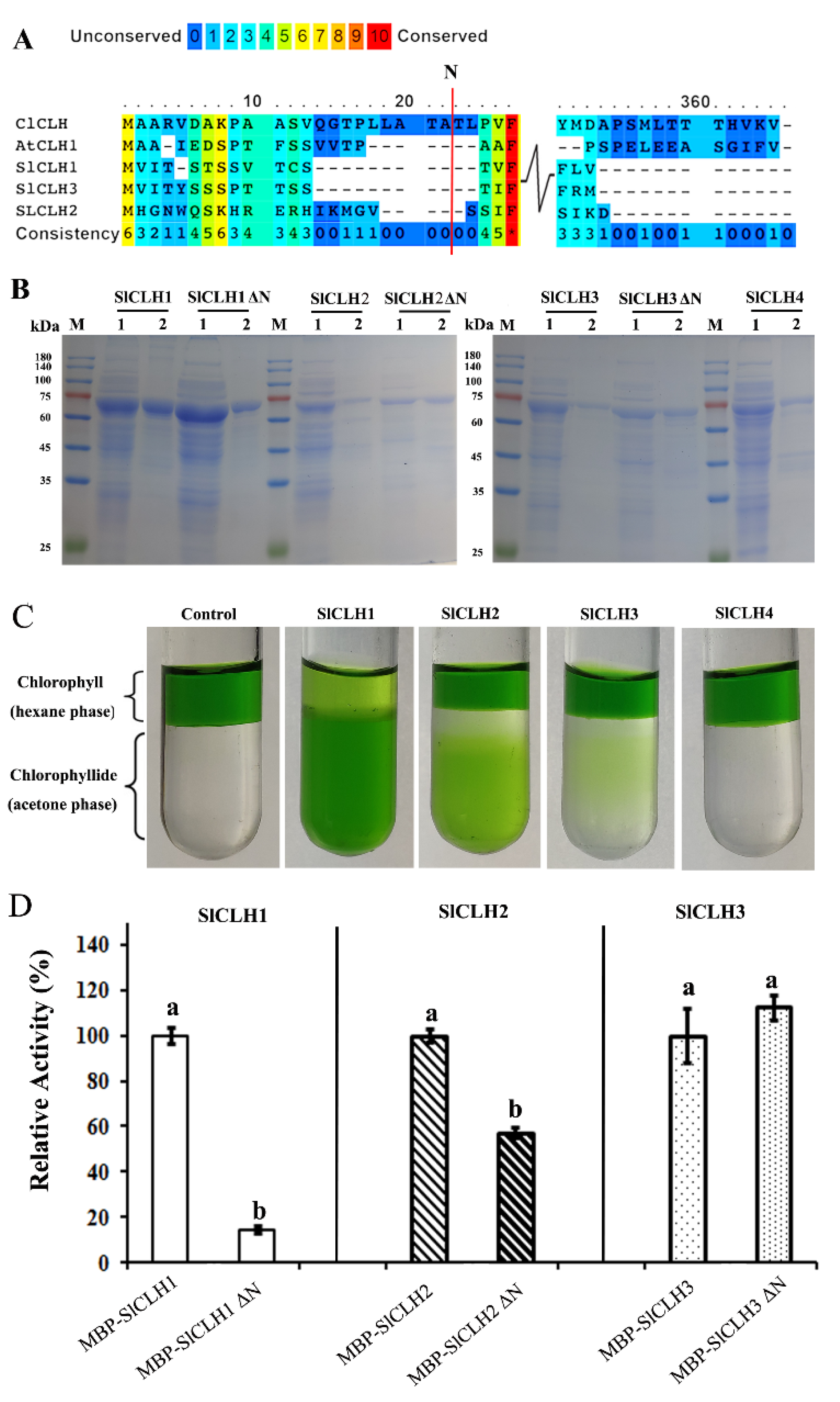
2.3. Analysis of Amino Acid Sequences of SlCLHs
2.4. Expression and Purification of Recombinant SlCLHs in E. coli
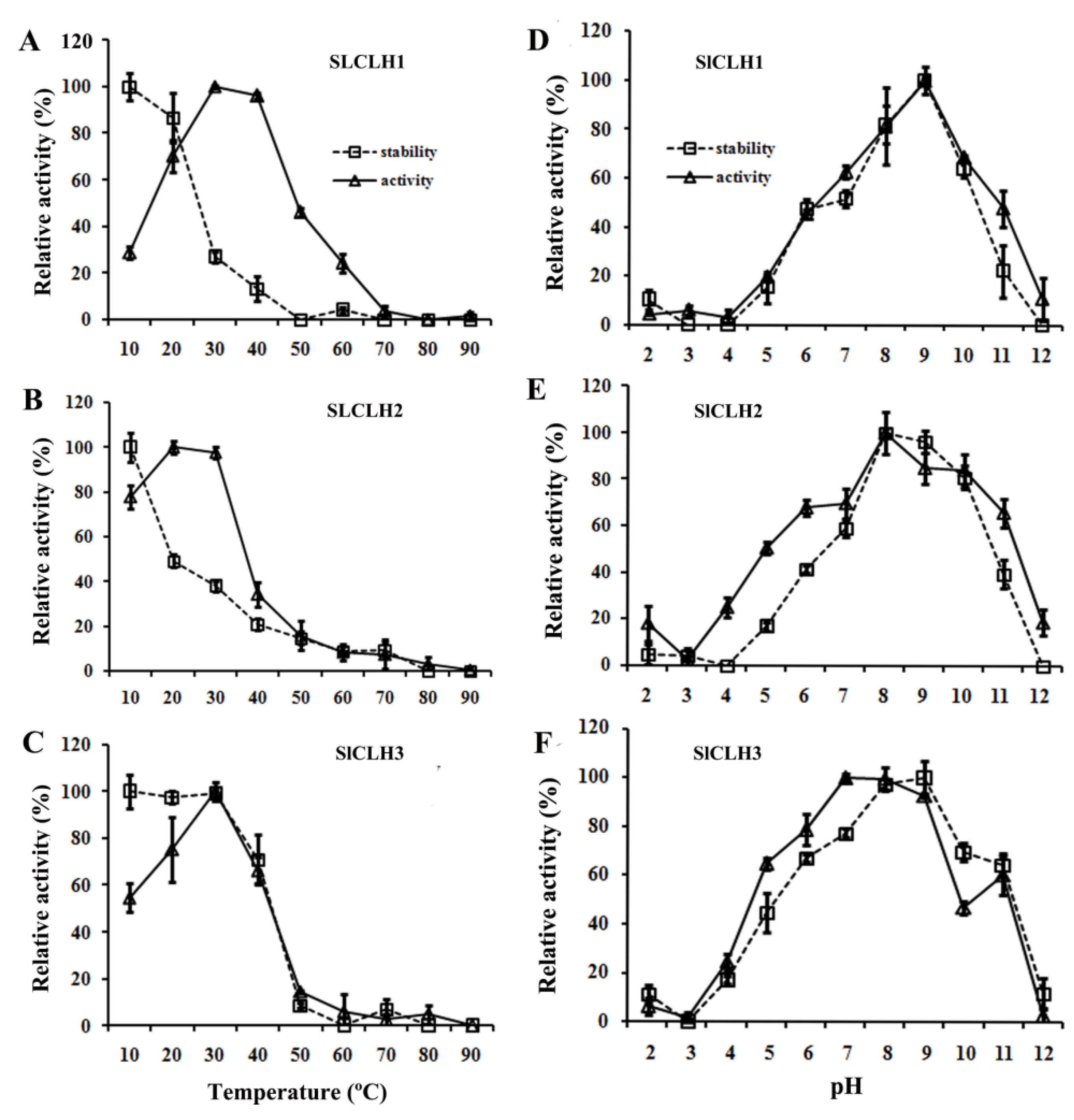
2.5. Optimal Temperature and pH for the Activity of the Recombinant SlCLHs
2.6. Enzyme Kinetics of Recombinant SlCLHs
2.7. Identification of Putative Catalytic Triad
3. Discussion
3.1. Different Expression Pattern of SlCLHs Suggest That They May Have Distinct Roles in Various Tissues
3.2. SlCLHs Belong to the α/β Hydrolase Superfamily, but Show Different Physiological and Biochemical Characteristics
3.3. The Role of the Predicted N-Terminal Processing Sequences in the Activity of Recombinant SlCLHs
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Isolation
4.3. Gene Expression Analysis
4.4. Protein Expression and Purification
4.5. Identification of the Putative Catalytic Triad
4.6. Sequence Analysis
4.7. In Silico Analysis of Cis-Elements in Promoters of SlCLH Genes
4.8. Chl Dephytylation Activity Assay
4.9. Analysis of Biochemical Characteristics of Recombinant SlCLHs
4.10. Enzyme Kinetic Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruno, A.K.; Wetzel, C.M. The early light-inducible protein (ELIP) gene is expressed during the chloroplast-to-chromoplast transition in ripening tomato fruit. J. Exp. Bot. 2004, 55, 2541–2548. [Google Scholar] [CrossRef] [Green Version]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.P.; Charng, Y.Y. Chlorophyll dephytylation in chlorophyll metabolism: A simple reaction catalyzed by various enzymes. Plant Sci. 2021, 302, 110682. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.P.; Wu, M.C.; Charng, Y.Y. Identification of a Chlorophyll Dephytylase Involved in Chlorophyll Turnover in Arabidopsis. Plant Cell 2016, 28, 2974–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchiya, T.; Ohta, H.; Masuda, T.; Mikami, B.; Kita, N.; Shioi, Y.; Takamiya, K.-I. Purification and Characterization of Two Isozymes of Chlorophyllase from Mature Leaves of Chenopodium album. Plant Cell Physiol. 1997, 38, 1026–1031. [Google Scholar] [CrossRef] [Green Version]
