Multi-Omics Analysis Revealed a Significant Alteration of Critical Metabolic Pathways Due to Sorafenib-Resistance in Hep3B Cell Lines
Abstract
1. Introduction
2. Results
2.1. Sorafenib-Resistant Hep3B Cells
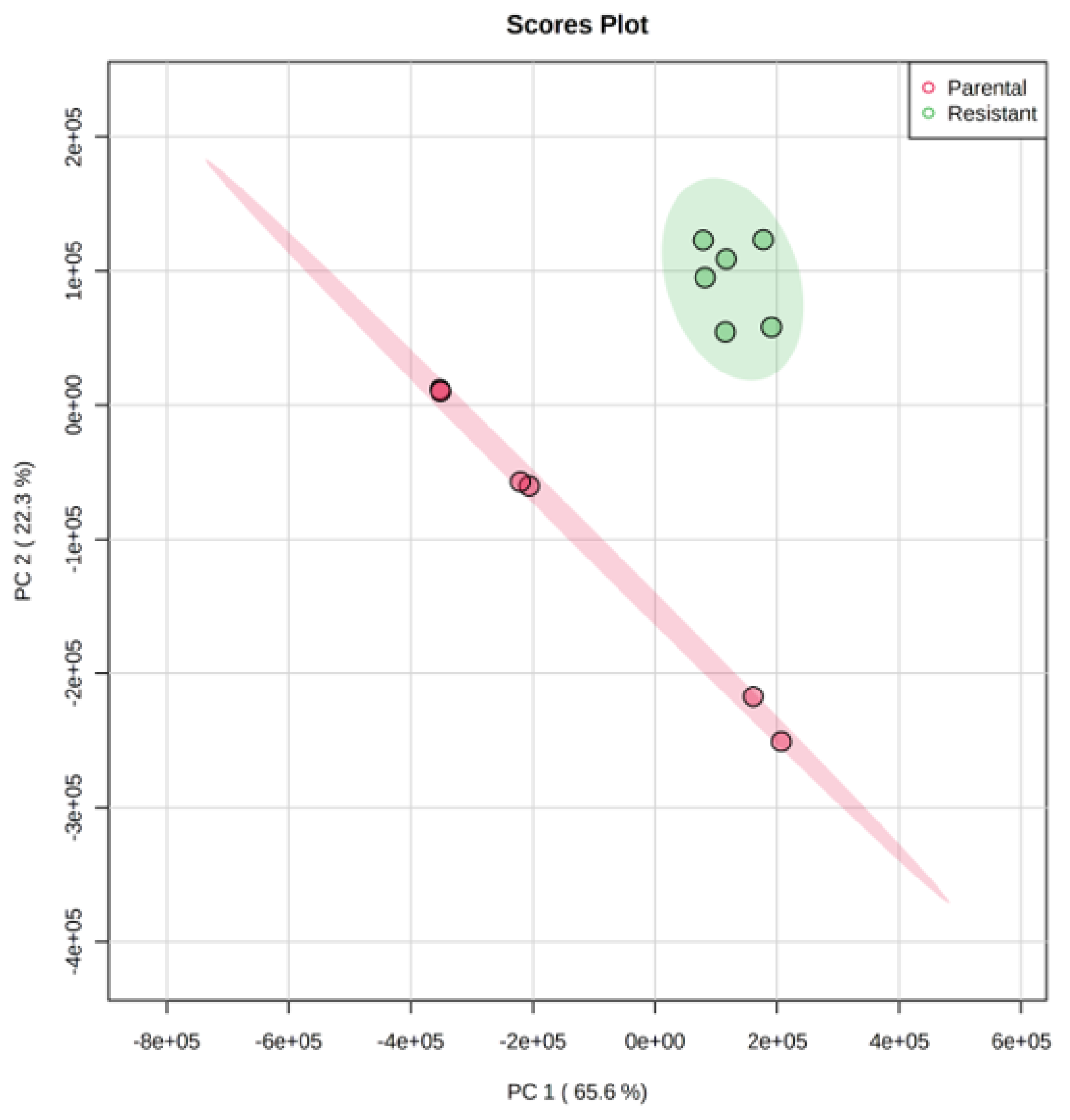
2.2. Metabolomics Analysis Revealed a Significant Change/Shift of Metabolic Pathways Due to Sorafenib Resistance
2.3. Proteomics Analysis Indicate a Unique Protein Profile Associated with Hep3 Drug Resistance
2.4. Multi-Omics Integrated Analysis Demonstrated the Involvement of Major Pathways in HCC Development and Drug Resistance
3. Discussion
3.1. Comparative Metabolomics Revealed That Sorafenib-Resistant Hep3B Cells Demonstrated a Significant Dysregulation in Amino Acid and Nucleotide Metabolic Pathways, Energy Production Pathways and Other Cancer Aggression, Migration, Proliferation and Drug Resistance-Related Pathways
3.2. Comparative Proteomics Indicated That Resistant Hep3B Cells Significantly Alter Specific Pathways That Have a Great Impact on the Development of Chemotherapeutic Resistance
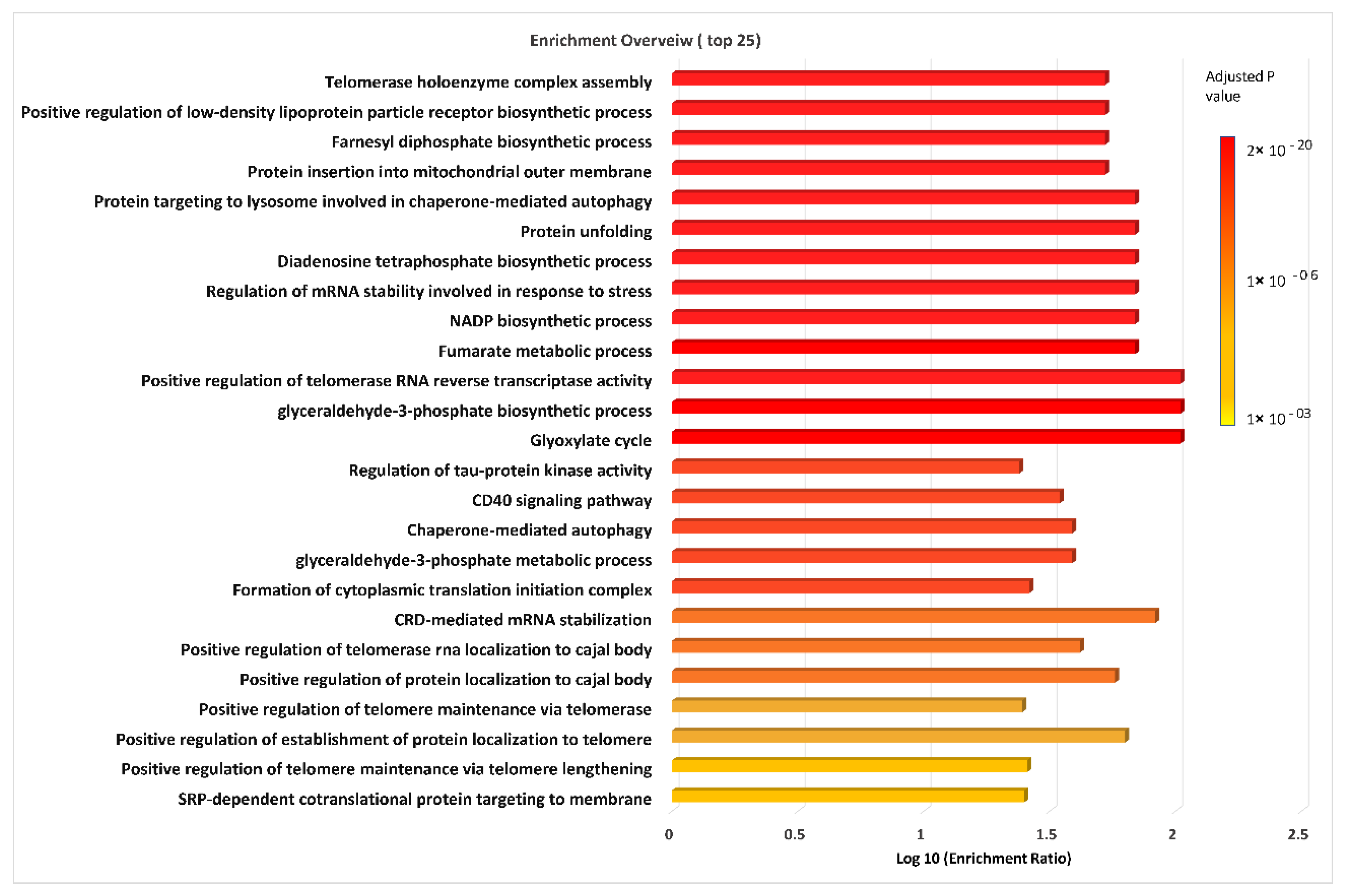
3.3. Top Significantly Enriched Gene Ontology Biological Process Terms Indicate Dysregulation in Processes Linked to Energy Production, Anabolism and Cancer Cell Survival and Growth
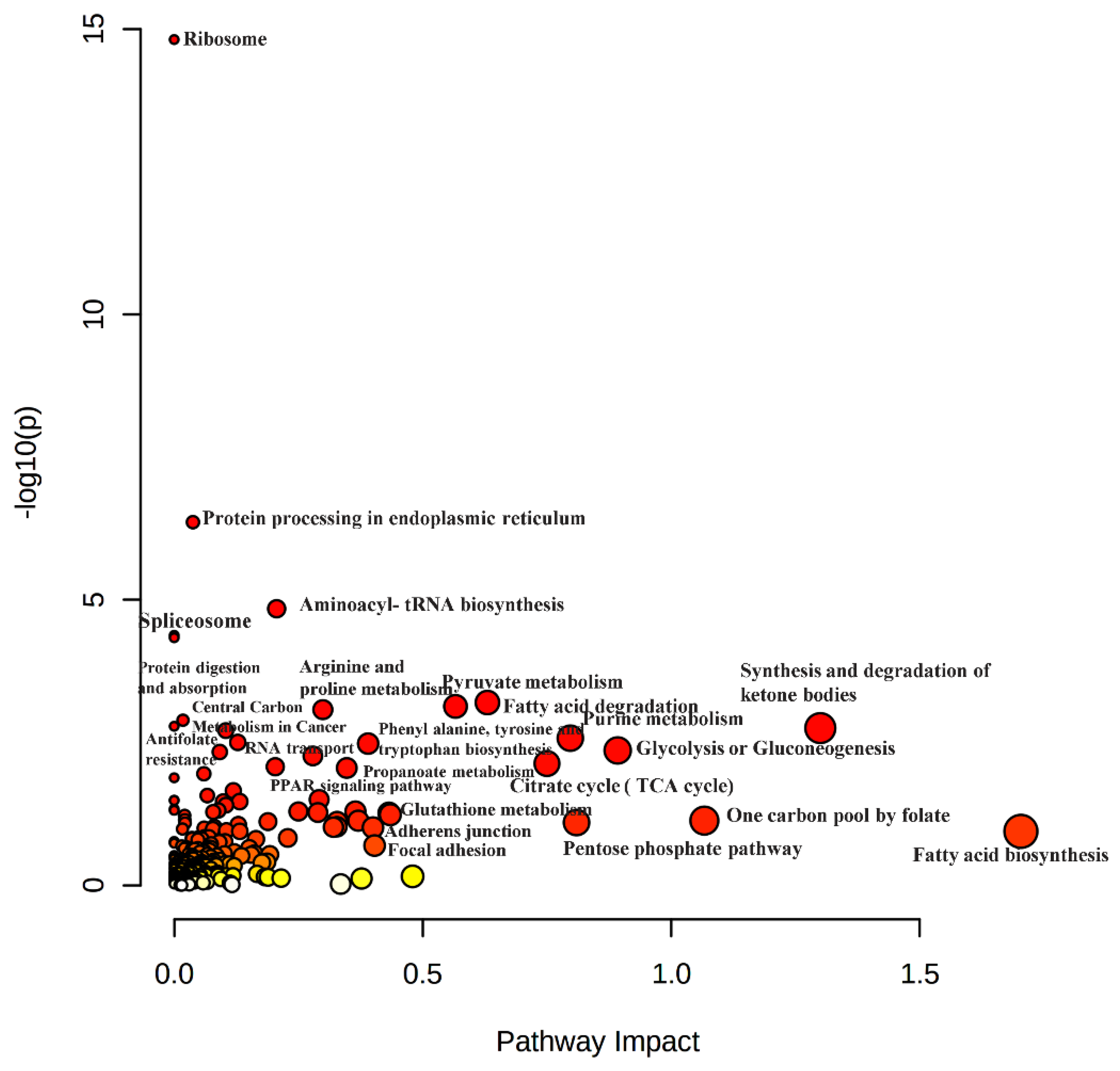
3.4. Multi-Omics Joint Pathway Enrichment Analysis Revealed Unique Pathways That Might be Druggable Targets in the Resistant HCC
4. Materials and Methods
4.1. Reagents
4.2. Cell Lines and Culture Conditions
4.3. Generation of Resistant Hep3B Cell Line
4.4. MTT Cell Viability Assay
4.5. Metabolite Extraction
4.6. Protein Extraction and Quantification
4.7. Bradford Assay
4.8. In-Solution Protein Digestion and Peptide Desalting
4.9. Ultra-High-Performance Liquid Chromatography-Tandem Mass Spectrometry (UHPLC-MS/MS)
4.10. Data Analysis and Statistical Approach
4.10.1. Metabolomics Data Processing
4.10.2. Proteomics Data Processing
4.10.3. Joint Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Golabi, P.; Fazel, S.; Otgonsuren, M.; Sayiner, M.; Younossi, Z.M.; Locklear, C.T. Mortality Assessment of Patients with Hepatocellular Carcinoma According to Underlying Disease and Treatment Modalities. Medicine 2017, 96, e5904. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- De Matteis, S.; Ragusa, A.; Marisi, G.; De Domenico, S.; Casadei Gardini, A.; Bonafè, M.; Giudetti, A.M. Aberrant Metabolism in Hepatocellular Carcinoma Provides Diagnostic and Therapeutic Opportunities. Oxidative Med. Cell. Longev. 2018, 2018, 7512159. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tan, H.Y.; Wang, N.; Wang, X.; Feng, Y. Deciphering Hepatocellular Carcinoma through Metabolomics: From Biomarker Discovery to Therapy Evaluation. Cancer Manag. Res. 2018, 10, 715–734. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhang, A.; Wang, X. Metabolomic Applications in Hepatocellular Carcinoma: Toward the Exploration of Therapeutics and Diagnosis through Small Molecules. RSC Adv. 2017, 7, 17217. [Google Scholar] [CrossRef]
- Nie, W.; Yan, L.; Lee, Y.H.; Guha, C.; Kurland, I.J.; Lu, H. Advanced Mass Spectrometry-Based Multi-Omics Technologies for Exploring the Pathogenesis of Hepatocellular Carcinoma. Mass Spectrom. Rev. 2016, 35, 331–349. [Google Scholar] [CrossRef]
- Su, B.; Luo, P.; Zhao, Y.; Yu, P.; Li, Z.; Yin, P.; Zhou, L.; Fan, J.; Huang, X.; Lin, X.; et al. A Novel Analysis Method for Biomarker Identification Based on Horizontal Relationship: Identifying Potential Biomarkers from Large-Scale Hepatocellular Carcinoma Metabolomics Data. Anal. Bioanal. Chem. 2019, 411, 6377–6386. [Google Scholar] [CrossRef]
- Hesketh, R.L.; Zhu, A.X.; Oklu, R. Hepatocellular Carcinoma: Can Circulating Tumor Cells and Radiogenomics Deliver Personalized Care? Am. J. Clin. Oncol. 2015, 38, 431–436. [Google Scholar] [CrossRef]
- Lünse, S.; Heidecke, C.; Partecke, L.I. Current Topics and Perspectives in Surgical Management of Hepatocellular Carcinoma. In Hepatocellular Carcinoma; Codon Publications: Brisbane, Australia, 2019. [Google Scholar]
