Natural Variation of Fatty Acid Desaturase Gene Affects Linolenic Acid Content and Starch Pasting Viscosity in Rice Grains
Abstract
:1. Introduction
2. Results
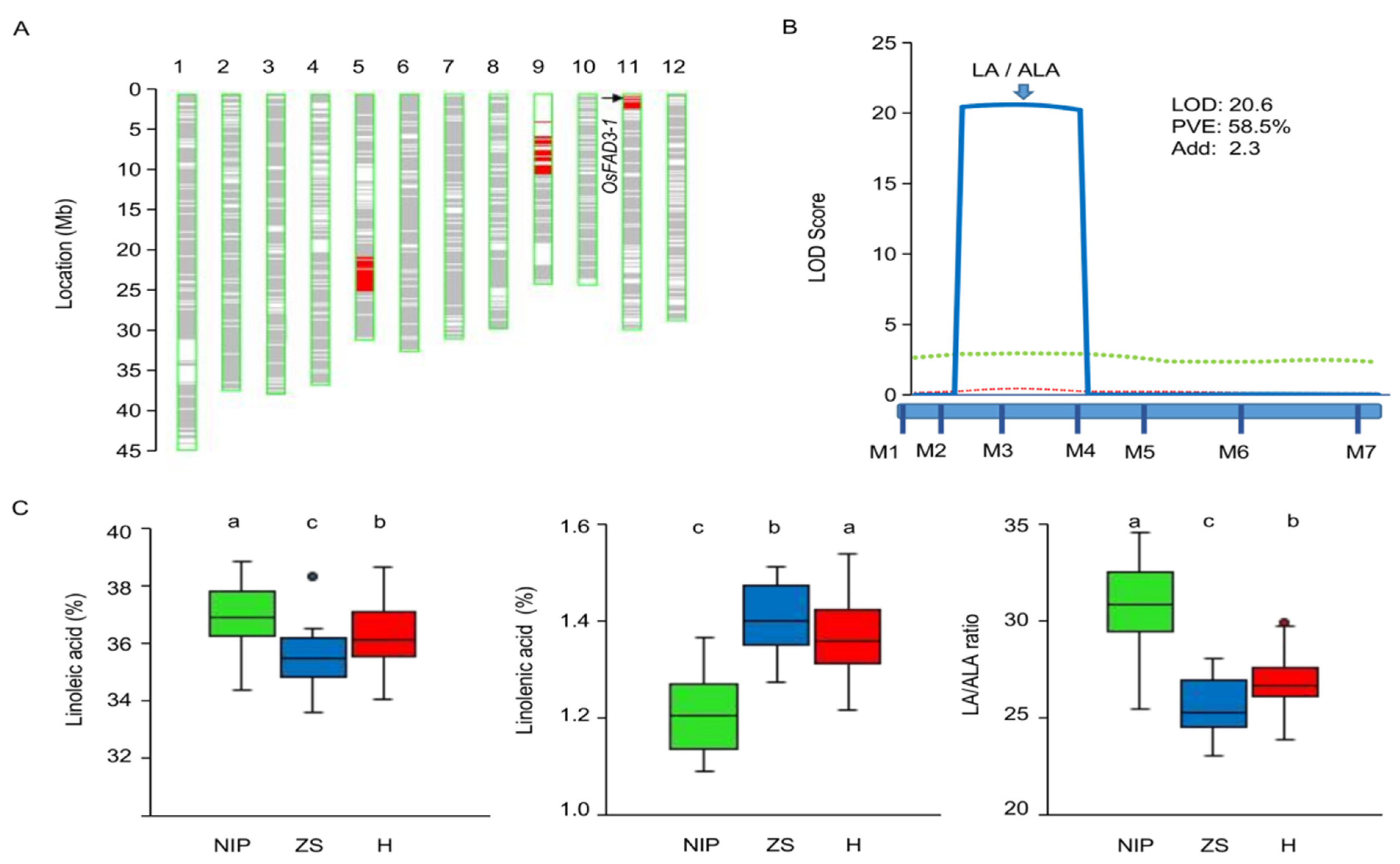
2.1. Effects of OsFAD3-1 and OsFAD3-2 on Unsaturated Fatty Acids
2.2. Validate of Genetic Effect of OsFAD3-1 on the LA/ALA Ratio
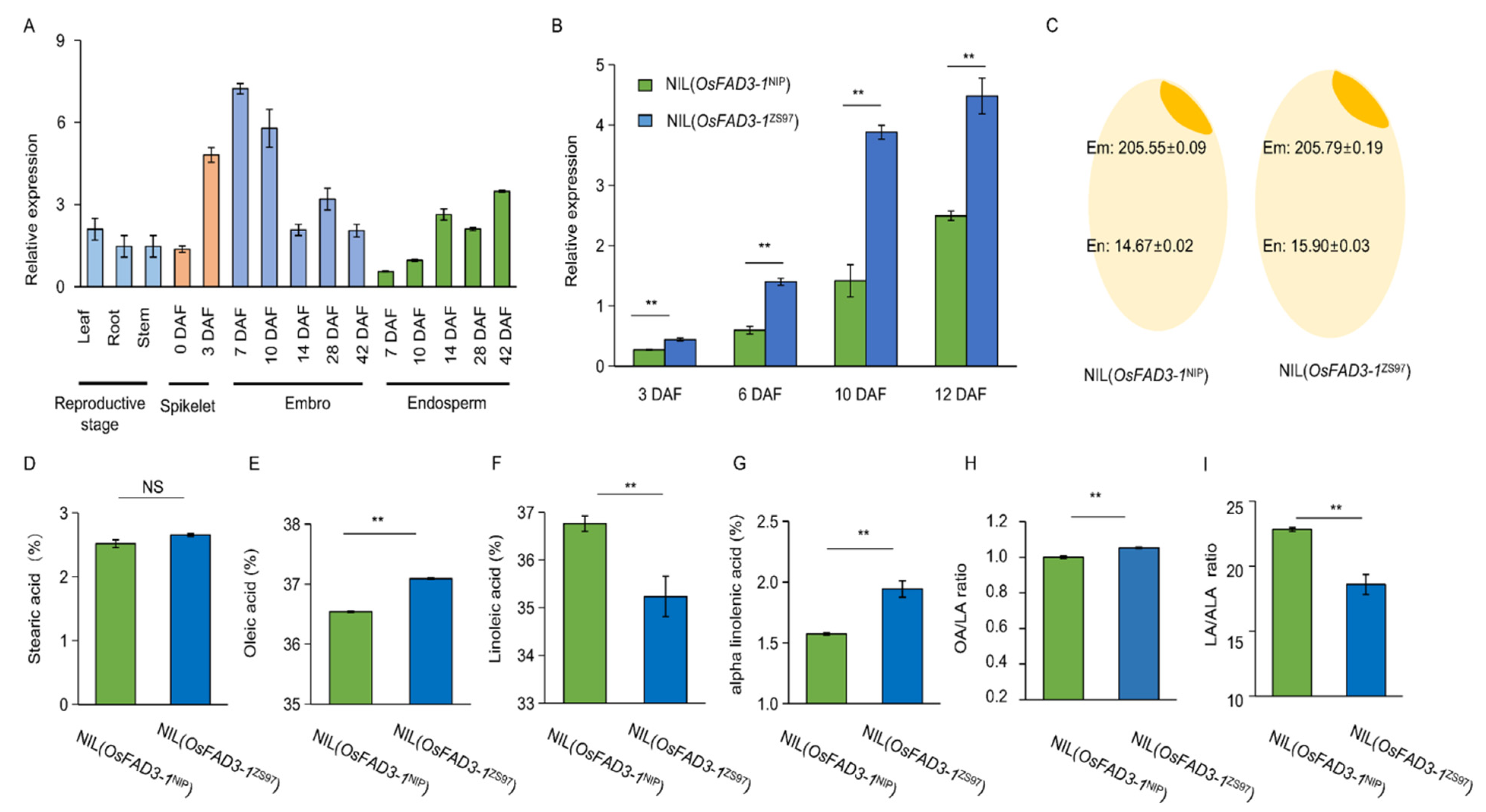
2.3. Expression Variation of OsFAD3-1 in Rice Seed
2.4. Overexpression of OsFAD3-1 Increases Alpha-Linolenic Acid Content in Rice
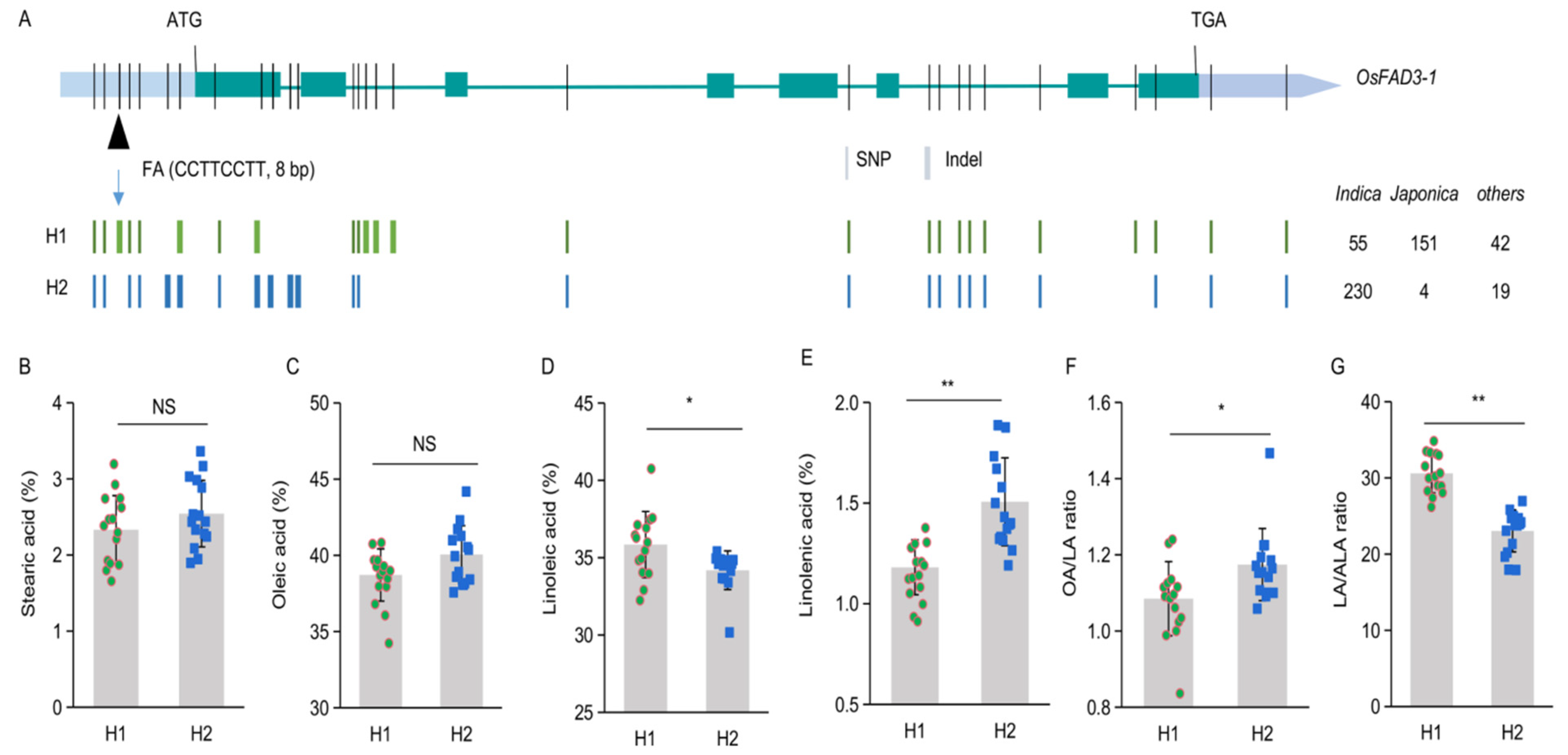
2.5. Allelic Variation of OsFAD3-1 in Rice Germplasm
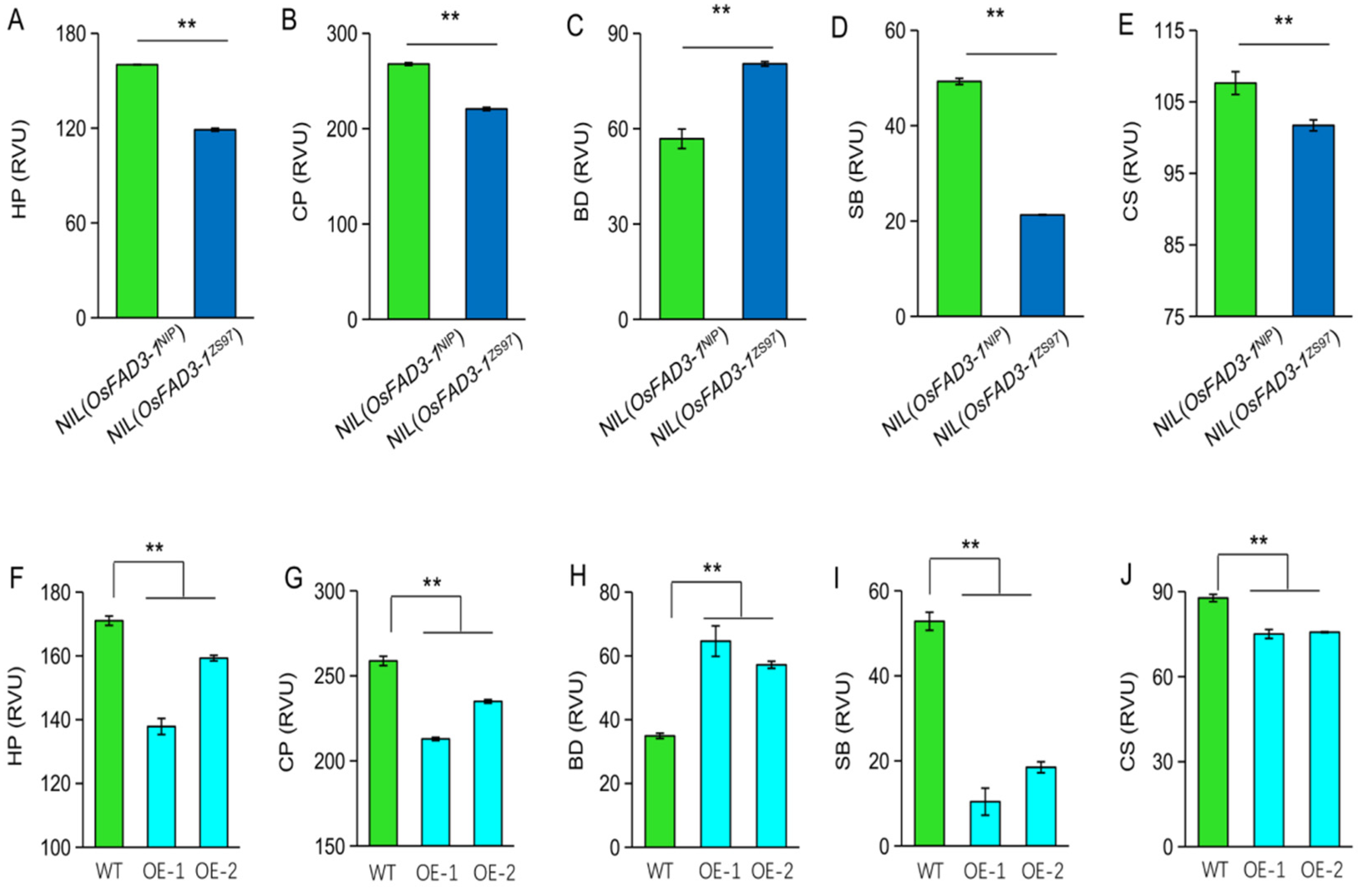
2.6. OsFAD3-1 Alters the Cooking and Eating Quality of Rice
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Genotype and QTL Analyses
4.3. Expression Analysis
4.4. Vector Construction and Transformation
4.5. Measurement of Fatty Acids
4.6. Pasting Viscosity Value Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parengam, M.; Judprasong, K.; Srianujata, S.; Jittinandana, S.; Laoharojanaphand, S.; Busamongko, A. Study of nutrients and toxic minerals in rice and legumes by instrumental neutron activation analysis and graphite furnace atomic absorption spectrophotometry. J. Food Compos. Anal. 2010, 23, 340–345. [Google Scholar] [CrossRef]
- Huang, Y.P.; Lai, H.M. Bioactive compounds and antioxidative activity of colored rice bran. J. Food Drug. Anal. 2016, 24, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Nakamura, K.; Kayahara, H. Analysis of phenolic compounds in white rice, brown rice, and germinated brown rice. J. Agric. Food Chem. 2004, 52, 4808–4813. [Google Scholar] [CrossRef] [PubMed]
