Caspase 3 and Cleaved Caspase 3 Expression in Tumorogenesis and Its Correlations with Prognosis in Head and Neck Cancer: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Material and Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Data Extraction
2.4. Quality Assessment
2.5. Statistical Analysis
3. Results
3.1. Study Selection Process and Study Features
3.2. Quality Assessment within Studies
3.3. Quantitative Evaluation (Meta-Analysis)
3.3.1. Comparative Evaluation between Caspase 3 and Cleaved Caspase 3 Expression in OPMD and HNC
3.3.2. Quantitative Evaluation (Meta-Analysis)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehanna, H.; Paleri, V.; West, C.M.L.; Nutting, C. Head and neck cancer—Part 1: Epidemiology, presentation, and prevention. BMJ 2010, 341, c4684. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.D.; Grandis, J.R. The Molecular Pathogenesis of Head and Neck Cancer. Cancer Biol. Ther. 2010, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sturgis, E.M.; Cinciripini, P.M. Trends in head and neck cancer incidence in relation to smoking prevalence: An emerging epidemic of human papillomavirus-associated cancers? Cancer 2007, 110, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.; Fedewa, S.; Chen, A.Y. Epidemiology and Demographics of the Head and Neck Cancer Population. Oral Maxillofac. Surg. Clin. N. Am. 2018, 30, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Andressakis, D.; Lazaris, A.C.; Tsiambas, E.; Kavantzas, N.; Rapidis, A.; Patsouris, E. Evaluation of caspase-3 and caspase-8 deregulation in tongue squamous cell carcinoma, based on immunohistochemistry and computerised image analysis. J. Laryngol. Otol. 2008, 122, 1213–1218. [Google Scholar] [CrossRef]
- Xu, B.; Salama, A.M.; Valero, C.; Yuan, A.; Khimraj, A.; Saliba, M.; Zanoni, D.K.; Ganly, I.; Patel, S.G.; Katabi, N.; et al. The prognostic role of histologic grade, worst pattern of invasion, and tumor budding in early oral tongue squamous cell carcinoma: A comparative study. Virchows Arch. 2021, 479, 597–606. [Google Scholar] [CrossRef]
- Kovtunenko, O.V.; Bakaiev, A.A.; Shponka, I.S. Analysis of expression of p63 and caspase-3 and their predictive value in patients with squamous cell carcinoma of maxillary sinus. Wiad Lek. 2019, 72, 2305–2314. [Google Scholar] [CrossRef]
- Lo Muzio, L.; Santarelli, A.; Pannone, G.; Staibano, S.; De Rosa, G. Survivin in head neck cancerogenesis: A valid prognosis marker and a possible therapeutic target. Anticancer. Res. 2004, 24, 3620. [Google Scholar]
- Schenker, H.; Büttner-Herold, M.; Fietkau, R.; Distel, L.V. Cell-in-cell structures are more potent predictors of outcome than senescence or apoptosis in head and neck squamous cell carcinomas. Radiat. Oncol. 2017, 12, 19. [Google Scholar] [CrossRef]
- Bascones-Martinez, A.; Rodriguez-Gutierrez, R.; Rodriguez-Gomez, E.; Gil-Montoya, J.; Gomez-Font, R.; Gonzalez-Moles, M. Evaluation of p53, Caspase-3, Bcl-2, and Ki-67 markers in oral squamous cell carcinoma and premalignant epithelium in a sample from Alava Province (Spain). Med. Oral. Patol. Oral. Cir. Bucal. 2013, 18, e846–e850. [Google Scholar] [CrossRef]