- Lurje, I.; Czigany, Z.; Bednarsch, J.; Lurje, G.; Neumann, U.P.; Roderburg, C.; Isfort, P. Treatment Strategies for Hepatocellular Carcinoma—A Multidisciplinary Approach. Int. J. Mol. Sci. 2019, 20, 1465. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Z.; Qu, X. The Adverse Effects of Sorafenib in Patients with Advanced Cancers. Basic Clin. Pharmacol. Toxicol. 2015, 116, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Lu, J.; Wang, X.; Guo, W.; Zhang, J. Comparative Metabolomic Profiling of Hepatocellular Carcinoma Cells Treated with Sorafenib Monotherapy vs. Sorafenib-Everolimus Combination Therapy. Med. Sci. Monit. 2015, 21, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Chen, Z.; Zhang, W.; Cheng, Y.; Zhang, B.; Wu, F.; Wang, Q.; Wang, S.; Rong, D.; Reiter, F.P.; et al. The Mechanisms of Sorafenib Resistance in Hepatocellular Carcinoma: Theoretical Basis and Therapeutic Aspects. Signal Transduct. Target. Ther. 2020, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Lohitesh, K.; Chowdhury, R.; Mukherjee, S. Resistance a Major Hindrance to Chemotherapy in Hepatocellular Carcinoma: An Insight. Cancer Cell Int. 2018, 18, 44. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Yuan, W.; Li, H.; Li, S.; Chen, Z.; Yang, H. Identification of Key Pathways and Biomarkers in Sorafenib-Resistant Hepatocellular Carcinoma using Bioinformatics Analysis. Exp. Ther. Med. 2018, 16, 1850–1858. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zheng, B.; Wang, H.; Chen, L. New Knowledge of the Mechanisms of Sorafenib Resistance in Liver Cancer. Acta Pharmacol. Sin. 2017, 38, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Hosen, M.I.; Ahmed, M.; Shekhar, H.U. Onco-Multi-OMICS Approach: A New Frontier in Cancer Research. BioMed Res. Int. 2018, 2018, 9836256. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lu, S.; Zheng, H.; Xu, M.; Song, J.; Yang, W.; Weng, Q.; Zheng, L.; Fan, X.; Cheng, X.; et al. Identification of the Potential Metabolic Pathways Involved in the Hepatic Tumorigenesis of Rat Diethylnitrosamine-Induced Hepatocellular Carcinoma Via 1H NMR-Based Metabolomic Analysis. BioMed. Res. Int. 2019, 2019, 9367082. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Xu, G. Metabolomics for Tumor Marker Discovery and Identification Based on Chromatography-Mass Spectrometry. Expert Rev. Mol. Diagn. 2013, 13, 339–348. [Google Scholar] [CrossRef]
- Smith, R.; Mathis, A.D.; Ventura, D.; Prince, J.T. Proteomics, Lipidomics, Metabolomics: A Mass Spectrometry Tutorial from a Computer Scientist’s Point of View. BMC Bioinform. 2014, 15 (Suppl. 7), S9. [Google Scholar] [CrossRef]
- Wang, S.; Yang, X.; Liu, F.; Wang, X.; Zhang, X.; He, K.; Wang, H. Comprehensive Metabolomic Analysis Reveals Dynamic Metabolic Reprogramming in Hep3B Cells with Aflatoxin B1 Exposure. Toxins 2021, 13, 384. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhuang, H.; Zhang, X.; Li, Y.; Liu, Y.; Yi, X.; Qin, G.; Wei, W.; Chen, R. Multiomics Integration Reveals the Landscape of Prometastasis Metabolism in Hepatocellular Carcinoma. Mol. Cell. Proteom. 2018, 17, 607–618. [Google Scholar] [CrossRef]
- Li, B.; Zhang, Z.; Wan, C. Identification of Microproteins in Hep3B Cells at Different Cell Cycle Stages. J. Proteome Res. 2022, 21, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.; Tripathi, Y.B.; Singh, P.; Kesharwani, R.K.; Keservani, R.K. 2-Roles of AMP, ADP, ATP, and AMPK in Healthy Energy Boosting and Prolonged Life Span. In Sustained Energy for Enhanced Human Functions and Activity; Academic Press: Cambridge, MA, USA, 2017; pp. 31–51. ISBN 9780128054130. [Google Scholar]
- Kurth, I.; Yamaguchi, N.; Andreu-Agullo, C.; Tian, H.S.; Sridhar, S.; Takeda, S.; Gonsalves, F.C.; Loo, J.M.; Barlas, A.; Manova-Todorova, K.; et al. Therapeutic Targeting of SLC6A8 Creatine Transporter Suppresses Colon Cancer Progression and Modulates Human Creatine Levels. Sci. Adv. 2021, 7, eabi7511. [Google Scholar] [CrossRef] [PubMed]
- Sciacovelli, M.; Frezza, C. Oncometabolites: Unconventional Triggers of Oncogenic Signalling Cascades. Free Radic. Biol. Med. 2016, 100, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bachhawat, A.K. Pyroglutamic Acid: Throwing Light on a Lightly Studied Metabolite. Curr. Sci. 2012, 102, 288–297. [Google Scholar]
