Bioinformatics and Experimental Analyses Reveal Immune-Related LncRNA–mRNA Pair AC011483.1-CCR7 as a Biomarker and Therapeutic Target for Ischemic Cardiomyopathy
Abstract
:1. Introduction
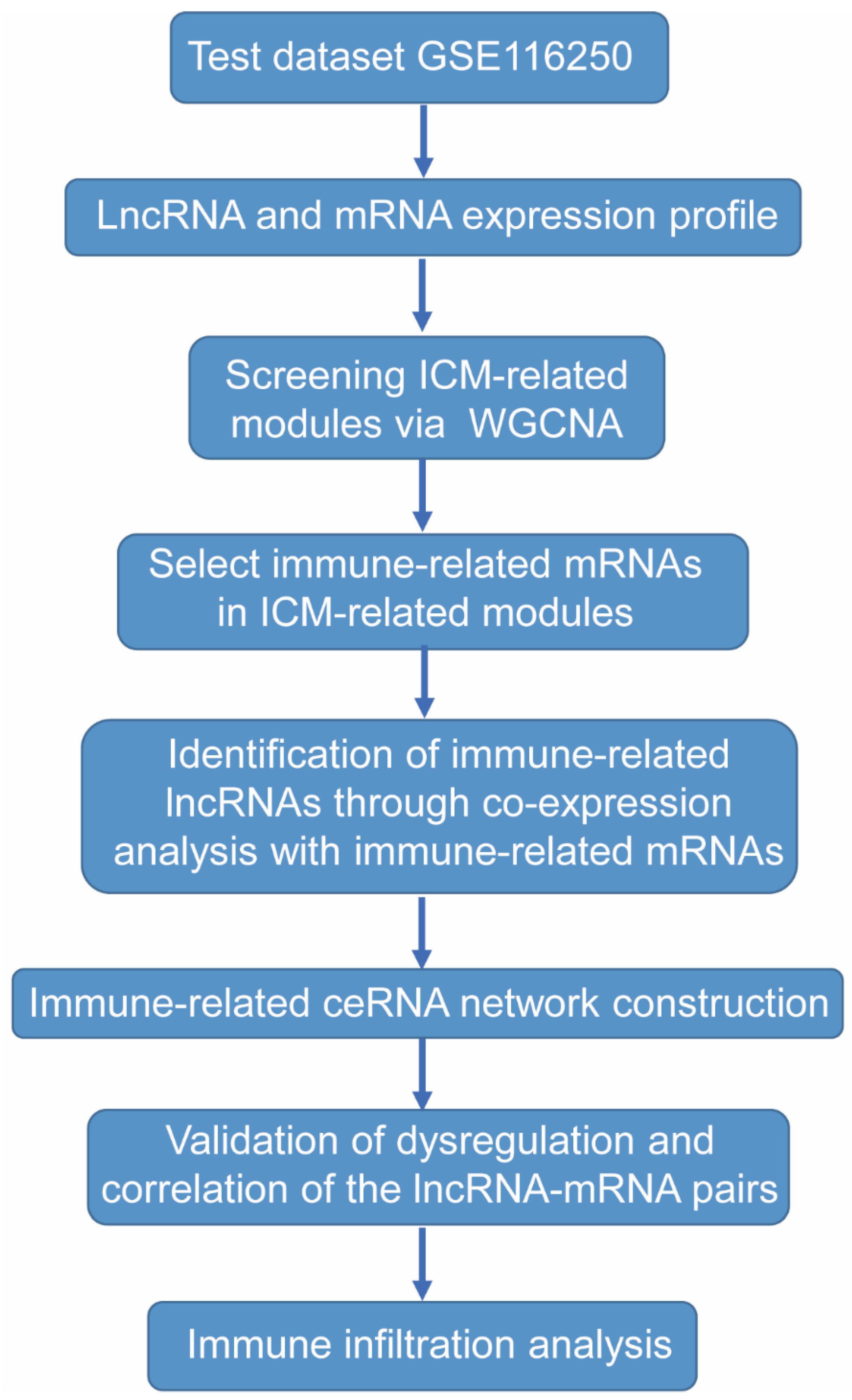
2. Results
2.1. Data Preprocessing and DEGs Screening
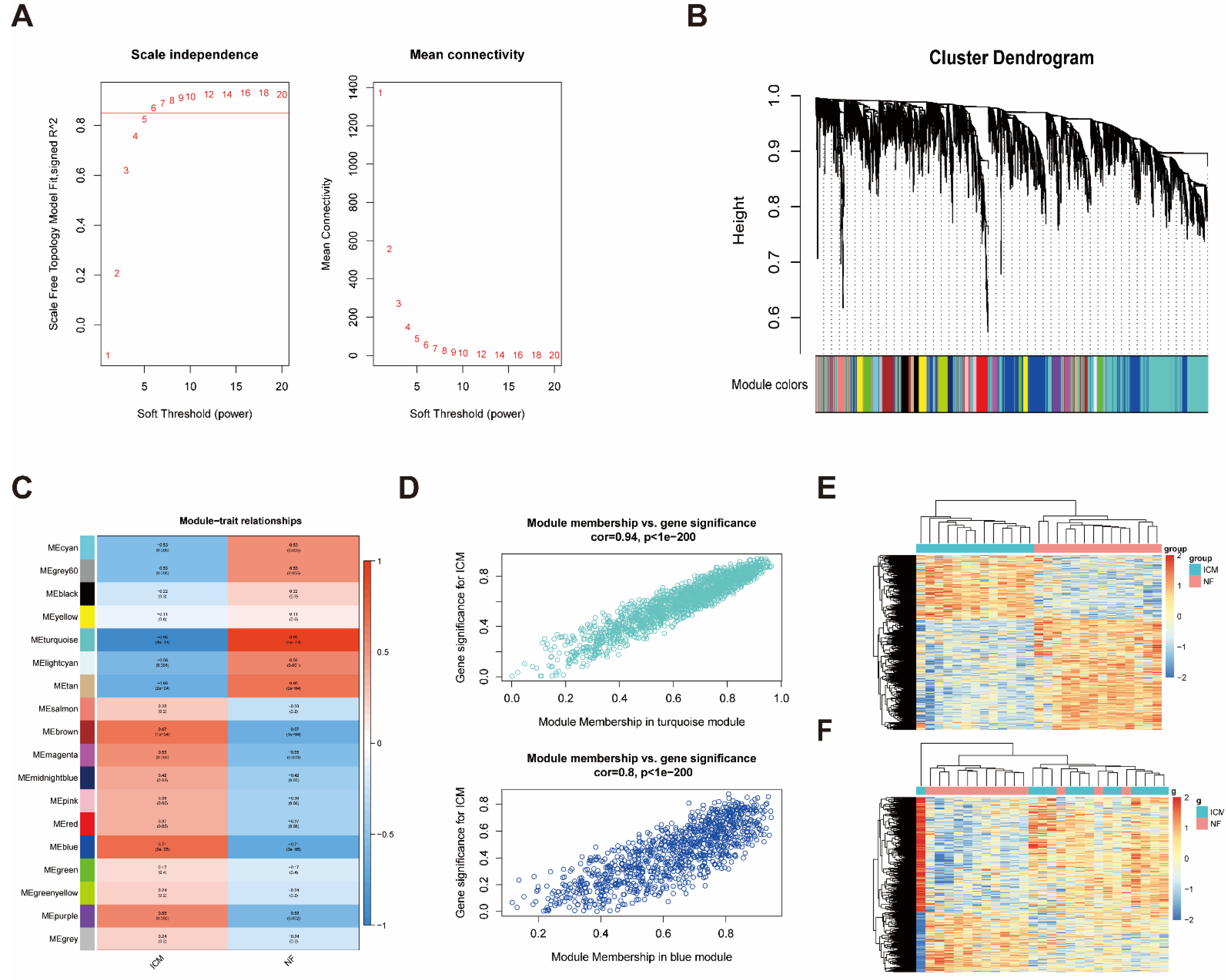
2.2. Construction of Weighted Co-Expression Network and Identification of ICM-Related Key Modules
2.3. Identification of Immune-Related LncRNAs in the Key Modules
2.4. The ceRNA Network Construction
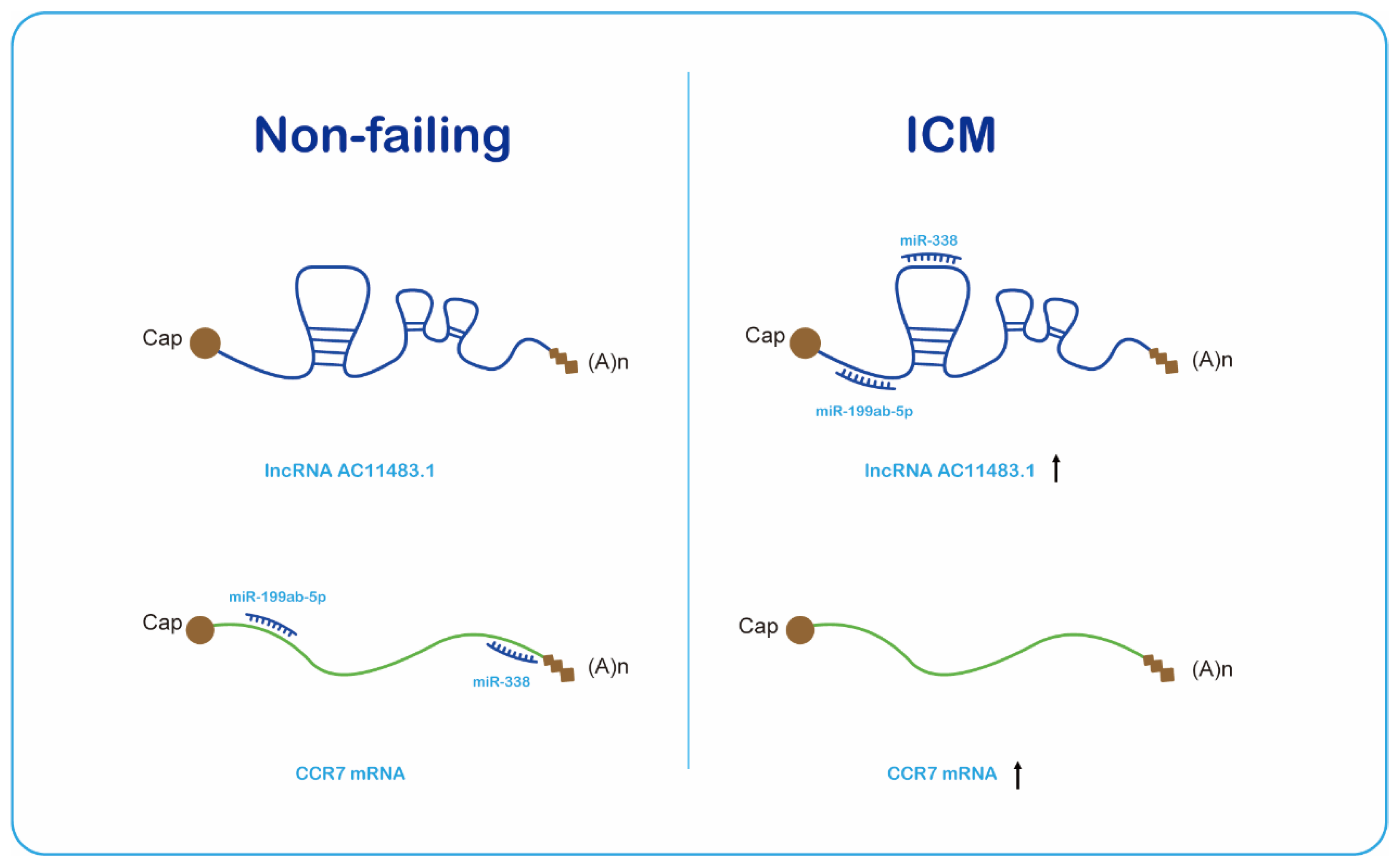
2.5. Validation of the Immune-Related lncRNA–mRNA Pairs
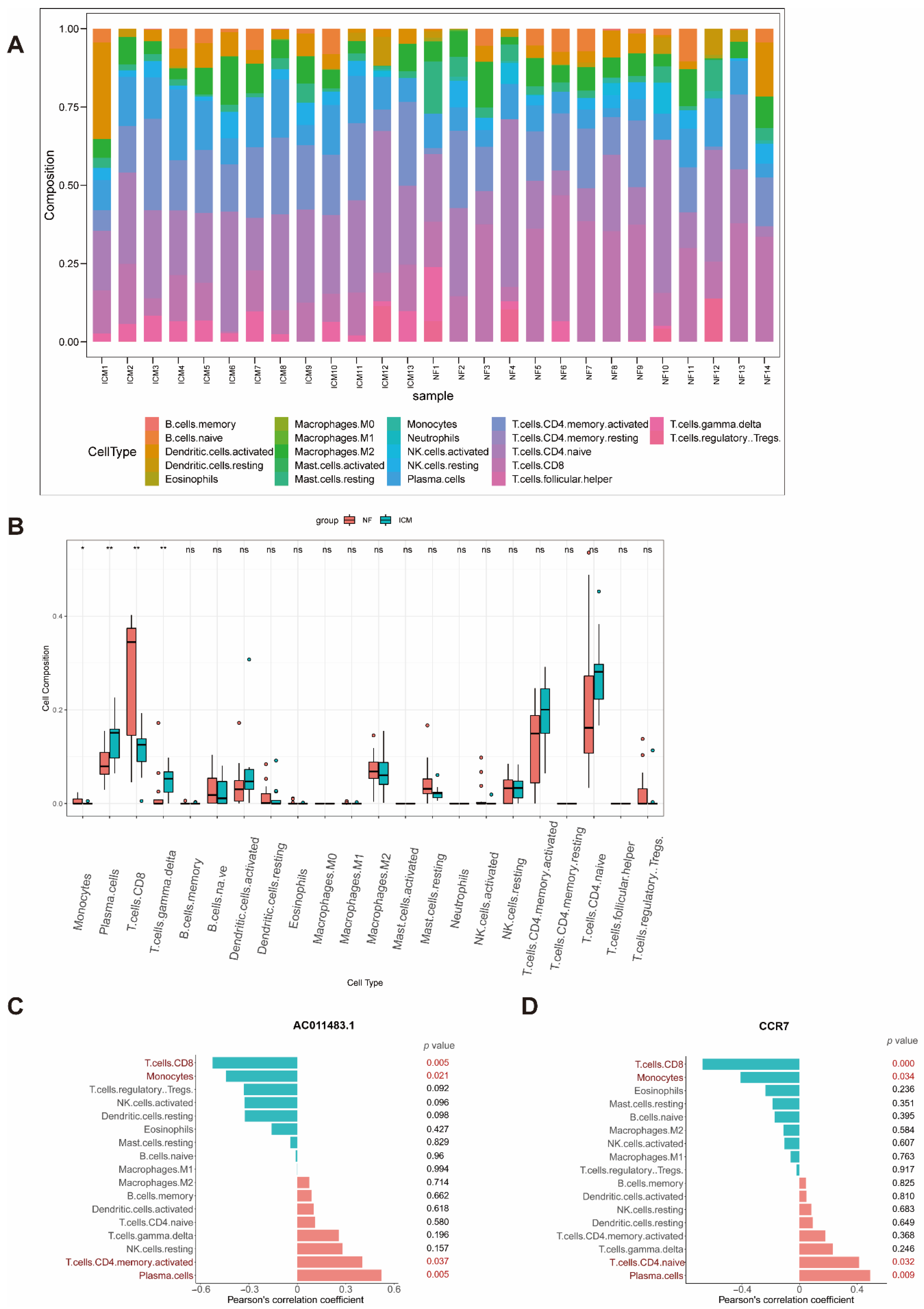
2.6. Immune Cell Infiltration
3. Discussion
4. Materials and Methods
4.1. Data Selection
4.2. Data Preprocessing and DEGs Screening
4.3. Co-Expression Network Analysis
4.4. Identification of Immune-Related LncRNAs
4.5. Construction and Analysis of an Immune-Related LncRNA-Associated Competing Endogenous RNA Network
4.6. Construction of Ischemic Cell Model and Cell Culture
4.7. Quantitative Real-Time PCR (qRT-PCR)
4.8. CIBERSORTx
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moroni, F.; Gertz, Z.; Azzalini, L. Relief of Ischemia in Ischemic Cardiomyopathy. Curr. Cardiol. Rep. 2021, 23, 80. [Google Scholar] [CrossRef] [PubMed]
- Rosamond, W.; Flegal, K.; Friday, G.; Furie, K.; Go, A.; Greenlund, K.; Haase, N.; Ho, M.; Howard, V.; Kissela, B.; et al. Heart disease and stroke statistics--2007 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2007, 115, e69–e171. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.H.; Rathore, S.S.; Radford, M.J.; Wang, Y.; Wang, Y.; Krumholz, H.M. Acute myocardial infarction in the elderly: Differences by age. J. Am. Coll. Cardiol. 2001, 38, 736–741. [Google Scholar] [CrossRef] [Green Version]
