Dynamic Contrast-Enhanced Magnetic Resonance Imaging for the Prediction of Monoclonal Antibody Tumor Disposition
Abstract
:1. Introduction
2. Results
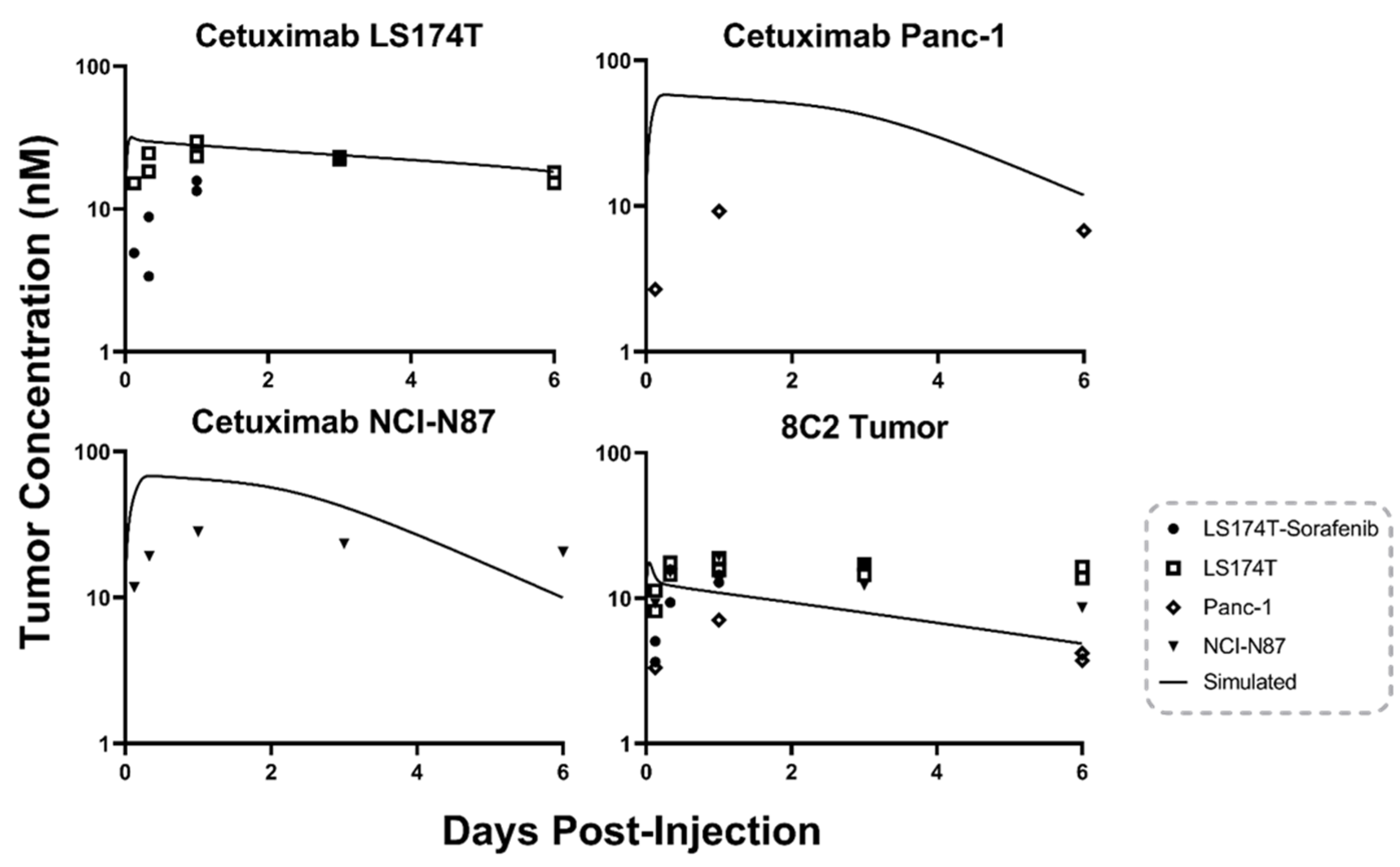
2.1. Antibody Tumor Pharmacokinetics and a Priori PBPK Model Predictions
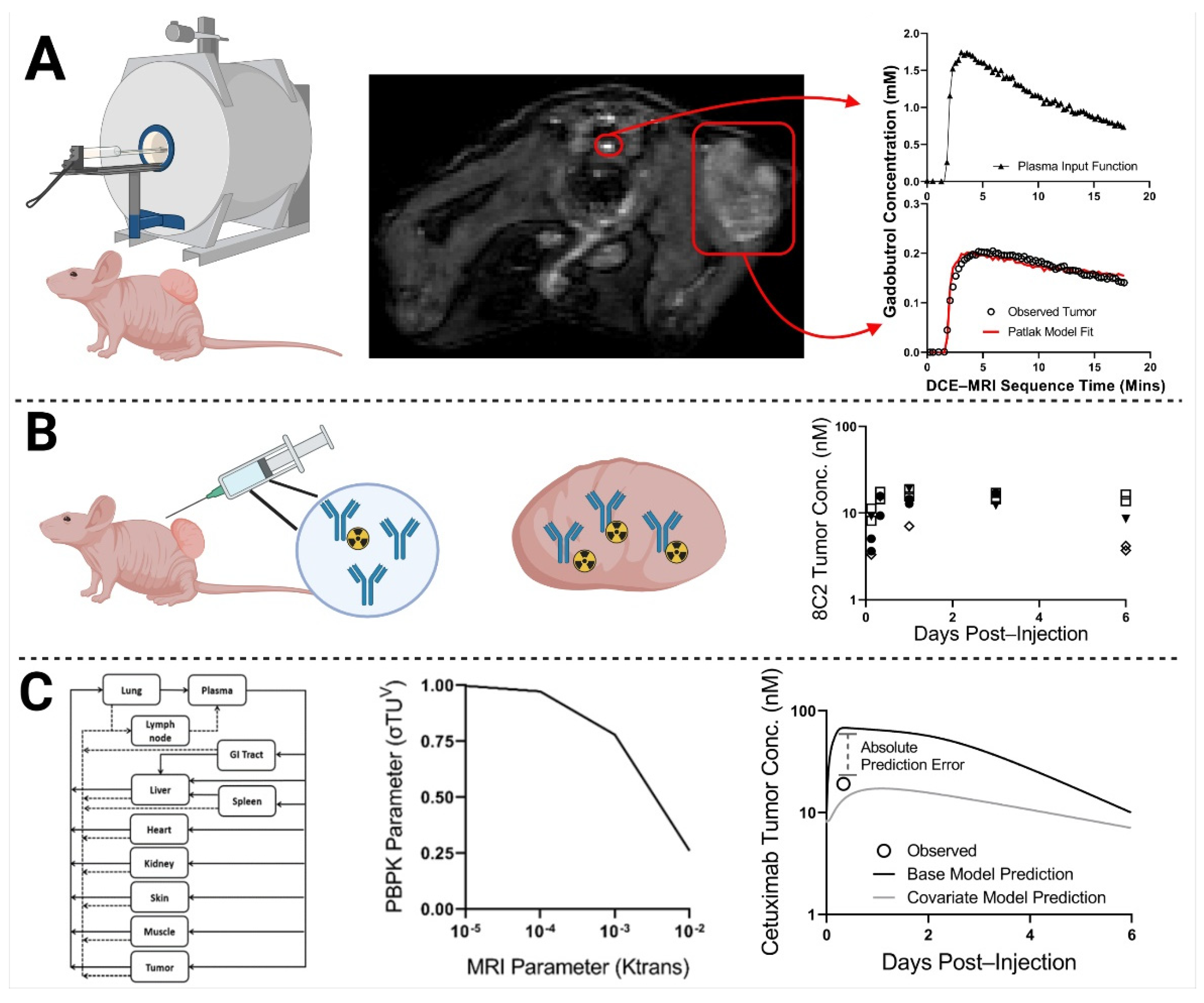
2.2. DCE-MRI Fitting
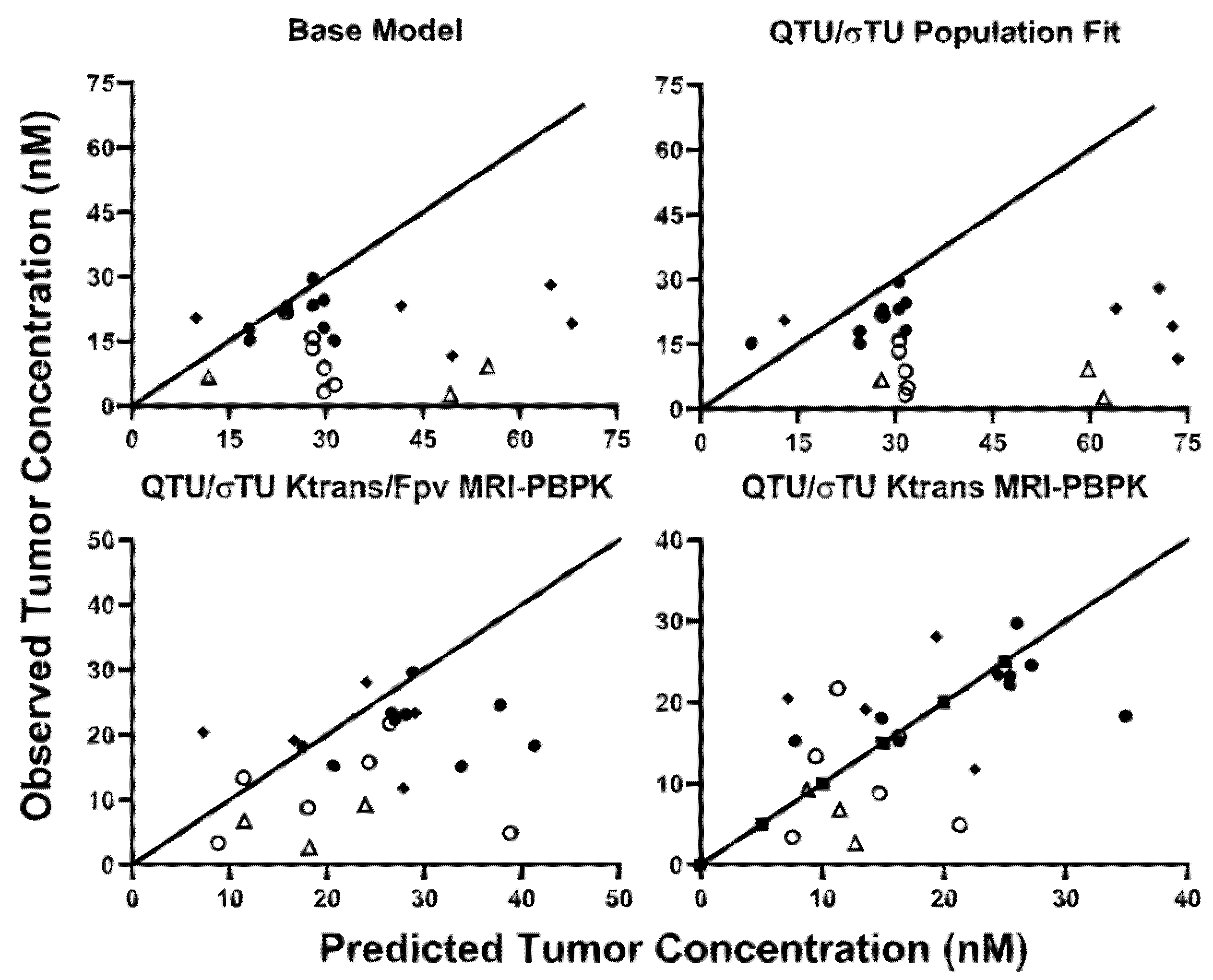
2.3. MRI-PBPK Covariate Modeling
3. Discussion
4. Materials and Methods
4.1. Antibody
4.2. Anti-Angiogenesis Agent
4.3. Xenograft Cell Lines
4.4. Animals
4.5. Establishment of Xenografts
4.6. MR Imaging
4.7. Plasma and Tumor Pharmacokinetics
4.8. Pharmacokinetic Modeling of DCE-MRI Images