- Budczies, J.; Pfitzner, B.M.; Györffy, B.; Winzer, K.; Radke, C.; Dietel, M.; Fiehn, O.; Denkert, C. Glutamate Enrichment as New Diagnostic Opportunity in Breast Cancer. Int. J. Cancer 2015, 136, 1619–1628. [Google Scholar] [CrossRef]
- Villa, E.; Ali, E.S.; Sahu, U.; Ben-Sahra, I. Cancer Cells Tune the Signaling Pathways to Empower De Novo Synthesis of Nucleotides. Cancers 2019, 11, 688. [Google Scholar] [CrossRef] [PubMed]
- Pranzini, E.; Pardella, E.; Paoli, P.; Fendt, S.; Taddei, M.L. Metabolic Reprogramming in Anticancer Drug Resistance: A Focus on Amino Acids. Trends Cancer 2021, 7, 682–699. [Google Scholar] [CrossRef]
- Saito, R.d.F.; de Sousa, A.L.N.; Bustos, S.O.; Chammas, R. Phosphatidylcholine-Derived Lipid Mediators: The Crosstalk between Cancer Cells and Immune Cells. Front. Immunol. 2022, 13, 768606. [Google Scholar] [CrossRef]
- Häfner, S.; Adler, H.S.; Mischak, H.; Janosch, P.; Heidecker, G.; Wolfman, A.; Pippig, S.; Lohse, M.; Ueffing, M.; Kolch, W. Mechanism of Inhibition of Raf-1 by Protein Kinase A. Mol. Cell. Biol. 1994, 14, 6696–6703. [Google Scholar] [PubMed]
- Lee, J.; Choi, Y.H.; Nguyen, P.; Kim, J.; Lee, S.J.; Trepel, J.B. Cyclic AMP Induces Inhibition of Cyclin A Expression and Growth Arrest in Human Hepatoma Cells. BBA Mol. Cell Res. 1999, 1449, 261–268. [Google Scholar] [CrossRef]
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the Fire: Emerging Role of the Hexosamine Biosynthetic Pathway in Cancer. BMC Biol. 2019, 17, 52. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of Telomeres and Telomerase in Cancer, and Advances in Telomerase-Targeted Therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef]
- Roslan, Z.; Muhamad, M.; Selvaratnam, L.; Ab-Rahim, S. The Roles of Low-Density Lipoprotein Receptor-Related Proteins 5, 6, and 8 in Cancer: A Review. J. Oncol. 2019, 2019, 4536302. [Google Scholar] [CrossRef]
- Ha, N.T.; Lee, C.H. Roles of Farnesyl-Diphosphate Farnesyltransferase 1 in Tumour and Tumour Microenvironments. Cells 2020, 9, 2352. [Google Scholar] [CrossRef]
- Palmer, C.S.; Anderson, A.J.; Stojanovski, D. Mitochondrial Protein Import Dysfunction: Mitochondrial Disease, Neurodegenerative Disease and Cancer. FEBS Lett. 2021, 595, 1107–1131. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.; Cuervo, A.M. Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology. Trends Endocrinol. Metab. 2020, 31, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Madden, E.; Logue, S.E.; Healy, S.J.; Manie, S.; Samali, A. The Role of the Unfolded Protein Response in Cancer Progression: From Oncogenesis to Chemoresistance. Biol. Cell. 2019, 111, 1–17. [Google Scholar] [CrossRef]
- Ferguson, F.; McLennan, A.G.; Urbaniak, M.D.; Jones, N.J.; Copeland, N.A. Re-Evaluation of Diadenosine Tetraphosphate (Ap4A) from a Stress Metabolite to Bona Fide Secondary Messenger. Front. Mol. Biosci. 2020, 7, 606807. [Google Scholar] [CrossRef]
- Griseri, P.; Pagès, G. Regulation of the mRNA Half-Life in Breast Cancer. World J. Clin. Oncol. 2014, 5, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.; Lin, J.; Tian, T.; Xie, D.; Xu, R. NADPH Homeostasis in Cancer: Functions, Mechanisms and Therapeutic Implications. Signal Transduct. Target. Ther. 2020, 5, 231. [Google Scholar] [CrossRef]
- Yang, M.; Soga, T.; Pollard, P.J.; Adam, J. The Emerging Role of Fumarate as an Oncometabolite. Front. Oncol. 2012, 2, 85. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy-Kanniappan, S.; Kunjithapatham, R.; Geschwind, J. Glyceraldehyde-3-Phosphate Dehydrogenase: A Promising Target for Molecular Therapy in Hepatocellular Carcinoma. Oncotarget 2012, 3, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Vettore, L.; Westbrook, R.L.; Tennant, D.A. New Aspects of Amino Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef]
- Lu’o’ng, K.V.Q.; Nguyễn, L.T.H. The Role of Thiamine in Cancer: Possible Genetic and Cellular Signaling Mechanisms. Cancer Genom. Proteom. 2013, 10, 169–185. [Google Scholar]
- Urra, F.A.; Weiss-López, B.; Araya-Maturana, R. Determinants of Anti-Cancer Effect of Mitochondrial Electron Transport Chain Inhibitors: Bioenergetic Profile and Metabolic Flexibility of Cancer Cells. Curr. Pharm. Des. 2016, 22, 5998–6008. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef]