- Alexander, K.P.; Newby, L.K.; Cannon, C.P.; Armstrong, P.W.; Gibler, W.B.; Rich, M.W.; Van de Werf, F.; White, H.D.; Weaver, W.D.; Naylor, M.D.; et al. Acute coronary care in the elderly, part I: Non-ST-segment-elevation acute coronary syndromes: A scientific statement for healthcare professionals from the American Heart Association Council on Clinical Cardiology: In collaboration with the Society of Geriatric Cardiology. Circulation 2007, 115, 2549–2569. [Google Scholar] [CrossRef] [Green Version]
- Alexander, K.P.; Newby, L.K.; Armstrong, P.W.; Cannon, C.P.; Gibler, W.B.; Rich, M.W.; Van de Werf, F.; White, H.D.; Weaver, W.D.; Naylor, M.D.; et al. Acute coronary care in the elderly, part II: ST-segment-elevation myocardial infarction: A scientific statement for healthcare professionals from the American Heart Association Council on Clinical Cardiology: In collaboration with the Society of Geriatric Cardiology. Circulation 2007, 115, 2570–2589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekulic, M.; Zacharias, M.; Medalion, B. Ischemic Cardiomyopathy and Heart Failure. Circ. Heart Fail. 2019, 12, e006006. [Google Scholar] [CrossRef]
- Abdolmaleki, F.; Gheibi Hayat, S.M.; Bianconi, V.; Johnston, T.P.; Sahebkar, A. Atherosclerosis and immunity: A perspective. Trends Cardiovasc. Med. 2019, 29, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, S.D.; Frangogiannis, N.G. The Biological Basis for Cardiac Repair After Myocardial Infarction: From Inflammation to Fibrosis. Circ. Res. 2016, 119, 91–112. [Google Scholar] [CrossRef] [PubMed]
- van der Laan, A.M.; Nahrendorf, M.; Piek, J.J. Healing and adverse remodelling after acute myocardial infarction: Role of the cellular immune response. Heart 2012, 98, 1384–1390. [Google Scholar] [CrossRef] [PubMed]