- Azoulay Shemer, T.; Harpaz-Saad, S.; Belausov, E.; Lovat, N.; Krokhin, O.; Spicer, V.; Standing, K.G.; Goldschmidt, E.E.; Eyal, Y. Citrus chlorophyllase dynamics at ethylene-induced fruit color-break: A study of chlorophyllase expression, posttranslational processing kinetics, and in situ intracellular localization. Plant Physiol. 2008, 148, 108–118. [Google Scholar] [CrossRef] [Green Version]
- Azoulay-Shemer, T.; Harpaz-Saad, S.; Cohen-Peer, R.; Mett, A.; Spicer, V.; Lovat, N.; Krokhin, O.; Brand, A.; Gidoni, D.; Standing, K.G.; et al. Dual N- and C-terminal processing of citrus chlorophyllase precursor within the plastid membranes leads to the mature enzyme. Plant Cell Physiol. 2011, 52, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.M.; Yang, J.H.; Liu, C.H.; Lin, K.H.; Yang, C.M. Molecular, structural, and phylogenetic characterization of two chlorophyllase isoforms in Pachira macrocarpa. Plant Syst. Evol. 2014, 300, 633–643. [Google Scholar] [CrossRef]
- Chou, Y.L.; Ko, C.Y.; Yen, C.C.; Chen, L.F.; Shaw, J.F. A Novel Recombinant Chlorophyllase1 from Chlamydomonas reinhardtii for the Production of Chlorophyllide Derivatives. J. Agric. Food Chem. 2015, 63, 9496–9503. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Dai, X.; Xu, Z.; Niu, Q.; Jiang, J.; Liu, Y. Molecular, structural and biochemical characterization of a novel recombinant chlorophyllase from cyanobacterium Oscillatoria acuminata PCC 6304. Microb. Cell Fact. 2021, 20, 14. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.N.; Zhong, R.H.; Wei, J.B.; Luo, H.H.; Eyal, Y.; Jin, H.L.; Wu, L.J.; Liang, K.Y.; Li, Y.M.; Chen, S.Z.; et al. Arabidopsis CHLOROPHYLLASE 1 protects young leaves from long-term photodamage by facilitating FtsH-mediated D1 degradation in photosystem II repair. Mol. Plant 2021, 14, 1149–1167. [Google Scholar] [CrossRef]
- Ardao, C.; Vennesland, B. Chlorophyllase Activity of Spinach Chloroplastin. Plant Physiol. 1960, 35, 368–371. [Google Scholar] [CrossRef] [Green Version]
- Terpstra, W. Influence of lecithin liposomes on chlorophyllase-catalyzed chlorophyll hydrolysis: Comparison of intramembraneous and solubilized Phaeodactylum chlorophyllase. Biochim. Biophys. Acta 1980, 600, 36–47. [Google Scholar] [CrossRef]
- Schoch, S.; Brown, J. The Action of Chlorophyllase on Chlorophyll-Protein Complexes. J. Plant Physiol. 1987, 126, 483–494. [Google Scholar] [CrossRef]