- Doktorova, H.; Hrabeta, J.; Khalil, M.A.; Eckschlager, T. Hypoxia-Induced Chemoresistance in Cancer Cells: The Role of Not Only HIF-1. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Repub. 2015, 159, 166–177. [Google Scholar] [CrossRef]
- Livingstone, E.; Swann, S.; Lilla, C.; Schadendorf, D.; Roesch, A. Combining BRAFV600E Inhibition with Modulators of the Mitochondrial Bioenergy Metabolism to Overcome Drug Resistance in Metastatic Melanoma. Exp. Dermatol. 2015, 24, 709–710. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Pontious, C.; Kovalenko, I.; Lapienyte, L.; Dreyer, S.; Lee, H.; Thurston, G.; Zhang, Y.; Lazarus, J.; Sajjakulnukit, P.; et al. Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab. 2019, 29, 1390–1399.e6. [Google Scholar] [CrossRef] [PubMed]
- Saiki, Y.; Yoshino, Y.; Fujimura, H.; Manabe, T.; Kudo, Y.; Shimada, M.; Mano, N.; Nakano, T.; Lee, Y.; Shimizu, S.; et al. DCK is Frequently Inactivated in Acquired Gemcitabine-Resistant Human Cancer Cells. Biochem. Biophys. Res. Commun. 2012, 421, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Choi, Y.; Park, S.Y.; Jang, S.Y.; Lee, J.Y.; Ham, H.J.; Kim, B.; Jeon, H.; Kim, J.; Kim, J.; et al. PPARδ Reprograms Glutamine Metabolism in Sorafenib-Resistant HCC. Mol. Cancer Res. 2017, 15, 1230–1242. [Google Scholar] [CrossRef]
- Cotte, A.K.; Cottet, V.; Aires, V.; Mouillot, T.; Rizk, M.; Vinault, S.; Binquet, C.; de Barros, J.P.; Hillon, P.; Delmas, D. Phospholipid Profiles and Hepatocellular Carcinoma Risk and Prognosis in Cirrhotic Patients. Oncotarget 2019, 10, 2161–2172. [Google Scholar] [CrossRef]
- Cotte, A.K.; Aires, V.; Fredon, M.; Limagne, E.; Derangère, V.; Thibaudin, M.; Humblin, E.; Scagliarini, A.; de Barros, J.P.; Hillon, P.; et al. Lysophosphatidylcholine Acyltransferase 2-Mediated Lipid Droplet Production Supports Colorectal Cancer Chemoresistance. Nat. Commun. 2018, 9, 322. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Takizawa, Y.; Mineta, H.; Hayasaka, T.; Masaki, N.; Setou, M.; Kusama, Y.; Su, J. Increased Phosphatidylcholine (16:0/16:0) in the Folliculus Lymphaticus of Warthin Tumor. Anal. Bioanal. Chem. 2014, 406, 5815–5825. [Google Scholar] [CrossRef] [PubMed]
- Oishi, Y.; Nagai, S.; Yoshida, M.; Fujisawa, S.; Sazawa, A.; Shinohara, N.; Nonomura, K.; Matsuno, K.; Shimizu, C. Mutation Analysis of the SDHB and SDHD Genes in Pheochromocytomas and Paragangliomas: Identification of a Novel Nonsense Mutation (Q168X) in the SDHB Gene. Endocr. J. 2010, 57, 745–750. [Google Scholar] [CrossRef]
- Pollard, P.J.; Brière, J.J.; Alam, N.A.; Barwell, J.; Barclay, E.; Wortham, N.C.; Hunt, T.; Mitchell, M.; Olpin, S.; Moat, S.J.; et al. Accumulation of Krebs Cycle Intermediates and Over-Expression of HIF1alpha in Tumours which Result from Germline FH and SDH Mutations. Hum. Mol. Genet. 2005, 14, 2231–2239. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Mu, X.; You, Q. Succinate: An Initiator in Tumorigenesis and Progression. Oncotarget 2017, 8, 53819–53828. [Google Scholar] [CrossRef] [PubMed]
- King, A.; Selak, M.A.; Gottlieb, E. Succinate Dehydrogenase and Fumarate Hydratase: Linking Mitochondrial Dysfunction and Cancer. Oncogene 2006, 25, 4675–4682. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond Aerobic Glycolysis: Transformed Cells can Engage in Glutamine Metabolism that Exceeds the Requirement for Protein and Nucleotide Synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, J.B.; Yoon, H.; Ringel, A.E.; Jeanfavre, S.; Clish, C.B.; Haigis, M.C. Metabolic Recycling of Ammonia Via Glutamate Dehydrogenase Supports Breast Cancer Biomass. Science 2017, 358, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Wang, H.; Dong, D.; Li, T.; Yu, Z.; Guo, J.; Zhou, W.; Li, D.; Yan, R.; Wang, L.; et al. Urea as a by-Product of Ammonia Metabolism can be a Potential Serum Biomarker of Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2021, 9, 650748. [Google Scholar] [CrossRef] [PubMed]