- Latet, S.C.; Hoymans, V.Y.; Van Herck, P.L.; Vrints, C.J. The cellular immune system in the post-myocardial infarction repair process. Int. J. Cardiol. 2015, 179, 240–247. [Google Scholar] [CrossRef]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Sun, L.; Xuan, L.; Pan, Z.; Hu, X.; Liu, H.; Bai, Y.; Jiao, L.; Li, Z.; Cui, L.; et al. Long non-coding RNA CCRR controls cardiac conduction via regulating intercellular coupling. Nat. Commun. 2018, 9, 4176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017, 9, eaai9118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Kataoka, M.; Wang, Y.; Pu, L.; Dong, X.; Fu, X.; Zhang, F.; Gao, F.; Liang, T.; Pei, J.; et al. LncRNA LncHrt preserves cardiac metabolic homeostasis and heart function by modulating the LKB1-AMPK signaling pathway. Basic Res. Cardiol. 2021, 116, 48. [Google Scholar] [CrossRef]
- Greco, S.; Zaccagnini, G.; Perfetti, A.; Fuschi, P.; Valaperta, R.; Voellenkle, C.; Castelvecchio, S.; Gaetano, C.; Finato, N.; Beltrami, A.P.; et al. Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 2016, 14, 183. [Google Scholar] [CrossRef] [Green Version]
- van ’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [Green Version]
- Kolur, V.; Vastrad, B.; Vastrad, C.; Kotturshetti, S.; Tengli, A. Identification of candidate biomarkers and therapeutic agents for heart failure by bioinformatics analysis. BMC Cardiovasc. Disord. 2021, 21, 329. [Google Scholar] [CrossRef]
- Ye, Y.; Jin, Q.; Gong, Q.; Li, A.; Sun, M.; Jiang, S.; Jin, Y.; Zhang, Z.; He, J.; Zhuang, L. Bioinformatics and Experimental Analyses Reveal NFIC as an Upstream Transcriptional Regulator for Ischemic Cardiomyopathy. Genes 2022, 13, 1051. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Jeggari, A.; Marks, D.S.; Larsson, E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012, 28, 2062–2063. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Wang, L.; Bei, Y.; Wu, X.; Wang, J.; Zhou, Q.; Tao, L.; Das, S.; Li, X.; Xiao, J. Long Noncoding RNA Cardiac Physiological Hypertrophy-Associated Regulator Induces Cardiac Physiological Hypertrophy and Promotes Functional Recovery After Myocardial Ischemia-Reperfusion Injury. Circulation 2021, 144, 303–317. [Google Scholar] [CrossRef]