- Okazawa, A.; Tango, L.; Itoh, Y.; Fukusaki, E.; Kobayashi, A. Characterization and subcellular localization of chlorophyllase from Ginkgo biloba. Z. Naturforsch. C 2006, 61, 111–117. [Google Scholar] [CrossRef]
- Schenk, N.; Schelbert, S.; Kanwischer, M.; Goldschmidt, E.E.; Dörmann, P.; Hörtensteiner, S. The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett. 2007, 581, 5517–5525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Makita, S.; Schelbert, S.; Sano, S.; Ochiai, M.; Tsuchiya, T.; Hasegawa, S.F.; Hörtensteiner, S.; Tanaka, A.; Tanaka, R. Reexamination of chlorophyllase function implies its involvement in defense against chewing herbivores. Plant Physiol. 2015, 167, 660–670. [Google Scholar] [CrossRef] [Green Version]
- Jacob-Wilk, D.; Holland, D.; Goldschmidt, E.E.; Riov, J.; Eyal, Y. Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J. 1999, 20, 653–661. [Google Scholar] [CrossRef]
- Chen, L.-F.O.; Lin, C.-H.; Kelkar, S.M.; Chang, Y.-M.; Shaw, J.-F. Transgenic broccoli (Brassica oleracea var. italica) with antisense chlorophyllase (BoCLH1) delays postharvest yellowing. Plant Sci. 2008, 174, 25–31. [Google Scholar] [CrossRef]
- Guyer, L.; Hofstetter, S.S.; Christ, B.; Lira, B.S.; Rossi, M.; Hörtensteiner, S. Different Mechanisms Are Responsible for Chlorophyll Dephytylation during Fruit Ripening and Leaf Senescence in Tomato. Plant Physiol. 2014, 166, 44–56. [Google Scholar] [CrossRef]
- Lira, B.S.; de Setta, N.; Rosado, D.; Almeida, J.; Freschi, L.; Rossi, M. Plant degreening: Evolution and expression of tomato (Solanum lycopersicum) dephytylation enzymes. Gene 2014, 546, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Okazawa, A.; Itoh, Y.; Fukusaki, E.; Kobayashi, A. Expression of chlorophyllase is not induced during autumnal yellowing in Ginkgo biloba. Z. Naturforsch. C 2004, 59, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, C.E.; Arruda, P. Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol. 2002, 128, 1255–1263. [Google Scholar] [CrossRef] [Green Version]
- Kariola, T.; Brader, G.; Li, J.; Palva, E.T. Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 2005, 17, 282–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Gupta, S.M.; Sane, A.P.; Kumar, N. Chlorophyllase in Piper betle L. has a role in chlorophyll homeostasis and senescence dependent chlorophyll breakdown. Mol. Biol. Rep. 2012, 39, 7133–7142. [Google Scholar] [CrossRef]