- Keshet, R.; Szlosarek, P.; Carracedo, A.; Erez, A. Rewiring Urea Cycle Metabolism In cancer to Support Anabolism. Nat. Rev.Cancer 2018, 18, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kobayashi, E.; Kubota, D.; Suehara, Y.; Mukaihara, K.; Akaike, K.; Ito, A.; Kaneko, K.; Chuman, H.; Kawai, A.; et al. Reduced Argininosuccinate Synthetase Expression in Refractory Sarcomas: Impacts on Therapeutic Potential and Drug Resistance. Oncotarget 2016, 7, 70832–70844. [Google Scholar] [CrossRef] [PubMed]
- Haines, R.J.; Pendleton, L.C.; Eichler, D.C. Argininosuccinate Synthase: At the Center of Arginine Metabolism. Int. J. Biochem. Mol. Biol. 2011, 2, 8–23. [Google Scholar] [PubMed]
- Ensor, C.M.; Holtsberg, F.W.; Bomalaski, J.S.; Clark, M.A. Pegylated Arginine Deiminase (ADI-SS PEG20,000 Mw) Inhibits Human Melanomas and Hepatocellular Carcinomas in vitro and in vivo. Cancer Res. 2002, 62, 5443–5450. [Google Scholar] [PubMed]
- Larsen, C.N.; Price, J.S.; Wilkinson, K.D. Substrate Binding and Catalysis by Ubiquitin C-Terminal Hydrolases: Identification of Two Active Site Residues. Biochemistry 1996, 35, 6735–6744. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, Y.M.; Lim, S.; Nam, Y.K.; Jeong, J.; Kim, H.-J.; Lee, K.-J. Ubiquitin C-Terminal Hydrolase-L1 is a Key Regulator of Tumor Cell Invasion and Metastasis. Oncogene 2009, 28, 117–127. [Google Scholar] [CrossRef]
- Hussain, S.; Van Deursen, J.; Galardy, P.J.; Foreman, O.; Perkins, S.L.; Miles, R.R.; Witzig, T.E. The De-Ubiquitinase UCH-L1 is an Oncogene that Drives the Development of Lymphoma in Vivo by Deregulating PHLPP1 and Akt Signaling. Leukemia 2010, 24, 1641–1655. [Google Scholar] [CrossRef] [PubMed]
- Hurst-Kennedy, J.; Chin, L.-S.; Li, L. Ubiquitin C-Terminal Hydrolase L1 in Tumorigenesis. Biotechnol. Res. Int. 2012, 2012, 123706. [Google Scholar] [CrossRef]
- Wang, W.; Zou, L.; Zhou, D.; Zhou, Z.; Tang, F.; Xu, Z.; Liu, X. Overexpression of Ubiquitin Carboxyl Terminal Hydrolase-L1 Enhances Multidrug Resistance and Invasion/Metastasis in Breast Cancer by Activating the MAPK/Erk Signaling Pathway. Mol. Carcinog. 2016, 55, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Reyimu, A.; Sun, A.; Duoji, Z.; Zhou, W.; Liang, S.; Hu, S.; Wang, X.; Dai, J.; Xu, X. UCHL1 Acts as a Potential Oncogene and Affects Sensitivity of Common Anti-Tumor Drugs in Lung Adenocarcinoma. World J. Surg. Oncol. 2022, 20, 153. [Google Scholar] [CrossRef]
- Yang, G.; Fan, G.; Zhang, T.; Ma, K.; Huang, J.; Liu, M.; Teng, X.; Xu, K.; Fan, P.; Cheng, D. Upregulation of Ubiquitin Carboxyl-Terminal Hydrolase L1 (UCHL1) Mediates the Reversal Effect of Verapamil on Chemo-Resistance to Adriamycin of Hepatocellular Carcinoma. Med. Sci. Monit. 2018, 24, 2072–2082. [Google Scholar] [CrossRef]
- Yusa, K.; Tsuruo, T. Reversal Mechanism of Multidrug Resistance by Verapamil: Direct Binding of Verapamil to P-Glycoprotein on Specific Sites and Transport of Verapamil Outward Across the Plasma Membrane of K562/ADM Cells. Cancer Res. 1989, 49, 5002–5006. [Google Scholar] [PubMed]
- Jin, Y.; Zhang, W.; Xu, J.; Wang, H.; Zhang, Z.; Chu, C.; Liu, X.; Zou, Q. UCH-L1 Involved in Regulating the Degradation of EGFR and Promoting Malignant Properties in Drug-Resistant Breast Cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12500–12508. [Google Scholar]
- Hu, Y.; Shu, Z.; Jiang, J.; Xie, Q.; Zheng, S. Torin2 Overcomes Sorafenib Resistance Via Suppressing mTORC2-AKT-BAD Pathway in Hepatocellular Carcinoma Cells. Hepatobiliary Pancreat. Dis. Int. 2020, 19, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Iwama, H.; Fujita, K.; Kobara, H.; Nishiyama, N.; Fujihara, S.; Goda, Y.; Yoneyama, H.; Morishita, A.; Tani, J.; et al. Evaluating the Effect of Lenvatinib on Sorafenib-Resistant Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2021, 22, 13071. [Google Scholar] [CrossRef]
- Hempel, N.; Carrico, P.M.; Melendez, J.A. Manganese Superoxide Dismutase (Sod2) and Redox-Control of Signaling Events that Drive Metastasis. Anticancer Agents Med. Chem. 2011, 11, 191–201. [Google Scholar] [CrossRef]
- Nishida, S.; Akai, F.; Iwasaki, H.; Hosokawa, K.; Kusunoki, T.; Suzuki, K.; Taniguchi, N.; Hashimoto, S.; Tamura, T.T. Manganese Superoxide Dismutase Content and Localization in Human Thyroid Tumours. J. Pathol. 1993, 169, 341–345. [Google Scholar] [CrossRef]