- Fernald, K.; Kurokawa, M. Evading apoptosis in cancer. Trends Cell Biol. 2013, 23, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.M.; Saavedra, V.; Ianez, R.C.F.; Sousa, E.A.; Gomes, N.; Kelner, N.; Nagai, M.A.; Kowalski, L.P.; Soares, F.A.; Lourenço, S.V.; et al. Apoptotic signaling in salivary mucoepidermoid carcinoma. Head Neck 2019, 41, 2904–2913. [Google Scholar] [CrossRef] [PubMed]
- Malheiros, C.C.C.; Vanessa, L.S.; Paulo, K.L.; Soares, A.F. Apoptotic signaling in oral squamous cell carcinoma and association with clinicopathological characteristics. VIRCHOWS Arch. 2009, 455, 333. [Google Scholar]
- Coutinho-Camillo, C.M.; Lourenço, S.V.; Nishimoto, I.N.; Kowalski, L.P.; Soares, F.A. Caspase expression in oral squamous cell carcinoma. Head Neck 2010, 33, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Bundela, S.; Tiwari, R.P.; Bisen, P.S. Oncoapoptotic Markers in Oral Cancer: Prognostics and Therapeutic Perspective. Mol. Diagn. Ther. 2014, 18, 483–494. [Google Scholar] [CrossRef]
- Nikitakis, N.G.; Sauk, J.; Papanicolaou, S.I. The role of apoptosis in oral disease: Mechanisms; aberrations in neoplastic, autoimmune, infectious, hematologic, and developmental diseases; and therapeutic opportunities. Oral Surgery Oral Med. Oral Pathol. Oral Radiol. Endodontology 2004, 97, 476–490. [Google Scholar] [CrossRef]
- Dutsch-Wicherek, M.; Sikora, J.; Tomaszewska, R. The possible biological role of metallothionein in apoptosis. Front. Biosci. 2008, 13, 4029–4038. [Google Scholar] [CrossRef]
- Li, S.; Yang, Y.; Ding, Y.; Tang, X.; Sun, Z. Impacts of survivin and caspase-3 on apoptosis and angiogenesis in oral cancer. Oncol. Lett. 2017, 14, 3774–3779. [Google Scholar] [CrossRef]
- Leite, A.F.S.D.A.; Bernardo, V.G.; Buexm, L.A.; FONSECA, E.C.D.; SILVA, L.E.D.; Barroso, D.R.C.; Lourenço, S.D.Q.C. Immunoexpression of cleaved caspase-3 shows lower apoptotic area indices in lip carcinomas than in intraoral cancer. J. Appl. Oral Sci. 2016, 24, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Bisen, P.S. Oncoapoptotic signaling and deregulated target genes in cancers: Special reference to oral cancer. Biochim. Biophys. Acta 2013, 1836, 123–145. [Google Scholar] [CrossRef] [PubMed]
- Peltanova, B.; Raudenska, M.; Masarik, M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: A systematic review. Mol. Cancer 2019, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Sagari, S.; Sanadhya, S.; Doddamani, M.; Rajput, R. Molecular markers in oral lichen planus: A systematic review. J. Oral Maxillofac. Pathol. 2016, 20, 115–121. [Google Scholar] [CrossRef]
- Boice, A.; Bouchier-Hayes, L. Targeting apoptotic caspases in cancer. Biochim. Biophys. Acta 2020, 1867, 118688. [Google Scholar] [CrossRef]
