Omega-3 Polyunsaturated Fatty Acids Protect against High-Fat Diet-Induced Morphological and Functional Impairments of Brown Fat in Transgenic Fat-1 Mice
Abstract
:1. Introduction
2. Results
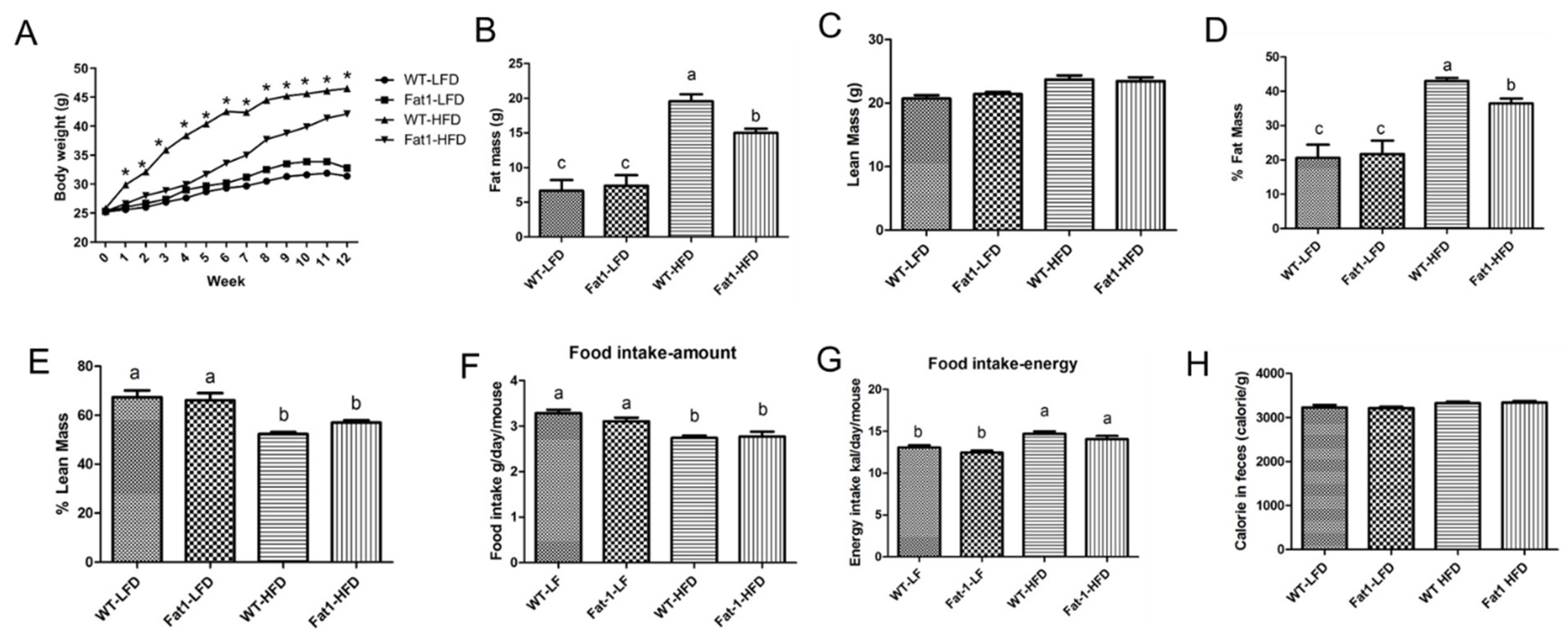
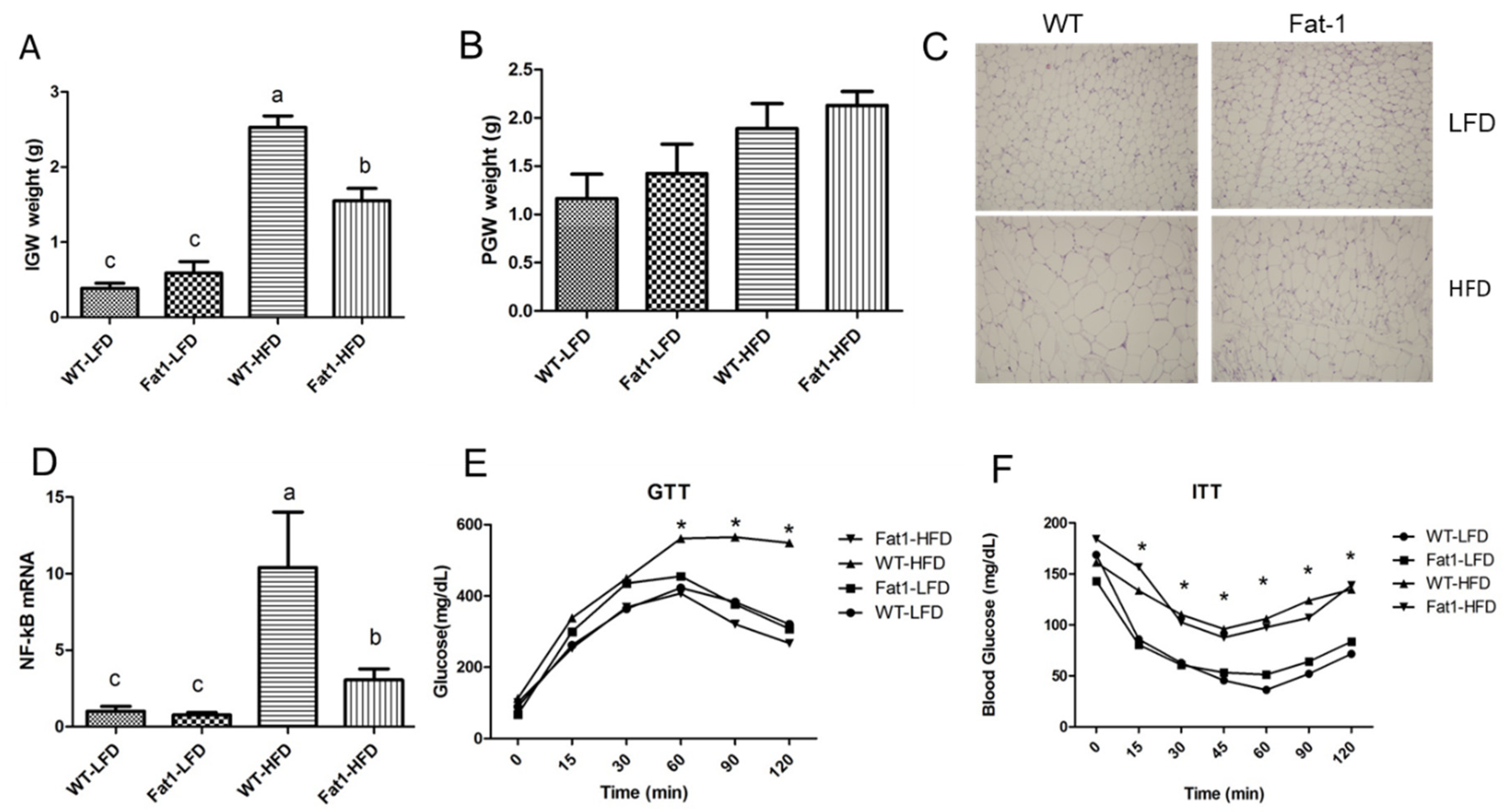
2.1. Transgenic Fat-1 Mice Were Resistant against Obesity Induced by a High-Fat Diet
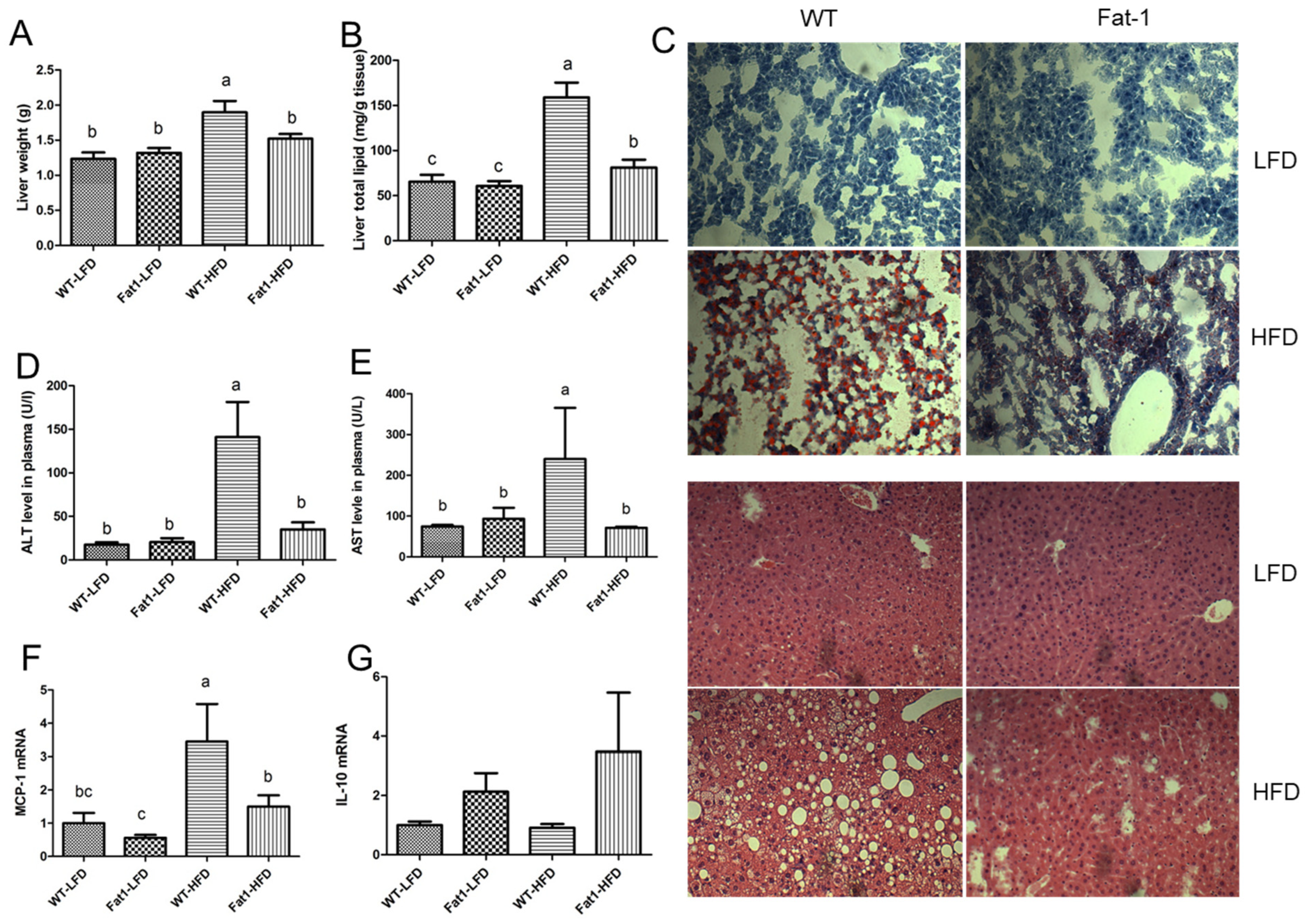
2.2. Fat-1 Mice Were Largely Protected from HFD-Induced Fatty Liver
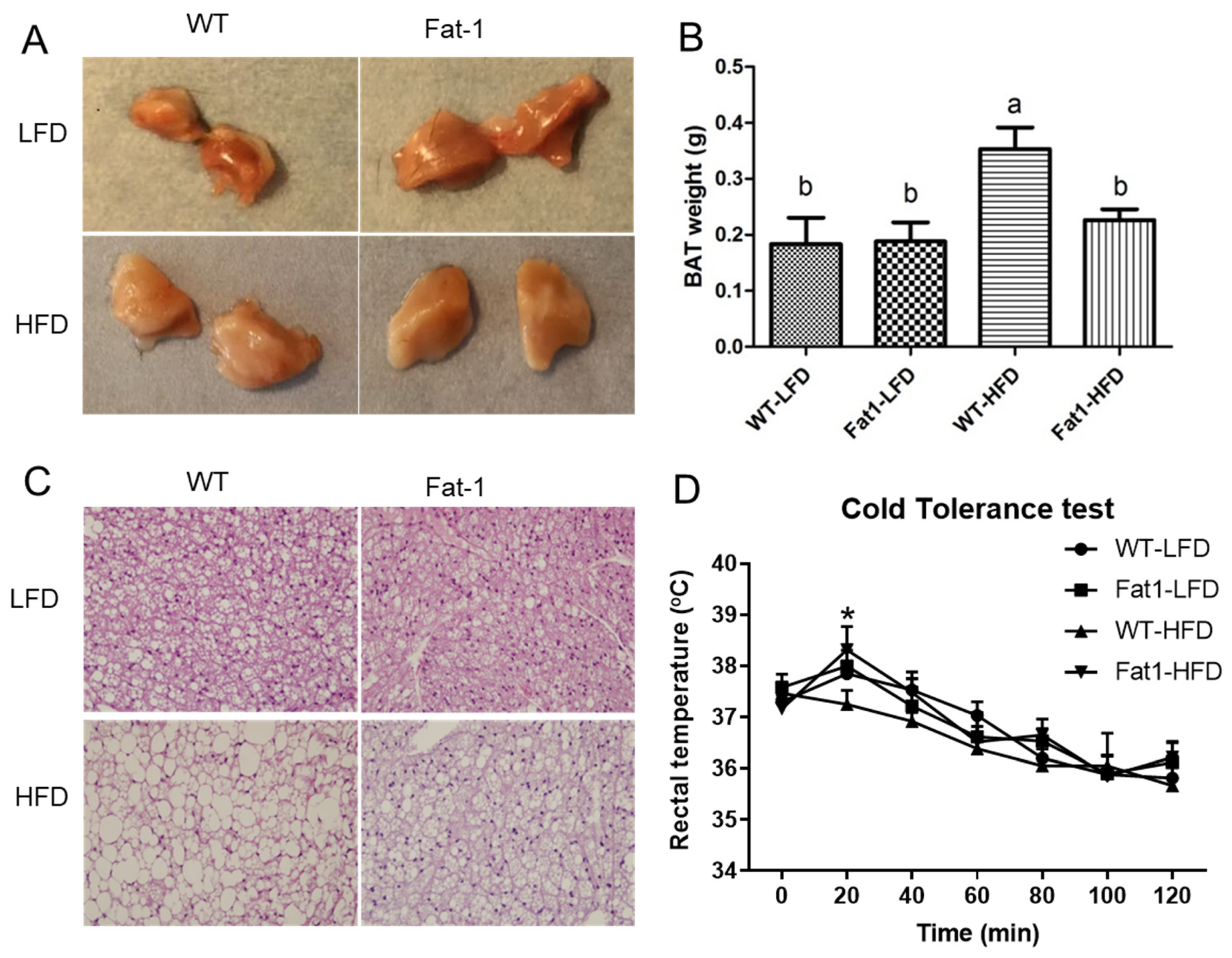
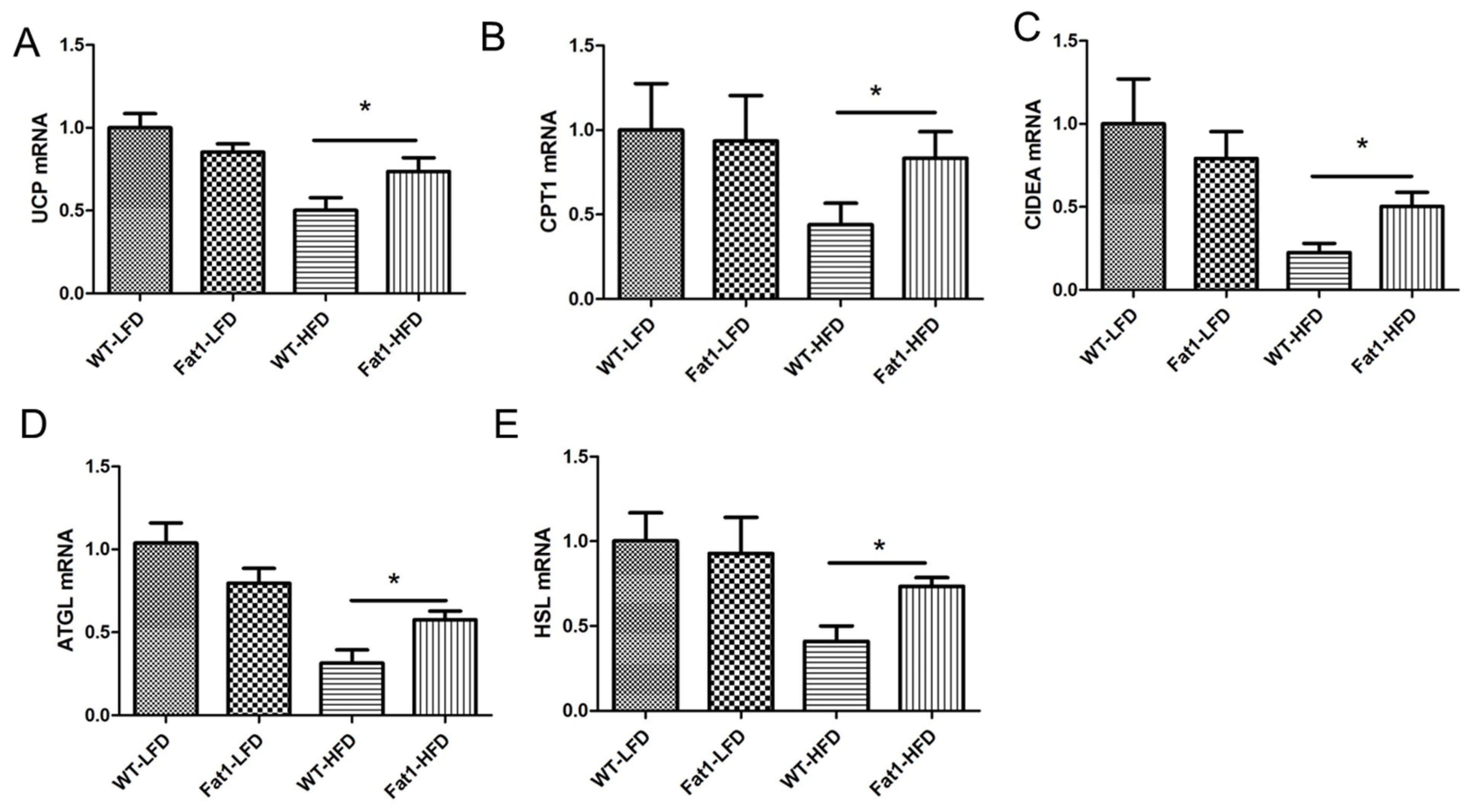
2.3. Endogenous n-3-PUFAs in Fat-1 Mice Protected against HFD-Induced Morphological and Functional Impairments of Brown Fat
2.4. Fat-1 Mice Were Resistant to LPS-Induced Suppression of UCP1 and PGC-1 Expression and Lipid Deposits in BAT
2.5. LPS Suppressed the Upregulation of UCP1 Induced by the β3-Adrenergic Agonist, CL-316243, in Mouse Brown Adipocytes In Vitro
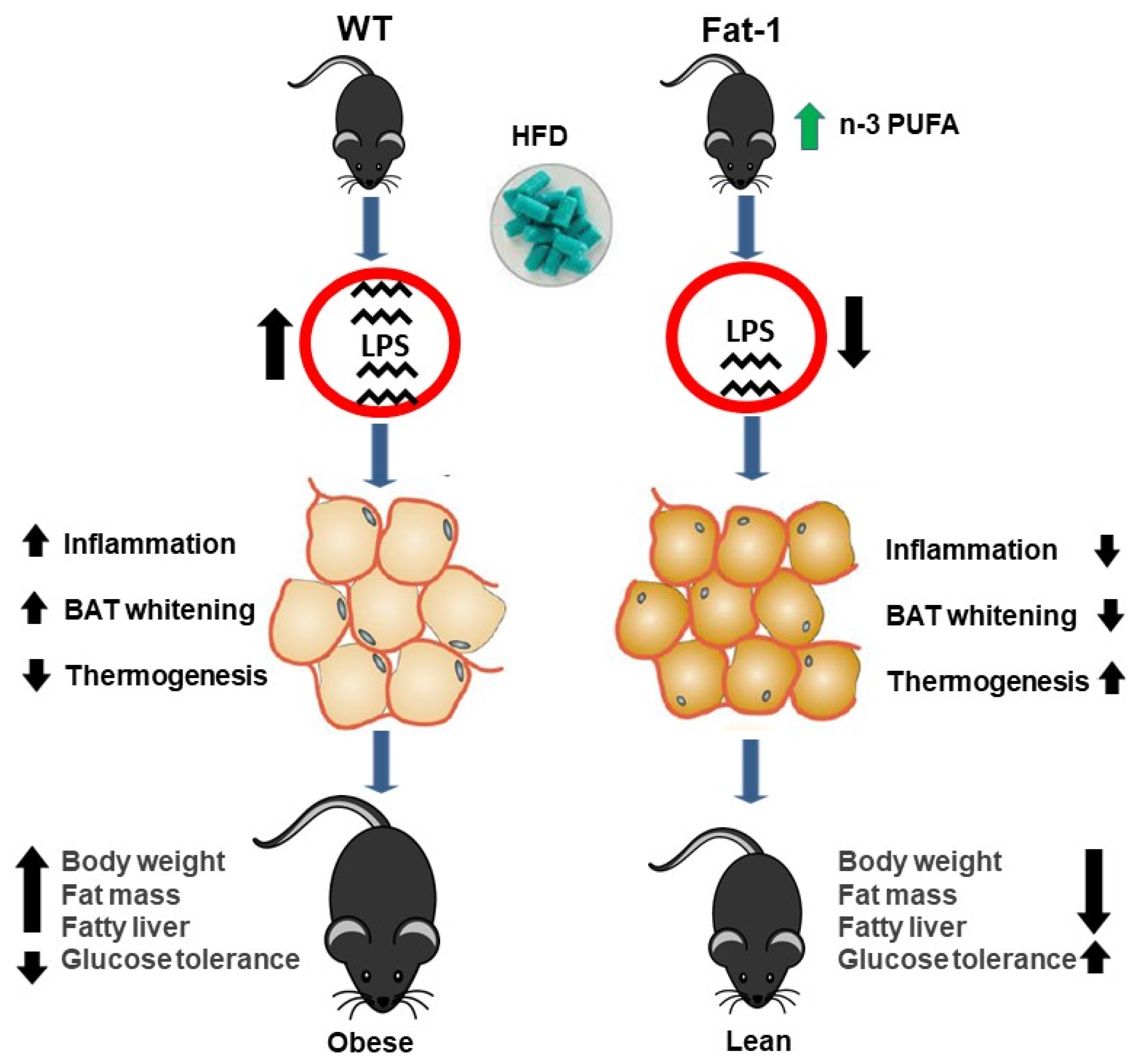
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animal Experiments
5.2. RNA Extraction and Real Time PCR
5.3. Histology
5.4. Liver Total Lipid Quantification
5.5. Cold Tolerance Test
5.6. Intraperitoneal Insulin Tolerance Test (IPITT)
5.7. Intraperitoneal Glucose Tolerance Test (IGTT)
5.8. Cell Culture
5.9. Measurement of LPS Concentration
5.10. Total Fecal Energy Content Analysis