- Zou, Y.; Wang, Y.; Rong, Z.; Wei, B.; Liu, Y.; Song, Z.; Li, W.; Hu, E.; Deng, G.; He, Y.; et al. A susceptibility biomarker identification strategy based on significantly differentially expressed ceRNA triplets for ischemic cardiomyopathy. Biosci Rep. 2020, 40, BSR20191731. [Google Scholar] [CrossRef]
- Xi, R.; Fan, Q.; Yan, X.; Zhang, H.; Xie, H.; Gu, G.; Xu, Y.; Wang, F.; Tao, R. Increased Serum Interleukin-34 Levels Are Related to the Presence and Severity of Cardiac Dysfunction in Patients With Ischemic Cardiomyopathy. Front. Physiol. 2018, 9, 904. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhou, Y.; Gu, X.; Zhang, X.; Jia, Z. NLRX1/FUNDC1/NIPSNAP1-2 axis regulates mitophagy and alleviates intestinal ischaemia/reperfusion injury. Cell Prolif. 2021, 54, e12986. [Google Scholar] [CrossRef]
- Zhang, H.; Xiao, Y.; Nederlof, R.; Bakker, D.; Zhang, P.; Girardin, S.E.; Hollmann, M.W.; Weber, N.C.; Houten, S.M.; van Weeghel, M.; et al. NLRX1 Deletion Increases Ischemia-Reperfusion Damage and Activates Glucose Metabolism in Mouse Heart. Front. Immunol. 2020, 11, 591815. [Google Scholar] [CrossRef]
- Li, Z.; Jin, D.; Wu, Y.; Zhang, K.; Hu, P.; Cao, X.; Chen, Z. Increased serum interleukin-34 in patients with coronary artery disease. J. Int. Med. Res. 2012, 40, 1866–1870. [Google Scholar] [CrossRef] [Green Version]
- Fan, Q.; Yan, X.; Zhang, H.; Lu, L.; Zhang, Q.; Wang, F.; Xi, R.; Hu, J.; Chen, Q.; Niu, W.; et al. IL-34 is associated with the presence and severity of renal dysfunction and coronary artery disease in patients with heart failure. Sci. Rep. 2016, 6, 39324. [Google Scholar] [CrossRef]