- Poomsawat, S.; Punyasingh, J.; Vejchapipat, P. Overexpression of Survivin and Caspase 3 in Oral Carcinogenesis. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 65–71. [Google Scholar] [CrossRef]
- Dozic, B.; Glumac, S.; Boricic, N.; Dozic, M.; Anicic, B.; Boricic, I. Immunohistochemical expression of caspases 9 and 3 in adenoid cystic carcinoma of salivary glands and association with clinicopathological parameters. J. Balk. Union Oncol. 2016, 21, 152–160. [Google Scholar]
- Wu, J.; Gao, H.; Ge, W.; He, J. Over expression of PTEN induces apoptosis and prevents cell proliferation in breast cancer cells. Acta Biochim. Pol. 2020, 67, 515–519. [Google Scholar] [CrossRef]
- Van Opdenbosch, N.; Lamkanfi, M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef]
- Chowdhury, I.; Tharakan, B.; Bhat, G.K. Caspases—An update. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 2008, 151, 10–27. [Google Scholar] [CrossRef]
- Salvesen, G.S. Caspases and apoptosis. Essays Biochem. 2002, 38, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Clark, A.C. Evolution of the folding landscape of effector caspases. J. Biol. Chem. 2021, 297, 101249. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, J. Icariin inhibits oral squamous cell carcinoma cell proliferation and induces apoptosis via inhibiting the NFκB and PI3K/AKT pathways. Exp. Ther. Med. 2021, 22, 3. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Duan, X.H.; Wang, X.L.; Yang, Q.Y.; Feng, Y.; Zhang, J.X. Effect of NLK on the proliferation and invasion of laryngeal carcinoma cells by regulating CDCP1. Eur. Rev. Med. Pharmacol Sci. 2019, 23, 6226–6233. [Google Scholar] [PubMed]
- Li, J.; Yuan, J. Caspases in apoptosis and beyond. Oncogene 2008, 27, 6194–6206. [Google Scholar] [CrossRef]
- Fan, T.-J.; Han, L.-H.; Cong, R.-S.; Liang, J. Caspase Family Proteases and Apoptosis. Acta Biochim. Biophys. Sin. 2005, 37, 719–727. [Google Scholar] [CrossRef]
- Oudejans, J.J.; Harijadi, A.; Cillessen, S.A.G.M.; Busson, P.; Tan, I.B.; Dukers, D.F.; Vos, W.; Hariwiyanto, B.; Middeldorp, J.; Meijer, C.J.L.M. Absence of caspase 3 activation in neoplastic cells of nasopharyngeal carcinoma biopsies predicts rapid fatal outcome. Mod. Pathol. 2005, 18, 877–885. [Google Scholar] [CrossRef]
- Sasabe, E.; Tatemoto, Y.; Li, D.; Yamamoto, T.; Osaki, T. Mechanism of HIF-1alpha-dependent suppression of hypoxia-induced apoptosis in squamous cell carcinoma cells. Cancer Sci. 2005, 96, 394–402. [Google Scholar] [CrossRef]
- Tonissi, F.; Lattanzio, L.; Astesana, V.; Cavicchioli, F.; Ghiglia, A.; Monteverde, M.; Vivenza, D.; Gianello, L.; Russi, E.; Merlano, M.; et al. Reoxygenation Reverses Hypoxia-related Radioresistance in Head and Neck Cancer Cell Lines. Anticancer Res. 2016, 36, 2211–2215. [Google Scholar]
- Tian, L.; Tao, Z.; Ye, H.; Li, G.; Zhan, Z.; Tuo, H. Over-expression of MEOX2 promotes apoptosis through inhibiting the PI3K/Akt pathway in laryngeal cancer cells. Neoplasma 2018, 65, 745–752. [Google Scholar] [CrossRef]