5.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Targher, G.; Byrne, C.D. Obesity: Metabolically healthy obesity and NAFLD. Nat. Reviews. Gastroenterol. Hepatol. 2016, 13, 442–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.C., Jr. Multiple risk factors for cardiovascular disease and diabetes mellitus. Am. J. Med. 2007, 120, S3–S11. [Google Scholar] [CrossRef] [PubMed]
- Cava, E.; Neri, B.; Carbonelli, M.G.; Riso, S.; Carbone, S. Obesity pandemic during COVID-19 outbreak: Narrative review and future considerations. Clin. Nutr. 2021, 40, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.; Mejia-Guevara, I.; Estrada, K.; Liu, S.Y.; Glymour, M.M. Association of a Genetic Risk Score With Body Mass Index Across Different Birth Cohorts. JAMA 2016, 316, 63–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredriksson, R.; Hagglund, M.; Olszewski, P.K.; Stephansson, O.; Jacobsson, J.A.; Olszewska, A.M.; Levine, A.S.; Lindblom, J.; Schioth, H.B. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology 2008, 149, 2062–2071. [Google Scholar] [CrossRef] [Green Version]
- Kilpelainen, T.O.; Qi, L.; Brage, S.; Sharp, S.J.; Sonestedt, E.; Demerath, E.; Ahmad, T.; Mora, S.; Kaakinen, M.; Sandholt, C.H.; et al. Physical activity attenuates the influence of FTO variants on obesity risk: A meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011, 8, e1001116. [Google Scholar] [CrossRef]
- Apovian, C.M. Obesity: Definition, comorbidities, causes, and burden. Am. J. Manag. Care 2016, 22, s176–s185. [Google Scholar]
- Rolland-Cachera, M.F.; Akrout, M.; Peneau, S. Nutrient Intakes in Early Life and Risk of Obesity. Int. J. Environ. Res. Public Health 2016, 13, 564. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef]