- Kalwa, M.; Hanzelmann, S.; Otto, S.; Kuo, C.C.; Franzen, J.; Joussen, S.; Fernandez-Rebollo, E.; Rath, B.; Koch, C.; Hofmann, A.; et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016, 44, 10631–10643. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Jiang, Y.; Xie, Y.; Leng, X.; Song, F. Comprehensive Analysis of lncRNAs Associated with the Pathogenesis and Prognosis of Gastric Cancer. DNA Cell Biol. 2020, 39, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.F.; Brown, M.A.; Nicolai, H.; Chambers, J.A.; Griffiths, B.L.; Solomon, E. Isolation and characterisation of the NBR2 gene which lies head to head with the human BRCA1 gene. Hum. Mol. Genet. 1997, 6, 1057–1062. [Google Scholar] [CrossRef] [Green Version]
- Bai, J.; Xu, J.; Zhao, J.; Zhang, R. LncRNA NBR2 suppresses migration and invasion of colorectal cancer cells by downregulating miRNA-21. Hum. Cell 2020, 33, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, H.; Feng, X.; Wang, L.; Yang, X. NBR2 promotes the proliferation of glioma cells via inhibiting p15 expression. J BUON 2021, 26, 388–394. [Google Scholar]
- Sheng, J.Q.; Wang, M.R.; Fang, D.; Liu, L.; Huang, W.J.; Tian, D.A.; He, X.X.; Li, P.Y. LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. Biomed Pharmacother. 2021, 133, 111023. [Google Scholar] [CrossRef]
- Zhu, C.; Wang, M.; Yu, X.; Shui, X.; Tang, L.; Chen, Z.; Xiong, Z. lncRNA NBR2 attenuates angiotensin II-induced myocardial hypertrophy through repressing ER stress via activating LKB1/AMPK/Sirt1 pathway. Bioengineered 2022, 13, 13667–13679. [Google Scholar] [CrossRef]
- Shi, Y.; Ren, J.; Zhuang, Z.; Zhang, W.; Wang, Z.; Liu, Y.; Li, J.; Liang, T.; He, R.; Wang, K. Comprehensive Analysis of a ceRNA Network Identifies lncR-C3orf35 Associated with Poor Prognosis in Osteosarcoma. Biomed Res. Int. 2020, 2020, 3178037. [Google Scholar] [CrossRef]