- Suminami, Y.; Nagashima, S.; Vujanovic, N.L.; Hirabayashi, K.; Kato, H.; Whiteside, T.L. Inhibition of apoptosis in human tumour cells by the tumour-associated serpin, SCC antigen-1. Br. J. Cancer 2000, 82, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Li, F.; Liu, X.; Li, W.; Shi, W.; Liu, F.-F.; O’Sullivan, B.; He, Z.; Peng, Y.; Tan, A.C.; et al. Caspase 3–mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 2011, 17, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.; Zhao, M.; Qiu, H.; Shi, D.; Wang, J.; Tian, Y.; Lin, L.; Deng, W. Effusanin E Suppresses Nasopharyngeal Carcinoma Cell Growth by Inhibiting NF-κB and COX-2 Signaling. PLoS ONE 2014, 9, e109951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-H.; Zhang, Y.; Herman, B. Caspases, apoptosis and aging. Ageing Res. Rev. 2003, 2, 357–366. [Google Scholar] [CrossRef]
- Stennicke, H.R.; Salvesen, G.S. Properties of the caspases. Biochim. Biophys. Acta 1998, 1387, 17–31. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Festjens, N.; Declercq, W.; Berghe, T.V.; Vandenabeele, P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2006, 14, 44–55. [Google Scholar] [CrossRef]
- Green, D.R. Caspases and Their Substrates. Cold Spring Harb. Perspect. Biol. 2022, 14, a041012. [Google Scholar] [CrossRef]
- Hounsell, C.; Fan, Y. The Duality of Caspases in Cancer, as Told through the Fly. Int. J. Mol. Sci. 2021, 22, 8927. [Google Scholar] [CrossRef]
- Pop, C.; Salvesen, G.S. Human Caspases: Activation, Specificity, and Regulation. J. Biol. Chem. 2009, 284, 21777–21781. [Google Scholar] [CrossRef]
- Bascones-Ilundain, C.; González-Moles, M.; Campo-Trapero, J.; Gil-Montoya, J.; Esparza-Gómez, G.; Cano-Sánchez, J.; Bascones-Martínez, A. No differences in caspase-3 and Bax expression in atrophic-erosive vs. reticular oral lichen planus. J. Eur. Acad. Dermatol. Venereol. 2007, 22, 204–212. [Google Scholar] [CrossRef]
- Kuribayashi, K.; Mayes, P.A.; El-Deiry, W.S. What are caspases 3 and 7 doing upstream of the mitochondria? Cancer Biol. Ther. 2006, 5, 763–765. [Google Scholar] [CrossRef] [PubMed]
- Asadi, M.; Taghizadeh, S.; Kaviani, E.; Vakili, O.; Taheri-Anganeh, M.; Tahamtan, M.; Savardashtaki, A. Caspase-3: Structure, function, and biotechnological aspects. Biotechnol. Appl. Biochem. 2021, 69, 1633–1645. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Huang, X.; Zheng, L.; Zheng, F.; Dong, J.; Chen, F.; Zeng, W. Biosensors for Caspase-3: From chemical methodologies to biomedical applications. Talanta 2022, 240, 123198. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Yadav, R.; Jain, S.; Vaidya, A. Caspase-3: A primary target for natural and synthetic compounds for cancer therapy. Chem. Biol. Drug Des. 2021, 98, 144–165. [Google Scholar] [CrossRef]
- Eskandari, E.; Eaves, C.J. Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J. Cell Biol. 2022, 221, e202201159. [Google Scholar]
- Beroske, L.; Wyngaert, T.V.D.; Stroobants, S.; Van der Veken, P.; Elvas, F. Molecular Imaging of Apoptosis: The Case of Caspase-3 Radiotracers. Int. J. Mol. Sci. 2021, 22, 3948. [Google Scholar] [CrossRef]