- Schramm, R.; Abadie, A.; Hua, N.; Xu, Z.; Lima, M. Fractionation of the rice bran layer and quantification of vitamin E, oryzanol, protein, and rice bran saccharide. J. Biol. Eng. 2007, 1, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, M.; Hossain, S.; Matsuzaki, K.; Shido, O.; Yoshino, K. The journey from white rice to ultra-high hydrostatic pressurized brown rice: An excellent endeavor for ideal nutrition from staple food. Crit. Rev. Food Sci. Nutr. 2022, 62, 1502–1520. [Google Scholar] [CrossRef]
- Liu, L.; Waters, D.L.; Rose, T.J.; Bao, J.; King, G.J. Phospholipids in rice: Significance in grain quality and health benefits: A review. Food Chem. 2013, 139, 1133–1145. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Xia, D.; He, Y.Q. Rice grain quality traditional traits for high quality rice and health-plus substances. Mol. Breed. 2019, 40, 1. [Google Scholar] [CrossRef]
- Baker, E.J.; Miles, E.A.; Burdge, G.C.; Yaqoob, P.; Calder, P.C. Metabolism and functional effects of plant-derived omega-3 fatty acids in humans. Prog. Lipid Res. 2016, 64, 30–56. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, A.S.; Brown, T.J.; Brainard, J.S.; Biswas, P.; Thorpe, G.C.; Moore, H.J.; Deane, K.H.; Summerbell, C.D.; Worthington, H.V.; Song, F.; et al. Omega-3 fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2020, 3, CD003177. [Google Scholar] [CrossRef]
- Stark, K.D. The percentage of n-3 highly unsaturated fatty acids in total HUFA as a biomarker for omega-3 fatty acid status in tissues. Lipids 2008, 43, 45–53. [Google Scholar] [CrossRef]