- Simopoulos, A.P. An Increase in the Omega-6/Omega-3 Fatty Acid Ratio Increases the Risk for Obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.X. The importance of omega-6/omega-3 fatty acid ratio in cell function. The gene transfer of omega-3 fatty acid desaturase. World Rev. Nutr. Diet. 2003, 92, 23–36. [Google Scholar] [PubMed]
- Wang, L.; Manson, J.E.; Rautiainen, S.; Gaziano, J.M.; Buring, J.E.; Tsai, M.Y.; Sesso, H.D. A prospective study of erythrocyte polyunsaturated fatty acid, weight gain, and risk of becoming overweight or obese in middle-aged and older women. Eur. J. Nutr. 2016, 55, 687–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amri, E.Z.; Ailhaud, G.; Grimaldi, P.A. Fatty acids as signal transducing molecules: Involvement in the differentiation of preadipose to adipose cells. J. Lipid Res. 1994, 35, 930–937. [Google Scholar] [CrossRef]
- Jump, D.B.; Clarke, S.D.; Thelen, A.; Liimatta, M. Coordinate regulation of glycolytic and lipogenic gene expression by polyunsaturated fatty acids. J. Lipid Res. 1994, 35, 1076–1084. [Google Scholar] [CrossRef]
- Schwinkendorf, D.R.; Tsatsos, N.G.; Gosnell, B.A.; Mashek, D.G. Effects of central administration of distinct fatty acids on hypothalamic neuropeptide expression and energy metabolism. Int. J. Obes. 2011, 35, 336–344. [Google Scholar] [CrossRef] [Green Version]