- Huang, J.-S.; Yang, C.-M.; Wang, J.-S.; Liou, H.-H.; Hsieh, I.-C.; Li, G.-C.; Huang, S.-J.; Shu, C.-W.; Fu, T.-Y.; Lin, Y.-C.; et al. Caspase-3 expression in tumorigenesis and prognosis of buccal mucosa squamous cell carcinoma. Oncotarget 2017, 8, 84237–84247. [Google Scholar] [CrossRef]
- Hague, A.; Eveson, J.W.; MacFarlane, M.; Huntley, S.; Janghra, N.; Thavaraj, S. Caspase-3 expression is reduced, in the absence of cleavage, in terminally differentiated normal oral epithelium but is increased in oral squamous cell carcinomas and correlates with tumour stage. J. Pathol. 2004, 204, 175–182. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, D. A caspase-3 activatable photoacoustic probe for in vivo imaging of tumor apoptosis. Methods Enzymol. 2021, 657, 21–57. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, B.; Li, D.; Wang, G.; Han, X.; Sun, X. GSDME mediates caspase-3-dependent pyroptosis in gastric cancer. Biochem. Biophys. Res. Commun. 2018, 495, 1418–1425. [Google Scholar] [CrossRef]
- Tobón-Arroyave, S.; Villegas-Acosta, F.; Ruiz-Restrepo, S.; Vieco-Duran, B.; Restrepo-Misas, M.; Londono-Lopez, M. Expression of caspase-3 and structural changes associated with apoptotic cell death of keratinocytes in oral lichen planus. Oral Dis. 2004, 10, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Damiani, E.; Yuecel, R.; Wallace, H.M. Repurposing of idebenone as a potential anti-cancer agent. Biochem. J. 2019, 476, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-P.; Lee, Y.-T.; Wang, J.-Y.; Miller, S.A.; Chiou, S.-H.; Hung, M.-C.; Hung, S.-C. Survival of Cancer Stem Cells under Hypoxia and Serum Depletion via Decrease in PP2A Activity and Activation of p38-MAPKAPK2-Hsp27. PLoS ONE 2012, 7, e49605. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, S.; Zhao, R.; Zhao, Q.; Zhang, T. Upregulated miR-9-3p Promotes Cell Growth and Inhibits Apoptosis in Medullary Thyroid Carcinoma by Targeting BLCAP. Oncol. Res. 2017, 25, 1215–1222. [Google Scholar] [CrossRef]
- Cör, A.; Pižem, J.; Gale, N. Immunohistochemical analysis of pro- and active-caspase 3 in laryngeal squamous cell carcinoma. Virchows Arch. 2004, 444, 439–446. [Google Scholar] [CrossRef]
- Hwang, D.; Kim, S.A.; Yang, E.G.; Song, H.K.; Chung, H.S. A facile method to prepare large quantities of active caspase-3 overexpressed by auto-induction in the C41(DE3) strain. Protein Expr. Purif. 2016, 126, 104–108. [Google Scholar] [CrossRef]
- Perreault, M.; Białek, A.; Trottier, J.; Verreault, M.; Caron, P.; Milkiewicz, P.; Barbier, O. Role of Glucuronidation for Hepatic Detoxification and Urinary Elimination of Toxic Bile Acids during Biliary Obstruction. PLoS ONE 2013, 8, e80994. [Google Scholar] [CrossRef]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and Elaboration. PLoS Med. 2012, 9, e1001216. [Google Scholar] [CrossRef]
- Tierney, J.F.; Stewart, L.A.; Ghersi, D.; Burdett, S.; Sydes, M.R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007, 8, 16. [Google Scholar] [CrossRef]