- Kaliannan, K.; Li, X.Y.; Wang, B.; Pan, Q.; Chen, C.Y.; Hao, L.; Xie, S.F.; Kang, J.X. Multi-omic analysis in transgenic mice implicates omega-6/omega-3 fatty acid imbalance as a risk factor for chronic disease. Commun. Biol. 2019, 2, 276–294. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.L.; Yin, Z.J.; Xiao, L.; Xu, Y.N.; Qu, L.Q. Identification and evaluation of ω-3 fatty acid desaturase genes for hyperfortifying α-linolenic acid in transgenic rice seed. J. Exp. Bot. 2012, 63, 3279–3287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallis, J.G.; Browse, J. Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 2002, 41, 254–278. [Google Scholar] [CrossRef]
- Arondel, V.; Lemieux, B.; Hwang, I.; Gibson, S.; Goodman, H.M.; Somerville, C.R. Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 1992, 258, 1353–1355. [Google Scholar] [CrossRef] [PubMed]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994, 6, 147–158. [Google Scholar] [PubMed]
- Zaplin, E.S.; Liu, Q.; Li, Z.Y.; Vito, M.; Butardo, J.; Blanchard, C.L.; Rahman, S. Production of high oleic rice grains by suppressing the expression of the OsFAD2-1 gene. Funct. Plant Biol. 2013, 40, 996–1004. [Google Scholar] [CrossRef]
- Shi, Y.L.; Cao, Y.P.; Fan, X.R.; Li, M.; Wang, Y.F.; Ming, F. A rice microsomal delta-12 fatty acid desaturase can enhance resistance to cold stress in yeast and Oryza sativa. Mol. Breeding. 2012, 29, 743–757. [Google Scholar] [CrossRef]
- Tiwari, G.J.; Liu, Q.; Shreshtha, P.; Li, Z.Y.; Rahman, S. RNAi-mediated down-regulation of the expression of OsFAD2-1: Effect on lipid accumulation and expression of lipid biosynthetic genes in the rice grain. BMC Plant Biol. 2016, 16, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, K.; Araki, E.; Suzuki, Y.; Toki, S.; Saika, H. Production of high oleic/low linoleic rice by genome editing. Plant Physiol. Biochem. 