- James, M.J.; Gibson, R.A.; Cleland, L.G. Dietary polyunsaturated fatty acids and inflammatory mediator production. Am. J. Clin. Nutr. 2000, 71, 343S–348S. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, F.R.; Wei, D.; Jia, W.; Kang, J.X.; Stefanovic-Racic, M.; Dai, Y.; Zhao, A.Z. Endogenous omega-3 polyunsaturated fatty acid production confers resistance to obesity, dyslipidemia, and diabetes in mice. Mol. Endocrinol. 2014, 28, 1316–1328. [Google Scholar] [CrossRef] [Green Version]
- Poekes, L.; Lanthier, N.; Leclercq, I.A. Brown adipose tissue: A potential target in the fight against obesity and the metabolic syndrome. Clin. Sci. 2015, 129, 933–949. [Google Scholar] [CrossRef]
- Kim, S.H.; Plutzky, J. Brown Fat and Browning for the Treatment of Obesity and Related Metabolic Disorders. Diabetes Metab. J. 2016, 40, 12–21. [Google Scholar] [CrossRef]
- Pan, R.; Zhu, X.; Maretich, P.; Chen, Y. Combating Obesity With Thermogenic Fat: Current Challenges and Advancements. Front. Endocrinol. 2020, 11, 185. [Google Scholar] [CrossRef] [Green Version]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.J.; Enerback, S.; et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Sampath, S.C.; Sampath, S.C.; Bredella, M.A.; Cypess, A.M.; Torriani, M. Imaging of Brown Adipose Tissue: State of the Art. Radiology 2016, 280, 4–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosen, E.D.; Spiegelman, B.M. What we talk about when we talk about fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [Green Version]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef] [Green Version]