- Hu, J.; Wang, T.; Chen, Q. Competitive endogenous RNA network identifies four long non-coding RNA signature as a candidate prognostic biomarker for lung adenocarcinoma. Transl. Cancer Res. 2019, 8, 1046–1064. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.G.; Zhang, K.Q. LINC00452 promotes ovarian carcinogenesis through increasing ROCK1 by sponging miR-501-3p and suppressing ubiquitin-mediated degradation. Aging 2020, 12, 21129–21146. [Google Scholar] [CrossRef]
- Hintzen, G.; Ohl, L.; del Rio, M.L.; Rodriguez-Barbosa, J.I.; Pabst, O.; Kocks, J.R.; Krege, J.; Hardtke, S.; Forster, R. Induction of tolerance to innocuous inhaled antigen relies on a CCR7-dependent dendritic cell-mediated antigen transport to the bronchial lymph node. J. Immunol. 2006, 177, 7346–7354. [Google Scholar] [CrossRef]
- Forster, R.; Schubel, A.; Breitfeld, D.; Kremmer, E.; Renner-Muller, I.; Wolf, E.; Lipp, M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 1999, 99, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Ohl, L.; Mohaupt, M.; Czeloth, N.; Hintzen, G.; Kiafard, Z.; Zwirner, J.; Blankenstein, T.; Henning, G.; Forster, R. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 2004, 21, 279–288. [Google Scholar] [CrossRef] [Green Version]
- Sweet, M.E.; Cocciolo, A.; Slavov, D.; Jones, K.L.; Sweet, J.R.; Graw, S.L.; Reece, T.B.; Ambardekar, A.V.; Bristow, M.R.; Mestroni, L.; et al. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genom. 2018, 19, 812. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.C.; Yamada, K.A.; Patel, A.Y.; Topkara, V.K.; George, I.; Cheema, F.H.; Ewald, G.A.; Mann, D.L.; Nerbonne, J.M. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014, 129, 1009–1021. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Zhuang, L.; Shen, X.; Yang, L. Glucotoxicity Activation of IL6 and IL11 and Subsequent Induction of Fibrosis May Be Involved in the Pathogenesis of Islet Dysfunction. Front. Mol. Biosci. 2021, 8, 708127. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Dunn, P.; Thomas, C.G.; Smith, B.; Schaefer, H.; Chen, J.; Hu, Z.; Zalocusky, K.A.; Shankar, R.D.; Shen-Orr, S.S.; et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 2018, 5, 180015. [Google Scholar] [CrossRef] [PubMed]



| Immune-Related mRNA | Immune-Related lncRNA | Correlation Coefficient |
|---|---|---|
| KL | C3orf35 | 0.938991 |
| HSPA2 | CCDC144NL-AS1 | −0.92054 |
| IL17RB | FP700111.1 | 0.908619 |
| IL17RB | AL137230.2 | 0.904658 |
| HSPA2 | AL137230.2 | 0.900697 |
| HSPA2 | NBR2 | 0.898993 |
| OAS1 | AL355483.1 | 0.893618 |
| IL17RB | AL353576.1 | 0.891683 |
| CD8A | AL109924.2 | −0.89132 |
| SSTR2 | NUTM2B-AS1 | 0.891205 |
| HSPA2 | AC018462.1 | −0.88867 |
| HSPA2 | FP700111.1 | 0.888298 |
| SSTR2 | AL357033.1 | −0.88791 |
| PPBP | AL355483.1 | 0.886973 |
| PPBP | MYCL-AS1 | 0.886181 |
| JAG2 | AL445253.1 | 0.886144 |
| HSPA2 | AC006115.2 | −0.88587 |
| WFIKKN1 | AC099811.3 | 0.885222 |
| HSPA2 | AL139011.1 | −0.88446 |
| HSPA2 | AC005776.2 | −0.884 |
| SORT1 | AC135050.6 | −0.88333 |
| IL17RB | LINC00958 | 0.88294 |
| HSPA2 | HCG25 | −0.87913 |
| CD8A | NBR2 | 0.879123 |
| HSPA2 | AL357033.1 | −0.87882 |
| CD8A | AL109924.4 | −0.87795 |
| HSPA2 | LIPE-AS1 | −0.87617 |
| HSPA2 | AC100802.1 | −0.87556 |
| IL24 | NBR2 | −0.87349 |
| HSPA2 | AP002336.2 | −0.87336 |
| SORT1 | LINC01788 | 0.870983 |
| CD8A | AL136419.2 | 0.869864 |
| HSPA2 | STAM-AS1 | 0.869411 |
| HSPA2 | GLYCTK-AS1 | −0.86937 |
| CD8A | AL139011.1 | −0.86919 |
| SSTR2 | AC018462.1 | −0.86902 |
| HSPA2 | Z84492.1 | −0.86834 |
| HSPA2 | AC016727.1 | 0.86786 |
| HSPA2 | AC008124.1 | 0.867474 |
| IL24 | AL590666.2 | 0.867407 |
| A2M | AC117945.1 | −0.86682 |