2018, 131, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Kodama, H.; Akagi, H.; Kusumi, K.; Fujimura, T.; Iba, K. Structure, chromosomal location and expression of a rice gene encoding the microsome omega-3 fatty acid desaturase. Plant Mol. Biol. 1997, 33, 493–502. [Google Scholar] [CrossRef] [PubMed]
- E, Z.; Chen, C.; Yang, J.Y.; Tong, H.H.; Li, T.T.; Wang, L.; Chen, H.Q. Genome-wide analysis of fatty acid desaturase genes in rice (Oryza sativa L.). Sci. Rep. 2019, 9, 19445. [Google Scholar] [CrossRef] [Green Version]
- Yara, A.; Yaeno, T.; Hasegawa, M.; Seto, H.; Montillet, J.L.; Kusumi, K.; Seo, S.; Iba, K. Disease resistance against Magnaporthe grisea is enhanced in transgenic rice with suppression of omega-3 fatty acid desaturases. Plant Cell Physiol. 2007, 48, 1263–1274. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Xia, D.; Li, P.B.; Ao, Y.T.; Xu, X.D.; Wan, S.S.; Li, Y.H.; Wu, B.; Shi, H.; Wang, K.Y.; et al. Genetic architecture and key genes controlling the diversity of oil composition in rice grains. Mol. Plant 2021, 14, 456–469. [Google Scholar] [CrossRef]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef]
- Dyall, S.C. Interplay between n-3 and n-6 long-chain polyunsaturated fatty acids and the endocannabinoid system in brain protection and repair. Lipids 2017, 52, 885–900. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.S.M.; Wang, P.; Wang, N.; Yang, L.; Xiao, Z. Brown rice versus white rice: Nutritional quality, potential health benefits, development of food products, and preservation technologies. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1070–1096. [Google Scholar] [CrossRef] [Green Version]