- Ohno, T.; Ohinata, H.; Ogawa, K.; Kuroshima, A. Fatty acid profiles of phospholipids in brown adipose tissue from rats during cold acclimation and repetitive intermittent immobilization: With special reference to docosahexaenoic acid. Jpn. J. Physiol. 1996, 46, 265–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargut, T.C.; Silva-e-Silva, A.C.; Souza-Mello, V.; Mandarim-de-Lacerda, C.A.; Aguila, M.B. Mice fed fish oil diet and upregulation of brown adipose tissue thermogenic markers. Eur. J. Nutr. 2016, 55, 159–169. [Google Scholar] [CrossRef]
- Kim, M.; Goto, T.; Yu, R.; Uchida, K.; Tominaga, M.; Kano, Y.; Takahashi, N.; Kawada, T. Fish oil intake induces UCP1 upregulation in brown and white adipose tissue via the sympathetic nervous system. Sci. Rep. 2015, 5, 18013. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Ide, T. Dietary n-3 fatty acids affect mRNA level of brown adipose tissue uncoupling protein 1, and white adipose tissue leptin and glucose transporter 4 in the rat. Br. J. Nutr. 2000, 84, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Ohinata, H.; Saha, S.K.; Ohno, T.; Hata, N.; Misawa, Y.; Kuroshima, A. Effect of dietary docosahexaenoic acid on in vitro thermogenesis and fatty acid compositions of brown adipose tissue. Jpn. J. Physiol. 1998, 48, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.X.; Wang, J.; Wu, L.; Kang, Z.B. Transgenic mice: Fat-1 mice convert n-6 to n-3 fatty acids. Nature 2004, 427, 504. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.X. A transgenic mouse model for gene-nutrient interactions. J. Nutr. Nutr. 2008, 1, 172–177. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Arita, M.; Taguchi, R.; Kang, J.X.; Marette, A. Transgenic restoration of long-chain n-3 fatty acids in insulin target tissues improves resolution capacity and alleviates obesity-linked inflammation and insulin resistance in high-fat-fed mice. Diabetes 2010, 59, 3066–3073. [Google Scholar] [CrossRef] [Green Version]
- An, L.; Pang, Y.W.; Gao, H.M.; Tao, L.; Miao, K.; Wu, Z.H.; Tian, J.H. Heterologous expression of C. elegans fat-1 decreases the n-6/n-3 fatty acid ratio and inhibits adipogenesis in 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2012, 428, 405–410. [Google Scholar] [CrossRef]
- Kim, E.H.; Bae, J.S.; Hahm, K.B.; Cha, J.Y. Endogenously synthesized n-3 polyunsaturated fatty acids in fat-1 mice ameliorate high-fat diet-induced non-alcoholic fatty liver disease. Biochem. Pharmacol. 2012, 84, 1359–1365. [Google Scholar] [CrossRef]
- Lopez-Vicario, C.; Gonzalez-Periz, A.; Rius, B.; Moran-Salvador, E.; Garcia-Alonso, V.; Lozano, J.J.; Bataller, R.; Cofan, M.; Kang, J.X.; Arroyo, V.; et al. Molecular interplay between Delta5/Delta6 desaturases and long-chain fatty acids in the pathogenesis of non-alcoholic steatohepatitis. Gut 2014, 63, 344–355. [Google Scholar] [CrossRef]