- Hu, X.; Khan, I.; Jiao, Q.; Zada, A.; Jia, T. Chlorophyllase, a Common Plant Hydrolase Enzyme with a Long History, Is Still a Puzzle. Genes 2021, 12, 1871. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Banas, A.K.; Łabuz, J.; Sztatelman, O.; Gabrys, H.; Fiedor, L. Expression of enzymes involved in chlorophyll catabolism in Arabidopsis is light controlled. Plant Physiol. 2011, 157, 1497–1504. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, T.; Suzuki, T.; Yamada, T.; Shimada, H.; Masuda, T.; Ohta, H.; Takamiya, K. Chlorophyllase as a serine hydrolase: Identification of a putative catalytic triad. Plant Cell Physiol. 2003, 44, 96–101. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.C.; Chepyshko, H.; Chen, H.H.; Chu, C.C.; Chou, Y.F.; Akoh, C.C.; Shaw, J.F. Genes and biochemical characterization of three novel chlorophyllase isozymes from Brassica oleracea. J. Agric. Food Chem. 2010, 58, 8651–8657. [Google Scholar] [CrossRef]
- Rauwerdink, A.; Kazlauskas, R.J. How the Same Core Catalytic Machinery Catalyzes 17 Different Reactions: The Serine-Histidine-Aspartate Catalytic Triad of α/β-Hydrolase Fold Enzymes. ACS Catal. 2015, 5, 6153–6176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFeeters, R.F. Substrate specificity of chlorophyllase. Plant Physiol. 1975, 55, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Schoch, S.; Ihl, M. Substrate specificity of chlorophyllase from different plants. Z. Naturforsch. C 1998, 53, 21–26. [Google Scholar] [CrossRef]
- Tanaka, K.; Kakuno, T.; Yamashita, J.; Horio, T. Purification and properties of chlorophyllase from greened rye seedlings. J. Biochem. 1982, 92, 1763–1773. [Google Scholar] [CrossRef]
- Harpaz-Saad, S.; Azoulay, T.; Arazi, T.; Ben-Yaakov, E.; Mett, A.; Shiboleth, Y.M.; Hörtensteiner, S.; Gidoni, D.; Gal-On, A.; Goldschmidt, E.E.; et al. Chlorophyllase Is a Rate-Limiting Enzyme in Chlorophyll Catabolism and Is Posttranslationally Regulated. Plant Cell 2007, 19, 1007–1022. [Google Scholar] [CrossRef] [Green Version]