- Gayral, M.; Bakan, B.; Dalgalarrondo, M.; Elmorjani, K.; Delluc, C.; Brunet, S.; Linossier, L.; Morel, M.H.; Marion, D. Lipid partitioning in maize (Zea mays L.) endosperm highlights relationships among starch lipids, amylose, and vitreousness. J. Agric. Food Chem. 2015, 63, 3551–3558. [Google Scholar] [CrossRef]
- Yoon, M.R.; Lee, S.C.; Kang, M.Y. The lipid composition of rice cultivars with different eating qualities. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 291–295. [Google Scholar] [CrossRef]
- Luo, J.; Liu, L.; Konik-Rose, C.; Tian, L.; Singh, S.; Howitt, C.A.; Li, Z.; Liu, Q. Down-regulation of FAD2-1 gene expression alters lysophospholipid composition in the endosperm of rice grain and influences starch properties. Foods 2021, 10, 1169. [Google Scholar] [CrossRef]
- Chen, H.; Chen, D.; He, L.; Wang, T.; Lu, H.; Yang, F.; Deng, F.; Chen, Y.; Tao, Y.; Li, M.; et al. Correlation of taste values with chemical compositions and Rapid Visco Analyser profiles of 36 indica rice (Oryza sativa L.) varieties. Food Chem. 2021, 349, 129176. [Google Scholar] [CrossRef]
- Shi, S.J.; Wang, E.T.; Li, C.X.; Cai, M.L.; Cheng, B.; Cao, C.G.; Jiang, Y. Use of protein content, amylose content, and RVA parameters to evaluate the tste quality of rice. Front. Nutr. 2022, 8, 758547. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, Q.; Meng, D.; Isaac, G.; Zhao, R.; Fillmore, T.L.; Chu, R.K.; Zhou, J.; Tang, K.; Hu, Z.; et al. A reversed-phase capillary ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) method for comprehensive top-down/bottom-up lipid profiling. Anal. Bioanal. Chem. 2012, 402, 2923–2933. [Google Scholar] [CrossRef] [Green Version]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Astarita, G. Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat. Protoc. 2017, 12, 797–813. [Google Scholar] [CrossRef]