- Belchior, T.; Paschoal, V.A.; Magdalon, J.; Chimin, P.; Farias, T.M.; Chaves-Filho, A.B.; Gorjao, R.; St-Pierre, P.; Miyamoto, S.; Kang, J.X.; et al. Omega-3 fatty acids protect from diet-induced obesity, glucose intolerance, and adipose tissue inflammation through PPARgamma-dependent and PPARgamma-independent actions. Mol. Nutr. Food Res. 2015, 59, 957–967. [Google Scholar] [CrossRef]
- White, P.J.; Mitchell, P.L.; Schwab, M.; Trottier, J.; Kang, J.X.; Barbier, O.; Marette, A. Transgenic omega-3 PUFA enrichment alters morphology and gene expression profile in adipose tissue of obese mice: Potential role for protectins. Metab. Clin. Exp. 2015, 64, 666–676. [Google Scholar] [CrossRef]
- Song, M.Y.; Wang, J.; Lee, Y.; Lee, J.; Kwon, K.S.; Bae, E.J.; Park, B.H. Enhanced M2 macrophage polarization in high n-3 polyunsaturated fatty acid transgenic mice fed a high-fat diet. Mol. Nutr. Food Res. 2016, 60, 481–2492. [Google Scholar] [CrossRef]
- Shimizu, I.; Walsh, K. The Whitening of Brown Fat and Its Implications for Weight Management in Obesity. Curr. Obes. Rep. 2015, 4, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, I.; Aprahamian, T.; Kikuchi, R.; Shimizu, A.; Papanicolaou, K.N.; MacLauchlan, S.; Maruyama, S.; Walsh, K. Vascular rarefaction mediates whitening of brown fat in obesity. J. Clin. Investig. 2014, 124, 2099–2112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okla, M.; Wang, W.; Kang, I.; Pashaj, A.; Carr, T.; Chung, S. Activation of Toll-like receptor 4 (TLR4) attenuates adaptive thermogenesis via endoplasmic reticulum stress. J. Biol. Chem. 2015, 290, 