- Tanimoto, T.; Tsuda, H.; Imazeki, N.; Ohno, Y.; Imoto, I.; Inazawa, J.; Matsubara, O. Nuclear expression of cIAP-1, an apoptosis inhibiting protein, predicts lymph node metastasis and poor patient prognosis in head and neck squamous cell carcinomas. Cancer Lett. 2005, 224, 141–151. [Google Scholar] [CrossRef]
- Liu, P.-F.; Hu, Y.-C.; Kang, B.-H.; Tseng, Y.-K.; Wu, P.-C.; Liang, C.-C.; Hou, Y.-Y.; Fu, T.-Y.; Liou, H.-H.; Hsieh, I.-C.; et al. Expression levels of cleaved caspase-3 and caspase-3 in tumorigenesis and prognosis of oral tongue squamous cell carcinoma. PLoS ONE 2017, 12, e0180620. [Google Scholar] [CrossRef] [PubMed]
- Mattila, R.; Syrjänen, S. Caspase cascade pathways in apoptosis of oral lichen planus. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2010, 110, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Augustine, D.; Rao, R.S.; Patil, S.; Sowmya, S.V.; Haragannavar, V.C.; Nambiar, S. Interleukin-1beta and Caspase-3 expression serve as independent prognostic markers for metastasis and survival in oral squamous cell carcinoma. Cancer Biomark. 2019, 26, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007, 26, 1324–1337. [Google Scholar] [CrossRef]
- Mita, A.C.; Mita, M.M.; Nawrocki, S.T.; Giles, F.J. Survivin: Key Regulator of Mitosis and Apoptosis and Novel Target for Cancer Therapeutics. Clin. Cancer Res. 2008, 14, 5000–5005. [Google Scholar] [CrossRef]
- Raudenská, M.; Balvan, J.; Masařík, M. Cell death in head and neck cancer pathogenesis and treatment. Cell Death Dis. 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Wang, R.-A.; Li, Z.-S.; Yan, Q.-G.; Bian, X.-W.; Ding, Y.-Q.; Du, X.; Sun, B.-C.; Sun, Y.-T.; Zhang, X.-H. Resistance to apoptosis should not be taken as a hallmark of cancer. Chin. J. Cancer 2014, 33, 47–50. [Google Scholar] [CrossRef]
- Lipponen, P. Apoptosis in breast cancer: Relationship with other pathological parameters. Endocr.-Relat. Cancer 1999, 6, 13–16. [Google Scholar] [CrossRef]
- Lipponen, P.K.; Aaltomaa, S. Apoptosis in bladder cancer as related to standard prognostic factors and prognosis. J. Pathol. 1994, 173, 333–339. [Google Scholar] [CrossRef]
- Nishimura, R.; Nagao, K.; Miyayama, H.; Matsuda, M.; Baba, K.; Matsuoka, Y.; Higuchi, A. Apoptosis in breast cancer and its relationship to clinicopathological characteristics and prognosis. J. Surg. Oncol. 1999, 71, 226–234. [Google Scholar] [CrossRef]
- Krajewski, S.; Krajewska, M.; Turner, B.C.; Pratt, C.; Howard, B.; Zapata, J.M.; Frenkel, V.; Robertson, S.; Ionov, Y.; Yamamoto, H.; et al. Prognostic significance of apoptosis regulators in breast cancer. Endocr. Relat. Cancer 1999, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Callagy, G.M.; Webber, M.J.; Pharoah, P.D.; Caldas, C. Meta-analysis confirms BCL2 is an independent prognostic marker in breast cancer. BMC Cancer 2008, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Callagy, G.M.; Pharoah, P.D.; Pinder, S.E.; Hsu, F.D.; Nielsen, T.O.; Ragaz, J.; Ellis, I.O.; Huntsman, D.; Caldas, C. Bcl-2 Is a Prognostic Marker in Breast Cancer Independently of the Nottingham Prognostic Index. Clin. Cancer Res. 2006, 12, 2468–2475. [Google Scholar] [CrossRef]