- Gaikwad, K.B.; Rani, S.; Kumar, M.; Gupta, V.; Babu, P.H.; Bainsla, N.K.; Yadav, R. Enhancing the nutritional quality of major food crops through conventional and genomics-assisted breeding. Front. Nutr. 2020, 7, 533453. [Google Scholar] [CrossRef]
- Sun, W.; Zhou, Q.; Yao, Y.; Qiu, X.; Xie, K.; Yu, S. Identification of genomic regions and the isoamylase gene for reduced grain chalkiness in rice. PLoS ONE 2015, 10, e0122013. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, C.L.; Wendel, J.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Wang, J.; Sun, W.; Qiu, X.; Yuan, Z.; Yu, S. Verifying the breeding value of a rare haplotype of Chalk7, GS3, and Chalk5 to improve grain appearance quality in rice. Plants 2022, 11, 1470. [Google Scholar] [CrossRef]
- Zhao, H.; Yao, W.; Ouyang, Y.; Yang, W.; Wang, G.; Lian, X.; Xing, Y.; Chen, L.; Xie, W. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015, 43, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Gao, D.; Xiong, Y.; Tang, X.; Xiao, X.; Wang, C.; Yu, S. Hairy leaf 6, an AP2/ERF transcription factor, interacts with OsWOX3B and regulates trichome formation in rice. Mol. Plant 2017, 10, 1417–1433. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, M.; Miyahara, K.; Iida, S.; Fukuoka, H.; Takano, T.; Sassa, H.; Nishimura, M.; Nishio, T. Low glutelin content1: A dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell. 2003, 15, 14551467. [Google Scholar] [CrossRef] [Green Version]
- Ning, T.T.; Xie, T.T.; Qiu, Q.C.; Yang, W.; Zhou, S.Q.; Zhou, L.M.; Zheng, C.Y.; Zhu, Y.G.; Yang, D.C. Oral administration of recombinant human granulocyte-macrophage colony stimulating factor expressed in rice endosperm can increase leukocytes in mice. Biotechnol. Lett. 2008, 30, 1679–1686. [Google Scholar] [CrossRef]
