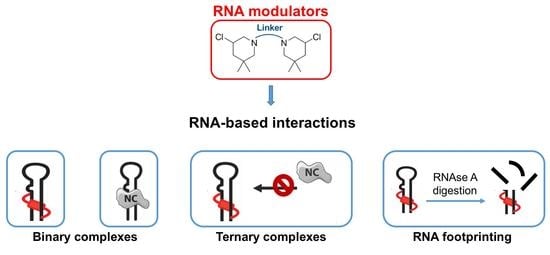
In Vitro Evaluation of Bis-3-Chloropiperidines as RNA Modulators Targeting TAR and TAR-Protein Interaction
Abstract
1. Introduction
2. Results and Discussion
2.1. Aliphatic Linker Improves B-CePs Reactivity towards TAR
2.2. B-CePs Adducts Inhibit NC Binding to TAR
2.3. Mapping the Aliphatic B-CePs Crosslinks on TAR
2.4. B-CePs Alkylation Affects NC-Mediated TAR Interactions with cTAR
3. Materials and Methods
3.1. Nucleic Acid Substrates and Protein
3.2. Chemical Reagents
3.3. Probing Reactions
3.4. Mass Spectrometric Analysis
3.5. Inhibition of NC Binding to TAR RNA
3.6. Enzymatic Digestion of TAR RNA
3.7. Gel Electrophoresis Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haniff, H.S.; Knerr, L.; Chen, J.L.; Disney, M.D.; Lightfoot, H.L. Target-Directed Approaches for Screening Small Molecules against RNA Targets. SLAS Discov. Adv. Life Sci. R&D 2020, 25, 869–894. [Google Scholar] [CrossRef]
- Meyer, S.M.; Williams, C.C.; Akahori, Y.; Tanaka, T.; Aikawa, H.; Tong, Y.; Childs-Disney, J.L.; Disney, M.D. Small molecule recognition of disease-relevant RNA structures. Chem. Soc. Rev. 2020, 49, 7167–7199. [Google Scholar] [CrossRef]
- Sheridan, C. First small-molecule drug targeting RNA gains momentum. Nat. Biotechnol. 2021, 39, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Costales, M.G.; Childs-Disney, J.L.; Haniff, H.; Disney, M.D. How We Think about Targeting RNA with Small Molecules. J. Med. Chem. 2020, 63, 8880–8900. [Google Scholar] [CrossRef]
- Disney, M.D. Targeting RNA with Small Molecules to Capture Opportunities at the Intersection of Chemistry, Biology, and Medicine. J. Am. Chem. Soc. 2019, 141, 6776–6790. [Google Scholar] [CrossRef]
- Donlic, A.; Hargrove, A.E. Targeting RNA in mammalian systems with small molecules. Wiley Interdiscip. Rev. RNA 2018, 9, e1477. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, A.E. Small molecule–RNA targeting: Starting with the fundamentals. Chem. Comm. 2020, 56, 14744–14756. [Google Scholar] [CrossRef]
- Sosic, A.; Gottlich, R.; Fabris, D.; Gatto, B. B-CePs as cross-linking probes for the investigation of RNA higher-order structure. Nucleic Acids Res. 2021, 49, 6660–6672. [Google Scholar] [CrossRef]
- Sosic, A.; Olivato, G.; Carraro, C.; Göttlich, R.; Fabris, D.; Gatto, B. Bis-3-Chloropiperidines Targeting TAR RNA as A Novel Strategy to Impair the HIV-1 Nucleocapsid Protein. Molecules 2021, 26, 1874. [Google Scholar] [CrossRef] [PubMed]
- Sosic, A.; Zuravka, I.; Schmitt, N.; Miola, A.; Göttlich, R.; Fabris, D.; Gatto, B. Direct and Topoisomerase II Mediated DNA Damage by Bis-3-chloropiperidines: The Importance of Being an Earnest G. ChemMedChem 2017, 12, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Zuravka, I.; Roesmann, R.; Sosic, A.; Göttlich, R.; Gatto, B. Bis-3-chloropiperidines containing bridging lysine linkers: Influence of side chain structure on DNA alkylating activity. Bioorg. Med. Chem. 2015, 23, 1241–1250. [Google Scholar] [CrossRef]
- Zuravka, I.; Roesmann, R.; Sosic, A.; Wende, W.; Pingoud, A.; Gatto, B.; Göttlich, R. Synthesis and DNA Cleavage Activity of Bis-3-chloropiperidines as Alkylating Agents. ChemMedChem 2014, 9, 2178–2185. [Google Scholar] [CrossRef]
- Zuravka, I.; Sosic, A.; Gatto, B.; Göttlich, R. Synthesis and evaluation of a bis-3-chloropiperidine derivative incorporating an anthraquinone pharmacophore. Bioorg. Med. Chem. Lett. 2015, 25, 4606–4609. [Google Scholar] [CrossRef]
- Belfetmi, A.; Zargarian, L.; Tisné, C.; Sleiman, D.; Morellet, N.; Lescop, E.; Maskri, O.; René, B.; Mély, Y.; Fossé, P.; et al. Insights into the mechanisms of RNA secondary structure destabilization by the HIV-1 nucleocapsid protein. RNA 2016, 22, 506–517. [Google Scholar] [CrossRef]