| CD8A | CCDC144NL-AS1 | −0.86612 |
| PPBP | KIF9-AS1 | 0.865588 |
| PPBP | AC015687.1 | 0.865373 |
| SORT1 | AL513285.1 | 0.864497 |
| PPBP | AC023282.1 | 0.864312 |
| HSPA2 | AL445253.1 | −0.86361 |
| HSPA2 | SEMA3F-AS1 | −0.86289 |
| A2M | AP003721.2 | −0.8627 |
| HSPA2 | AC021092.1 | −0.86154 |
| Immune-Related mRNA | Immune-Related lncRNA | Correlation Coefficient |
|---|---|---|
| HSPA2 | NBR2 | 0.898993 |
| CD8A | NBR2 | 0.879123 |
| NFYC | C3orf35 | 0.853143 |
| ITK | C3orf35 | 0.828428 |
| PTGFR | C3orf35 | 0.801013 |
| KL | C3orf35 | 0.938991 |
| SORT1 | C3orf35 | 0.822644 |
| PDK1 | C3orf35 | 0.800123 |
| IL17RB | LINC00452 | 0.801915 |
| HSPA2 | LINC00452 | 0.804171 |
| GZMB | UBE2Q1-AS1 | 0.819774 |
| HLA-DOB | UBE2Q1-AS1 | 0.807795 |
| CCR7 | AC011483.1 | 0.809768 |
| MICB | AC011483.1 | 0.812744 |
| HSPA2 | AC011407.1 | 0.818169 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Q.; Gong, Q.; Le, X.; He, J.; Zhuang, L. Bioinformatics and Experimental Analyses Reveal Immune-Related LncRNA–mRNA Pair AC011483.1-CCR7 as a Biomarker and Therapeutic Target for Ischemic Cardiomyopathy. Int. J. Mol. Sci. 2022, 23, 11994. https://doi.org/10.3390/ijms231911994
Jin Q, Gong Q, Le X, He J, Zhuang L. Bioinformatics and Experimental Analyses Reveal Immune-Related LncRNA–mRNA Pair AC011483.1-CCR7 as a Biomarker and Therapeutic Target for Ischemic Cardiomyopathy. International Journal of Molecular Sciences. 2022; 23(19):11994. https://doi.org/10.3390/ijms231911994
Chicago/Turabian StyleJin, Qiao, Qian Gong, Xuan Le, Jin He, and Lenan Zhuang. 2022. "Bioinformatics and Experimental Analyses Reveal Immune-Related LncRNA–mRNA Pair AC011483.1-CCR7 as a Biomarker and Therapeutic Target for Ischemic Cardiomyopathy" International Journal of Molecular Sciences 23, no. 19: 11994. https://doi.org/10.3390/ijms231911994