- Bao, J.; Xia, Y. Genetic control of the paste viscosity characteristics in indica rice (Oryza sativa L.). Theor. Appl. Genet. 1999, 98, 1120–1124. [Google Scholar] [CrossRef]
- Reddy, K.R.; Subramanian, R.; Ali, S.Z.; Bhattacharya, K.R. Viscoelastic properties of rice-flour pastes and their relationship to amylose content and rice quality. Cereal Chem. 1994, 71, 548–552. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Xia, Y.; Dong, Y.; Xie, T.; Sun, W.; Yu, S. Natural Variation of Fatty Acid Desaturase Gene Affects Linolenic Acid Content and Starch Pasting Viscosity in Rice Grains. Int. J. Mol. Sci. 2022, 23, 12055. https://doi.org/10.3390/ijms231912055
Zhang L, Xia Y, Dong Y, Xie T, Sun W, Yu S. Natural Variation of Fatty Acid Desaturase Gene Affects Linolenic Acid Content and Starch Pasting Viscosity in Rice Grains. International Journal of Molecular Sciences. 2022; 23(19):12055. https://doi.org/10.3390/ijms231912055
Chicago/Turabian StyleZhang, Liting, Yu Xia, Yage Dong, Tianyi Xie, Wenqiang Sun, and Sibin Yu. 2022. "Natural Variation of Fatty Acid Desaturase Gene Affects Linolenic Acid Content and Starch Pasting Viscosity in Rice Grains" International Journal of Molecular Sciences 23, no. 19: 12055. https://doi.org/10.3390/ijms231912055
APA StyleZhang, L., Xia, Y., Dong, Y., Xie, T., Sun, W., & Yu, S. (2022). Natural Variation of Fatty Acid Desaturase Gene Affects Linolenic Acid Content and Starch Pasting Viscosity in Rice Grains. International Journal of Molecular Sciences, 23(19), 12055. https://doi.org/10.3390/ijms231912055