- Connor, K.M.; Hempel, N.; Nelson, K.K.; Dabiri, G.; Gamarra, A.; Belarmino, J.; Van De Water, L.; Mian, B.M.; Melendez, J.A. Manganese Superoxide Dismutase Enhances the Invasive and Migratory Activity of Tumor Cells. Cancer Res. 2007, 67, 10260–10267. [Google Scholar] [CrossRef]
- Cui, Y.; She, K.; Tian, D.; Zhang, P.; Xin, X. miR-146a Inhibits Proliferation and Enhances Chemosensitivity in Epithelial Ovarian Cancer Via Reduction of SOD2. Oncol. Res. 2016, 23, 275–282. [Google Scholar] [CrossRef]
- Amano, T.; Chano, T.; Isono, T.; Kimura, F.; Kushima, R.; Murakami, T. Abundance of Mitochondrial Superoxide Dismutase is a Negative Predictive Biomarker for Endometriosis-Associated Ovarian Cancers. World J. Surg. Oncol. 2019, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ham, J.; Hong, T.; Song, G.; Lim, W. Fraxetin Suppresses Cell Proliferation and Induces Apoptosis through Mitochondria Dysfunction in Human Hepatocellular Carcinoma Cell Lines Huh7 and Hep3B. Pharmaceutics 2021, 13, 112. [Google Scholar] [CrossRef] [PubMed]
- Schwab, A.; Siddiqui, A.; Vazakidou, M.E.; Napoli, F.; Böttcher, M.; Menchicchi, B.; Raza, U.; Saatci, Ö.; Krebs, A.M.; Ferrazzi, F.; et al. Polyol Pathway Links Glucose Metabolism to the Aggressiveness of Cancer Cells. Cancer Res. 2018, 78, 1604–1618. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, D.; McArthur, D.L.; Boros, L.G.; Nissen, N.; Heaney, A.P. Fructose Induces Transketolase Flux to Promote Pancreatic Cancer Growth. Cancer Res. 2010, 70, 6368–6376. [Google Scholar] [CrossRef]
- Ismail, I.T.; Fiehn, O.; Elfert, A.; Helal, M.; Salama, I.; El-Said, H. Sugar Alcohols have a Key Role in Pathogenesis of Chronic Liver Disease and Hepatocellular Carcinoma in Whole Blood and Liver Tissues. Cancers 2020, 12, 484. [Google Scholar] [CrossRef]
- Jeon, D.; Choi, W.; Kim, J.; Jung, Y.; Lee, S.; Seo, H.R.; Kim, K.M. Serum Sorbitol Dehydrogenase as a Novel Prognostic Factor for Hepatocellular Carcinoma After Surgical Resection. Cancers 2021, 13, 6143. [Google Scholar] [CrossRef]
- Spicer, A.P.; Kaback, L.A.; Smith, T.J.; Seldin, M.F. Molecular Cloning and Characterization of the Human and Mouse UDP-Glucose Dehydrogenase Genes. J. Biol. Chem. 1998, 273, 25117–25124. [Google Scholar] [CrossRef]
- Wang, T.; Pan, Y.; Fu, C.; Chang, H. Down-Regulation of UDP-Glucose Dehydrogenase Affects Glycosaminoglycans Synthesis and Motility in HCT-8 Colorectal Carcinoma Cells. Exp. Cell Res. 2010, 316, 2893–2902. [Google Scholar] [CrossRef]
- Teoh, S.T.; Ogrodzinski, M.P.; Lunt, S.Y. UDP-Glucose 6-Dehydrogenase Knockout Impairs Migration and Decreases in Vivo Metastatic Ability of Breast Cancer Cells. Cancer Lett. 2020, 492, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Xu, X.; Shao, M.; Yang, X.; He, G.; Qi, K.; Gu, J.; Wang, L. UDP-Glucose 6-Dehydrogenase Lessens Sorafenib Sensitivity Via Modulating Unfolded Protein Response. Biochem. Biophys. Res. Commun. 2022, 613, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Nian, H.; Ma, B. Calpain–calpastatin System and Cancer Progression. Biol. Rev. 2021, 96, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Lampis, A.; Hahne, J.C.; Hedayat, S.; Valeri, N. MicroRNAs as Mediators of Drug Resistance Mechanisms. Curr. Opin. Pharmacol. 2020, 54, 44–50. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, W.; Ni, P.; Li, A.; Zhou, J.; Xu, S. MiR-99a and MiR-491 Regulate Cisplatin Resistance in Human Gastric Cancer Cells by Targeting CAPNS1. Int. J. Biol. Sci. 2016, 12, 1437–1447. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, R.; Handy, D.E.; Loscalzo, J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Ben-Sahra, I.; Lockwood, S.E.; Timson, R.C.; Byles, V.; Henning, G.T.; Gao, P.; Selfors, L.M.; Asara, J.M.; Manning, B.D. Direct Stimulation of NADP+ Synthesis through Akt-Mediated Phosphorylation of NAD Kinase. Science 2019, 363, 1088–1092. [Google Scholar] [CrossRef]
- Jackson, T.D.; Palmer, C.S.; Stojanovski, D. Mitochondrial Diseases Caused by Dysfunctional Mitochondrial Protein Import. Biochem. Soc. Trans. 2018, 46, 1225–1238. [Google Scholar] [CrossRef]