- Kanevsky, I.; Chaminade, F.; Ficheux, D.; Moumen, A.; Gorelick, R.; Negroni, M.; Darlix, J.-L.; Fossé, P. Specific Interactions Between HIV-1 Nucleocapsid Protein and the TAR Element. J. Mol. Biol. 2005, 348, 1059–1077. [Google Scholar] [CrossRef] [PubMed]
- Bernacchi, S.; Stoylov, S.; Piémont, E.; Ficheux, D.; Roques, B.P.; Darlix, J.L.; Mély, Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002, 317, 385–399. [Google Scholar] [CrossRef]
- Kanevsky, I.; Chaminade, F.; Chen, Y.; Godet, J.; René, B.; Darlix, J.-L.; Mély, Y.; Mauffret, O.; Fossé, P. Structural determinants of TAR RNA-DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2011, 39, 8148–8162. [Google Scholar] [CrossRef]
- Carraro, C.; Helbing, T.; Francke, A.; Zuravka, I.; Sosic, A.; De Franco, M.; Gandin, V.; Gatto, B.; Göttlich, D.R. Appended Aromatic Moieties in Flexible Bis-3-chloropiperidines Confer Tropism against Pancreatic Cancer Cells. ChemMedChem 2021, 16, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.B.; Hagan, N.A.; Fabris, D. Understanding the Isomerization of the HIV-1 Dimerization Initiation Domain by the Nucleocapsid Protein. J. Mol. Biol. 2007, 369, 812–828. [Google Scholar] [CrossRef]
- Turner, K.B.; Kohlway, A.S.; Hagan, N.A.; Fabris, D. Noncovalent probes for the investigation of structure and dynamics of protein-nucleic acid assemblies: The case of NC-mediated dimerization of genomic RNA in HIV-1. Biopolym. Orig. Res. Biomol. 2009, 91, 283–296. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hagan, N.A.; Fabris, D. Dissecting the Protein–RNA and RNA–RNA Interactions in the Nucleocapsid-mediated Dimerization and Isomerization of HIV-1 Stemloop 1. J. Mol. Biol. 2007, 365, 396–410. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sosic, A.; Saccone, I.; Carraro, C.; Kenderdine, T.; Gamba, E.; Caliendo, G.; Corvino, A.; Di Vaio, P.; Fiorino, F.; Magli, E.; et al. Non-Natural Linker Configuration in 2,6-Dipeptidyl-Anthraquinones Enhances the Inhibition of TAR RNA Binding/Annealing Activities by HIV-1 NC and Tat Proteins. Bioconjug. Chem. 2018, 29, 2195–2207. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.B.; Hagan, N.A.; Kohlway, A.S.; Fabris, D. Mapping noncovalent ligand binding to stemloop domains of the HIV-1 packaging signal by tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2006, 17, 1402–1411. [Google Scholar] [CrossRef] [PubMed]






| B-CePs | Linker | XL1 | XL2 |
|---|---|---|---|
| 1 | -(CH2)3- | ✓✓ | ✓✓ |
| 2 | -(CH2)5- | ✓✓ | ✓ |
| 3 | -(CH2)6- | ✓✓ | ✗ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sosic, A.; Olivato, G.; Carraro, C.; Göttlich, R.; Fabris, D.; Gatto, B. In Vitro Evaluation of Bis-3-Chloropiperidines as RNA Modulators Targeting TAR and TAR-Protein Interaction. Int. J. Mol. Sci. 2022, 23, 582. https://doi.org/10.3390/ijms23020582
Sosic A, Olivato G, Carraro C, Göttlich R, Fabris D, Gatto B. In Vitro Evaluation of Bis-3-Chloropiperidines as RNA Modulators Targeting TAR and TAR-Protein Interaction. International Journal of Molecular Sciences. 2022; 23(2):582. https://doi.org/10.3390/ijms23020582
Chicago/Turabian StyleSosic, Alice, Giulia Olivato, Caterina Carraro, Richard Göttlich, Dan Fabris, and Barbara Gatto. 2022. "In Vitro Evaluation of Bis-3-Chloropiperidines as RNA Modulators Targeting TAR and TAR-Protein Interaction" International Journal of Molecular Sciences 23, no. 2: 582. https://doi.org/10.3390/ijms23020582
APA StyleSosic, A., Olivato, G., Carraro, C., Göttlich, R., Fabris, D., & Gatto, B. (2022). In Vitro Evaluation of Bis-3-Chloropiperidines as RNA Modulators Targeting TAR and TAR-Protein Interaction. International Journal of Molecular Sciences, 23(2), 582. https://doi.org/10.3390/ijms23020582