- Ponchel, F.; Toomes, C.; Bransfield, K.; Leong, F.T.; Douglas, S.H.; Field, S.L.; Bell, S.M.; Combaret, V.; Puisieux, A.; Mighell, A.J.; et al. Real-time PCR based on SYBR-Green I fluorescence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Arkus, K.A.; Cahoon, E.B.; Jez, J.M. Mechanistic analysis of wheat chlorophyllase. Arch. Biochem. Biophys. 2005, 438, 146–155. [Google Scholar] [CrossRef] [PubMed]


| Source | Accession No. | Molecular Weight | pI | Estimated Half-Life (Hours) | Instability Index | Aliphatic Index | GRAVY | ||
|---|---|---|---|---|---|---|---|---|---|
| Mammalian Reticulocytes, In Vitro | Yeast, In Vivo | E. coli, In Vivo | |||||||
| SlCLH1 | Solyc06g053980.2.1 | 34,048.30 | 6.86 | 30 h | >20 h | >10 h | 39.87 | 93.08 | −0.019 |
| SlCLH2 | Solyc09g065620.2.1 | 33,706.42 | 9.02 | 30 h | >20 h | >10 h | 44.59 | 98.76 | 0.081 |
| SlCLH3 | Solyc09g082600.1.1 | 34,600.81 | 8.17 | 30 h | >20 h | >10 h | 40.00 | 90.98 | −0.028 |
| SlCLH4 | Solyc12g005300.1.1 | 37,257.12 | 9.14 | 30 h | >20 h | >10 h | 41.32 | 86.03 | −0.146 |
| Recombinant SlCLHs | Substrate | Vmax | Km (μM) | kcat (10−3 s−1) | kcat/Km |
|---|---|---|---|---|---|
| (10−3 μmoL mg−1 min−1) | (10−5 s−1μM−1) | ||||
| SlCLH1 | Chl a | 18.93 | 30.42 | 23.36 | 76.79 |
| Chl b | 23.86 | 48.52 | 29.45 | 60.68 | |
| Phein a | 2.61 | 302.50 | 3.22 | 1.06 | |
| SlCLH2 | Chl a | 1.96 | 140.17 | 2.41 | 1.72 |
| Chl b | 2.22 | 109.03 | 2.73 | 2.50 | |
| SlCLH3 | Chl a | 0.41 | 258.86 | 0.51 | 0.20 |
| Chl b | 0.59 | 295.32 | 0.73 | 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Meng, X.; Ren, Y.; Zhang, M.; Chen, Z.; Zhang, Z.; Pang, X.; Zhang, X. Genes, Structural, and Biochemical Characterization of Four Chlorophyllases from Solanum lycopersicum. Int. J. Mol. Sci. 2022, 23, 11716. https://doi.org/10.3390/ijms231911716
Liu G, Meng X, Ren Y, Zhang M, Chen Z, Zhang Z, Pang X, Zhang X. Genes, Structural, and Biochemical Characterization of Four Chlorophyllases from Solanum lycopersicum. International Journal of Molecular Sciences. 2022; 23(19):11716. https://doi.org/10.3390/ijms231911716
Chicago/Turabian StyleLiu, Guangyuan, Xue Meng, Yujun Ren, Min Zhang, Ziqing Chen, Zhaoqi Zhang, Xuequn Pang, and Xuelian Zhang. 2022. "Genes, Structural, and Biochemical Characterization of Four Chlorophyllases from Solanum lycopersicum" International Journal of Molecular Sciences 23, no. 19: 11716. https://doi.org/10.3390/ijms231911716