- Zheng, J. Energy Metabolism of Cancer: Glycolysis Versus Oxidative Phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef]
- Yang, Y.; Jin, M.; Dai, Y.; Shan, W.; Chen, S.; Cai, R.; Yang, H.; Tang, L.; Li, L. Involvement and Targeted Intervention of Mortalin-Regulated Proteome Phosphorylated-Modification in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 687871. [Google Scholar] [CrossRef]
- Ryu, J.; Kaul, Z.; Yoon, A.; Liu, Y.; Yaguchi, T.; Na, Y.; Ahn, H.M.; Gao, R.; Choi, I.; Yun, C.; et al. Identification and Functional Characterization of Nuclear Mortalin in Human Carcinogenesis. J. Biol. Chem. 2014, 289, 24832–24844. [Google Scholar] [CrossRef] [PubMed]
- Catez, F.; Dalla Venezia, N.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J. Ribosome Biogenesis: An Emerging Druggable Pathway for Cancer Therapeutics. Biochem. Pharmacol. 2019, 159, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, H.; You, C.; Leung, M.; Man, E.P.S.; Khoo, U. Targeting Ribosome Biogenesis to Combat Tamoxifen Resistance in ER+ve Breast Cancer. Cancers 2022, 14, 1251. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Gogonea, V.; Fox, P.L. Aminoacyl-tRNA Synthetases of the Multi-tRNA Synthetase Complex and their Role in Tumorigenesis. Transl. Oncol. 2022, 19, 101392. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef]
- Yang, H.; Beutler, B.; Zhang, D. Emerging Roles of Spliceosome in Cancer and Immunity. Protein Cell 2021, 13, 559–579. [Google Scholar] [CrossRef]
- Wang, B.; Lee, N.H. Aberrant RNA Splicing in Cancer and Drug Resistance. Cancers 2018, 10, 458. [Google Scholar] [CrossRef]
- Qiao, B.; Wang, J.; Xie, J.; Niu, Y.; Ye, S.; Wan, Q.; Ye, Q. Detection and Identification of Peroxiredoxin 3 as a Biomarker in Hepatocellular Carcinoma by a Proteomic Approach. Int. J. Mol. Med. 2012, 29, 832–840. [Google Scholar]
- Olson, K.A.; Schell, J.C.; Rutter, J. Pyruvate and Metabolic Flexibility: Illuminating a Path Toward Selective Cancer Therapies. Trends Biochem. Sci. 2016, 41, 219–230. [Google Scholar] [CrossRef]
- Zaal, E.A.; Berkers, C.R. The Influence of Metabolism on Drug Response in Cancer. Front. Oncol. 2018, 8, 500. [Google Scholar] [CrossRef]
- Wang, H.; Pochampalli, M.; Wang, L.; Zou, J.X.; Li, P.; Hsu, S.; Wang, B.; Huang, S.; Yang, P.; Yang, J.C.; et al. KDM8/JMJD5 as a Dual Coactivator of AR and PKM2 Integrates AR/EZH2 Network and Tumor Metabolism in CRPC. Oncogene 2019, 38, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Li, T.; Wang, L.; Zhang, L.; Yan, R.; Li, K.; Xing, S.; Wu, G.; Hu, L.; Gao, P.; et al. Hepatocellular Carcinoma Redirects to Ketolysis for Progression Under Nutrition Deprivation Stress. Cell Res. 2016, 26, 1112–1130. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Xie, X.; Xu, F.; Shi, X.; Wang, Y.; Deng, L. Distinctive Pharmacological Differences between Liver Cancer Cell Lines HepG2 and Hep3B. Cytotechnology 2015, 67, 1–12. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.; Eustace, A.J.; Breen, L.; Crown, J.; Clynes, M.; O’Donovan, N.; Stordal, B.; Busschots, S. In Vitro Development of Chemotherapy and Targeted Therapy Drug-Resistant Cancer Cell Lines: A Practical Guide with Case Studies. Front. Oncol. 2014, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Alhamad, D.W.; Elgendy, S.M.; Hersi, F.; El-Seedi, H.; Omar, H.A. The Inhibition of Autophagy by Spautin Boosts the Anticancer Activity of Fingolimod in Multidrug-Resistant Hepatocellular Carcinoma. Life Sci. 2022, 304, 120699. [Google Scholar] [CrossRef]
- Hersi, F.; Omar, H.A.; Al-Qawasmeh, R.; Ahmad, Z.; Jaber, A.M.; Zaher, D.M.; Al-Tel, T. Design and Synthesis of New Energy Restriction Mimetic Agents: Potent Anti-Tumor Activities of Hybrid Motifs of Aminothiazoles and Coumarins. Sci. Rep. 2020, 10, 2893. [Google Scholar] [CrossRef]
- Ramagli, L.S.; Rodriguez, L.V. Quantitation of Microgram Amounts of Protein in Two-Dimensional Polyacrylamide Gel Electrophoresis Sample Buffer. Electrophoresis 1985, 6, 559–563. [Google Scholar] [CrossRef]