- Tomita, M.; Matsuzaki, Y.; Edagawa, M.; Shimizu, T.; Hara, M.; Onitsuka, T. Prognostic significance of bcl-2 expression in resected pN2 non-small cell lung cancer. Eur. J. Surg. Oncol. (EJSO) 2003, 29, 654–657. [Google Scholar] [CrossRef]
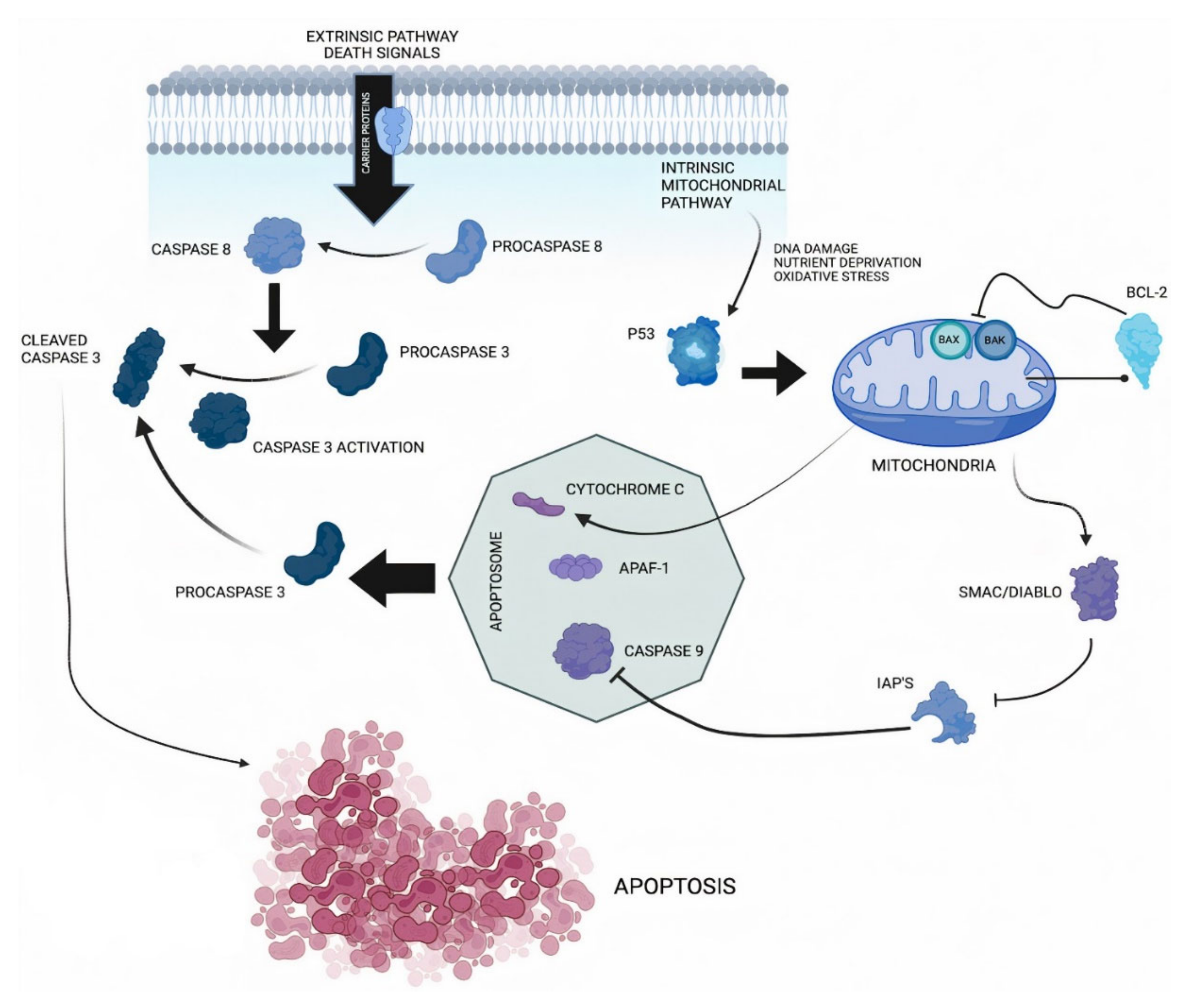


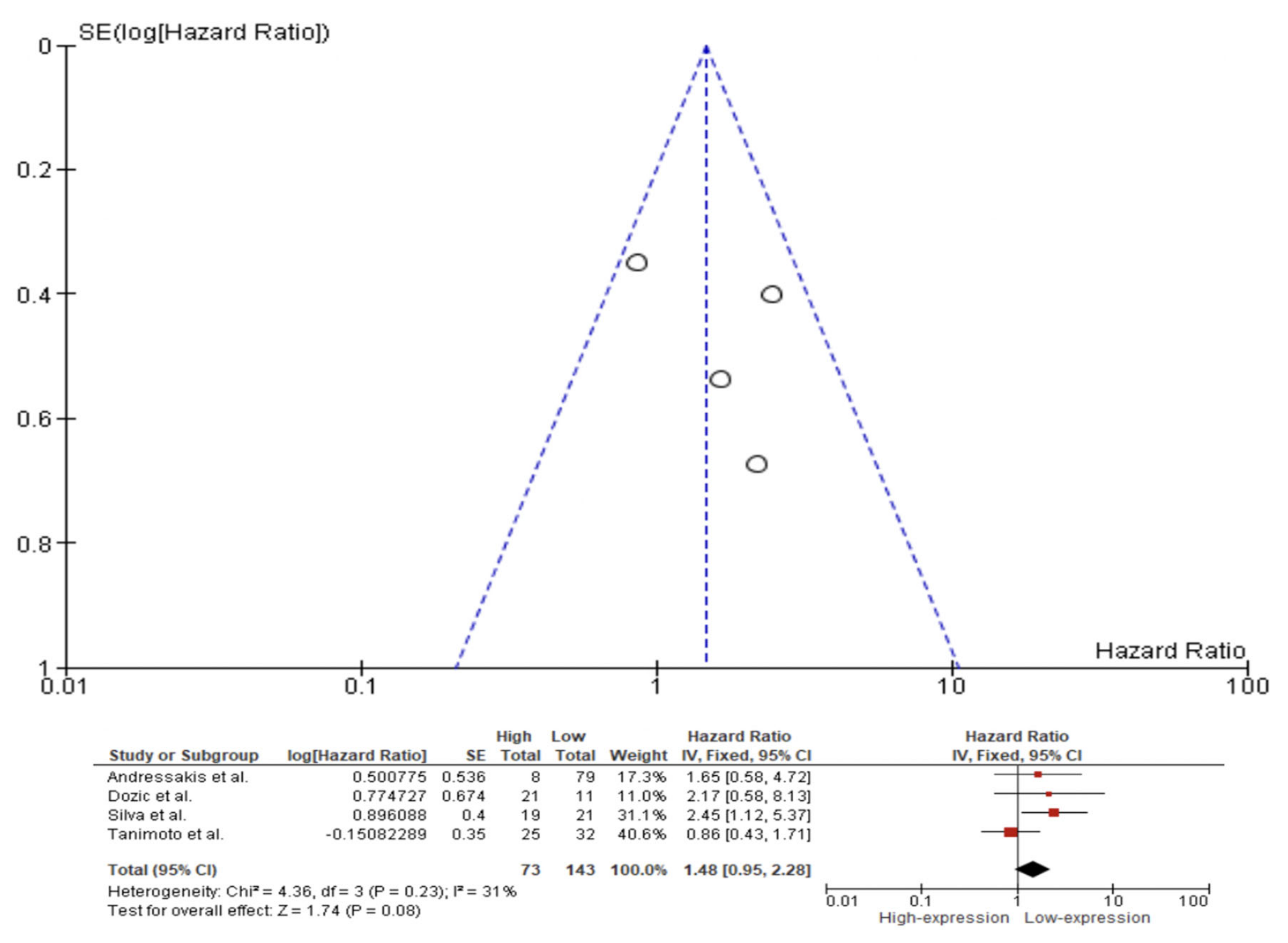
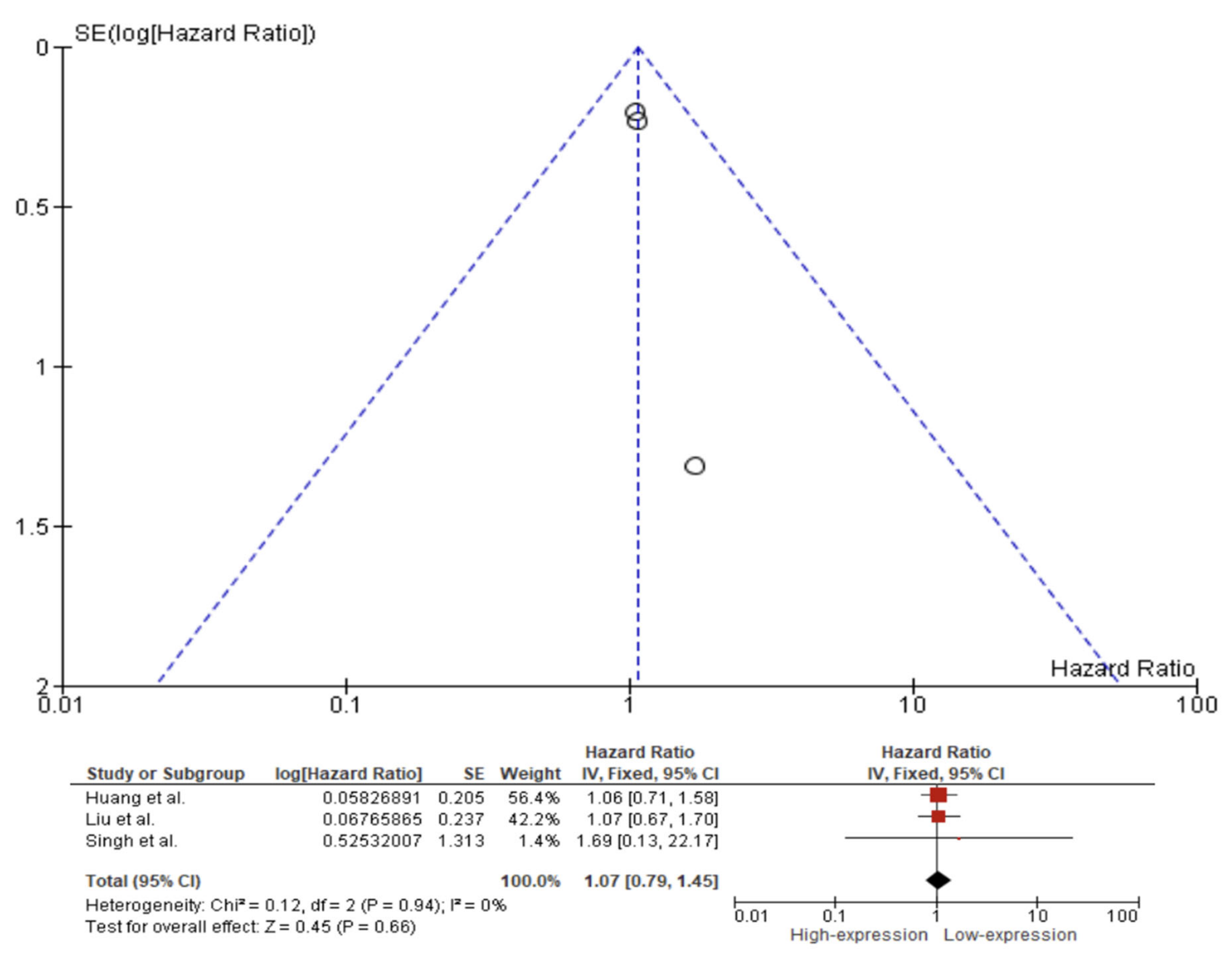

| Reference Number | Author | Year | Country | Sample Size | Staging Edition | Tumor Subsite | Recruitment Period | Type of Lesion | Survival Analysis | Caspase 3/Cleaved Caspase 3 (Monoclonal) | IHC Pattern | Cut-Off Point (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | Dozic et al. [27] | 2016 | Servia | 50 | NI | Salivary Glands | 1998–2008 | HNC | OS | CPP3 (monoclonal) | cytoplasmic membrane | NI |
| 10.1016/j.archoralbio.2005.02.005. | Bascones-Ilundan et al. [50] | 2006 | Spain | 52 | NI | Oral Mucosa, Gingiva, Lip, Tongue | 1999–2003 | OPMD | UN | Active-Caspase 3 (monoclonal) | cytoplasmic membrane | 10 |
| 10.1590/1678-775720160156. | Leite et al. [21] | 2016 | Brazil | 120 | NI | Oral Cavity, Lip | NI | HNC/OPMD | UN | Cleaved Caspase 3 (polyclonal) | cytoplasmic membrane | NI |
| 10.1038/modpathol.3800398. | Oudejans et al. [37] | 2005 | Netherlands | 36 | UICC | Nasopharynges | 1995–1996 | HNC | OS | Active Caspase 3 (polyclonal) | cytoplasmic membrane | NI |
| 10.1016/j.tripleo.2010.05.070. | Mattila et al. [72] | 2010 | Finland | 66 | NI | Oral Cavity | 1991–2002 | OPMD | UN | 3CSP03 (polyclonal) | cytoplasmic membrane | NI |
| 10.1017/S0022215108002636. | Andressakis et al. [5] | 2008 | Greece | 87 | NI | Tongue | 1998–2006 | HNC | OS | 3CSP03 (polyclonal) | cytoplasmic membrane | NI |
| 10.1002/hed.21602 | Coutinho-Camillo et al. [16] | 2010 | Brazil | 229 | NI | Oral Cavity | 1970–1992 | HNC | UN | Cleaved Caspase 3 (monoclonal) | cytoplasmic membrane | NI |
| 10.1002/hed.25763 | Silva et al. [14] | 2019 | Brazil | 40 | NI | Salivary Glands | 2014–2019 | HNC | DSS, OS | Caspase 3 (monoclonal) | cytoplasmic membrane | NI |
| 10.4317/medoral.18901. | Bascones-Martínez et al. [10] | 2013 | Spain | 41 | UICC | Oral Cavity | NI | HNC | UN | Cleaved Caspase 3 (polyclonal) | cytoplasmic membrane | 25 |
| 10.1016/j.canlet.2004.11.049 | Tanimoto et al. [70] | 2005 | Japan | 57 | UICC | Oral cavity, Oropharynx, Hipopharynx | 1989–2000 | HNC | OS | Caspase 3 (monoclonal) | cytoplasmic membrane | 3 |
| 10.3892/ol.2017.6626 | Li et al. [20] | 2017 | China | 45 | NI | Oral Cavity | 2005–2007 | HNC/OPMD | UN | Caspase 3 (monoclonal) | cytoplasmic membrane | NI |
| 10.18632/oncotarget.20494. | Huang et al. [57] | 2017 | Taiwan | 185 | AJCC | Buccal Mucosa | 1993–2006 | HNC | OS, DFS, DSS | Cleaved Caspase 3 and Caspase 3 (polyclonal) | cytoplasmic membrane | 25 |