26476–26490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, T.; Naknukool, S.; Yoshitake, R.; Hanafusa, Y.; Tokiwa, S.; Li, Y.; Sakamoto, T.; Nitta, T.; Kim, M.; Takahashi, N.; et al. Proinflammatory cytokine interleukin-1beta suppresses cold-induced thermogenesis in adipocytes. Cytokine 2016, 77, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Kaliannan, K.; Wang, B.; Li, X.Y.; Kim, K.J.; Kang, J.X. A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci. Rep. 2015, 5, 11276. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, L.; Nie, Y.-H.; Chen, C.-Y.; Li, X.-Y.; Kaliannan, K.; Kang, J.X. Omega-3 Polyunsaturated Fatty Acids Protect against High-Fat Diet-Induced Morphological and Functional Impairments of Brown Fat in Transgenic Fat-1 Mice. Int. J. Mol. Sci. 2022, 23, 11903. https://doi.org/10.3390/ijms231911903
Hao L, Nie Y-H, Chen C-Y, Li X-Y, Kaliannan K, Kang JX. Omega-3 Polyunsaturated Fatty Acids Protect against High-Fat Diet-Induced Morphological and Functional Impairments of Brown Fat in Transgenic Fat-1 Mice. International Journal of Molecular Sciences. 2022; 23(19):11903. https://doi.org/10.3390/ijms231911903
Chicago/Turabian StyleHao, Lei, Yong-Hui Nie, Chih-Yu Chen, Xiang-Yong Li, Kanakaraju Kaliannan, and Jing X. Kang. 2022. "Omega-3 Polyunsaturated Fatty Acids Protect against High-Fat Diet-Induced Morphological and Functional Impairments of Brown Fat in Transgenic Fat-1 Mice" International Journal of Molecular Sciences 23, no. 19: 11903. https://doi.org/10.3390/ijms231911903