- Borchert, N.; Krug, K.; Gnad, F.; Sinha, A.; Sommer, R.J.; Macek, B. Phosphoproteome of Pristionchus Pacificus Provides Insights into Architecture of Signaling Networks in Nematode Models. Mol. Cell. Proteom. 2012, 11, 1631–1639. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized P.P.B.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. MCP 2014, 13, 2513–2526. [Google Scholar] [CrossRef]

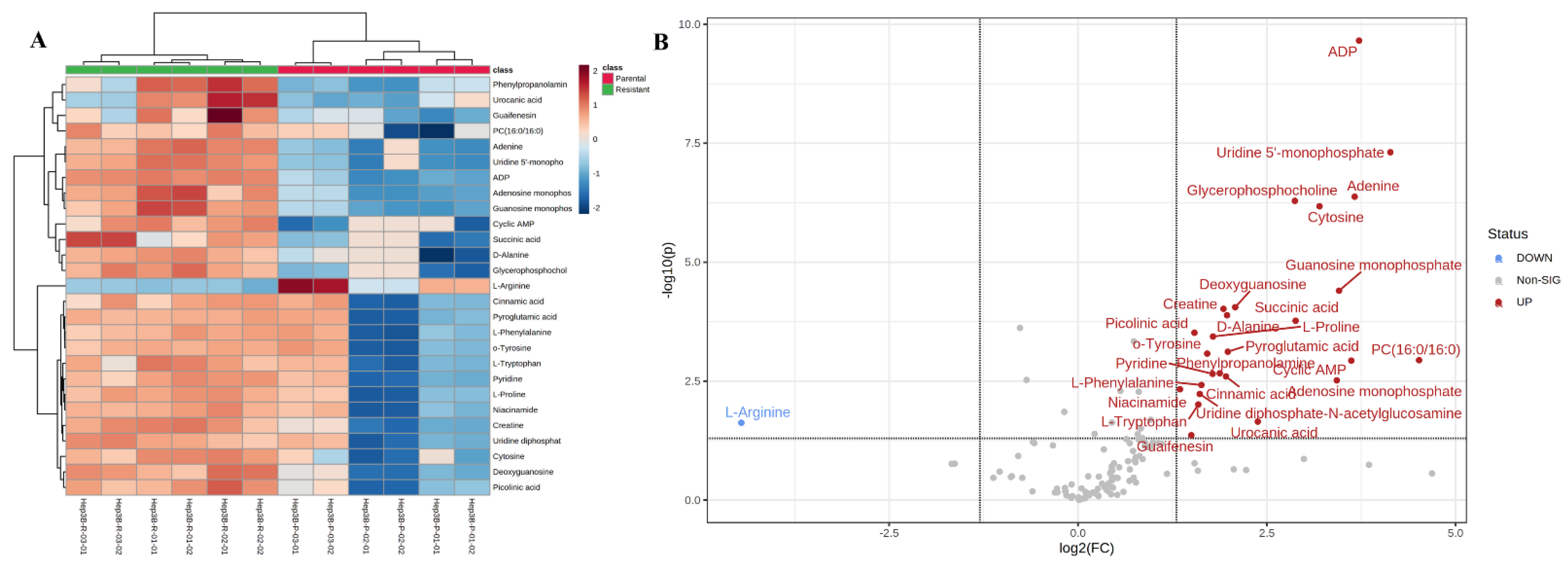


| Metabolite Name | p-Value | Fold Change |
|---|---|---|
| Uridine 5′-monophosphate | 0.000002 | 14.729 |
| ADP | 0.000001 | 13.186 |
| Adenosine monophosphate | 0.000778 | 12.894 |
| PC (16:0/16:0) | 0.000219 | 11.971 |
| Guanosine monophosphate | 0.000521 | 10.978 |
| Adenine | 0.000012 | 10.673 |
| Cyclic AMP | 0.000006 | 8.304 |
| Cytosine | 0.000071 | 6.264 |
| Urocanic acid | 0.037089 | 5.207 |
| Succinic acid | 0.001392 | 5.133 |
| Glycerophosphocholine | 0.000014 | 5.088 |
| Deoxyguanosine | 0.000114 | 4.233 |
| Creatine | 0.000253 | 3.801 |
| Phenylpropanolamine | 0.006535 | 3.672 |
| Guaifenesin | 0.043493 | 2.978 |
| Picolinic acid | 0.000545 | 2.911 |
| Pyridine | 0.007763 | 2.874 |
| D-Alanine | 0.019774 | 2.688 |
| Niacinamide | 0.012453 | 2.55 |
| L-Tryptophan | 0.015553 | 2.39 |
| L-Proline | 0.007556 | 2.372 |
| Uridine diphosphate-N-acetylglucosamine | 0.013429 | 2.253 |
| Pyroglutamic acid | 0.033094 | 2.238 |
| Cinnamic acid | 0.037616 | 2.159 |
| o-Tyrosine | 0.049019 | 2.098 |
| L-Phenylalanine | 0.039898 | 2.052 |
| L-Arginine | 0.04468 | −21.951 |
| Uniprot ID | Adjusted p Value | Effect Size | Significance |
|---|---|---|---|
| Q9C0H2 | 0.000315988 | −2.831839879 | Decreased |
| P07148 | 4.39 × 10−6 | −2.285889943 | Decreased |
| Q12769 | 0.009665038 | −2.158726692 | Decreased |
| O95573 | 0.000719872 | −1.838945548 | Decreased |
| P49591 | 0.001699466 | −1.737114747 | Decreased |
| Q9BWD1 | 3.18 × 10−10 | −1.703698476 | Decreased |
| P13473 | 0.009289266 | −1.515532017 | Decreased |
| P04632 | 0.005943185 | −1.197470983 | Decreased |
| Q01581 | 0.001058751 | −1.166633765 | Decreased |
| Q53GQ0 | 6.19 × 10−6 | −1.081565698 | Decreased |
| P49327 | 1.72 × 10−10 | −1.074158986 | Decreased |
| P27487 | 0.000121063 | −1.02650706 | Decreased |
| P17174 | 1.74 × 10−6 | −1.015660763 | Decreased |
| P04179 | 7.79 × 10−5 | 1.049084345 | Increased |
| Q9P035 | 0.003254458 | 1.252471606 | Increased |
| Q00796 | 0.000258132 | 1.474272092 | Increased |
| O60701 | 4.02 × 10−13 | 2.068751653 | Increased |
| P09936 | 2.41 × 10−10 | 2.986876806 | Increased |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abushawish, K.Y.I.; Soliman, S.S.M.; Giddey, A.D.; Al-Hroub, H.M.; Mousa, M.; Alzoubi, K.H.; El-Huneidi, W.; Abu-Gharbieh, E.; Omar, H.A.; Elgendy, S.M.; et al. Multi-Omics Analysis Revealed a Significant Alteration of Critical Metabolic Pathways Due to Sorafenib-Resistance in Hep3B Cell Lines. Int. J. Mol. Sci. 2022, 23, 11975. https://doi.org/10.3390/ijms231911975
Abushawish KYI, Soliman SSM, Giddey AD, Al-Hroub HM, Mousa M, Alzoubi KH, El-Huneidi W, Abu-Gharbieh E, Omar HA, Elgendy SM, et al. Multi-Omics Analysis Revealed a Significant Alteration of Critical Metabolic Pathways Due to Sorafenib-Resistance in Hep3B Cell Lines. International Journal of Molecular Sciences. 2022; 23(19):11975. https://doi.org/10.3390/ijms231911975
Chicago/Turabian StyleAbushawish, Kholoud Y. I., Sameh S. M. Soliman, Alexander D. Giddey, Hamza M. Al-Hroub, Muath Mousa, Karem H. Alzoubi, Waseem El-Huneidi, Eman Abu-Gharbieh, Hany A. Omar, Sara M. Elgendy, and et al. 2022. "Multi-Omics Analysis Revealed a Significant Alteration of Critical Metabolic Pathways Due to Sorafenib-Resistance in Hep3B Cell Lines" International Journal of Molecular Sciences 23, no. 19: 11975. https://doi.org/10.3390/ijms231911975
APA StyleAbushawish, K. Y. I., Soliman, S. S. M., Giddey, A. D., Al-Hroub, H. M., Mousa, M., Alzoubi, K. H., El-Huneidi, W., Abu-Gharbieh, E., Omar, H. A., Elgendy, S. M., Bustanji, Y., Soares, N. C., & Semreen, M. H. (2022). Multi-Omics Analysis Revealed a Significant Alteration of Critical Metabolic Pathways Due to Sorafenib-Resistance in Hep3B Cell Lines. International Journal of Molecular Sciences, 23(19), 11975. https://doi.org/10.3390/ijms231911975