| 10.1046/j.1601-0825.2003.00998.x. | Tobón-Arroyave et al. [61] | 2004 | Colombia | 30 | NI | Oral Cavity | NI | OPMD | UN | CPP32 (monoclonal) | cytoplasmic membrane | 25 |
| 10.1371/journal.pone.0180620. | Liu et al. [71] | 2017 | Taiwan | 246 | AJCC | Tongue | 1991–2010 | HNC | DFS, DSS | Caspase 3 and Cleaved-Caspase 3 (monoclonal) | cytoplasmic membrane | 25 |
| 10.1002/path.1630 | Hague et al. [58] | 2004 | UK | 54 | NI | Tongue, Labial Mucosa, Buccal Mucosa, Palate, Floor of Mouth, Alveolar Process/Gingiva | NI | HNC/OPMD | UN | Caspase 3 and Cleaved-Caspase 3 (monoclonal) | cytoplasmic and nuclear membrane | 25 |
| 10.1097/PAI.0b013e31828a0d0c. | Poomsawat, Punyasingh and Vejchapipat [26] | 2014 | Thailand | 104 | NI | Oral Cavity | 2000–2009 | HNC/OPMD | UN | Caspase 3 AF835 (monoclonal) | cytoplasmic membrane | NI |
| 10.3233/CBM-190149. | Singh et al. [73] | 2019 | India | 20 | AJCC | Oral Cavity | NI | HNC | DFS, DSS | Caspase 3 31A1067 (monoclonal) | cytoplasmic membrane | 10 |
| 10.36740/WLek201912108 | Kovtuneko, Bakaiev and Shponka [7] | 2019 | Ukraine | 80 | AJCC | Maxillary Sinus | 2011–2016 | HNC | UN | Caspase 3 (monoclonal) | cytoplasmic membrane | NI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, F.F.V.e.; Padín-Iruegas, M.E.; Caponio, V.C.A.; Lorenzo-Pouso, A.I.; Saavedra-Nieves, P.; Chamorro-Petronacci, C.M.; Suaréz-Peñaranda, J.; Pérez-Sayáns, M. Caspase 3 and Cleaved Caspase 3 Expression in Tumorogenesis and Its Correlations with Prognosis in Head and Neck Cancer: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 11937. https://doi.org/10.3390/ijms231911937
Silva FFVe, Padín-Iruegas ME, Caponio VCA, Lorenzo-Pouso AI, Saavedra-Nieves P, Chamorro-Petronacci CM, Suaréz-Peñaranda J, Pérez-Sayáns M. Caspase 3 and Cleaved Caspase 3 Expression in Tumorogenesis and Its Correlations with Prognosis in Head and Neck Cancer: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences. 2022; 23(19):11937. https://doi.org/10.3390/ijms231911937
Chicago/Turabian StyleSilva, Fábio França Vieira e, María Elena Padín-Iruegas, Vito Carlo Alberto Caponio, Alejandro I. Lorenzo-Pouso, Paula Saavedra-Nieves, Cintia Micaela Chamorro-Petronacci, José Suaréz-Peñaranda, and Mario Pérez-Sayáns. 2022. "Caspase 3 and Cleaved Caspase 3 Expression in Tumorogenesis and Its Correlations with Prognosis in Head and Neck Cancer: A Systematic Review and Meta-Analysis" International Journal of Molecular Sciences 23, no. 19: 11937. https://doi.org/10.3390/ijms231911937
APA StyleSilva, F. F. V. e., Padín-Iruegas, M. E., Caponio, V. C. A., Lorenzo-Pouso, A. I., Saavedra-Nieves, P., Chamorro-Petronacci, C. M., Suaréz-Peñaranda, J., & Pérez-Sayáns, M. (2022). Caspase 3 and Cleaved Caspase 3 Expression in Tumorogenesis and Its Correlations with Prognosis in Head and Neck Cancer: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences, 23(19), 11937. https://doi.org/10.3390/ijms231911937