4.9. Base Model PBPK Predictions
4.10. MRI-PBPK Covariate Modeling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, L.; Ehrlich, P.J.; Gibson, R.; Pickett, C.; Beckman, R.A. How can we improve antibody-based cancer therapy? MAbs 2009, 1, 67–70. [Google Scholar] [CrossRef] [Green Version]
- Kaplon, H.; Reichert, J.M. Antibodies to watch in 2021. MAbs 2021, 13, 1860476. [Google Scholar] [CrossRef]
- Baxter, L.T.; Zhu, H.; Mackensen, D.G.; Jain, R.K. Physiologically based pharmacokinetic model for specific and nonspecific monoclonal antibodies and fragments in normal tissues and human tumor xenografts in nude mice. Cancer Res. 1994, 54, 1517–1528. [Google Scholar]
- Glassman, P.M.; Balthasar, J.P. Physiologically-based pharmacokinetic modeling to predict the clinical pharmacokinetics of monoclonal antibodies. J. Pharm. Pharm. 2016, 43, 427–446. [Google Scholar] [CrossRef]
- Schmidt, M.M.; Wittrup, K.D. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol. Cancer Ther. 2009, 8, 2861–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Li, Y.; Chang, H.P.; Yu, X.; Shah, D.K. Two-pore physiologically based pharmacokinetic model validation using whole-body biodistribution of trastuzumab and different-size fragments in mice. J. Pharm. Pharm. 2021, 48, 743–762. [Google Scholar] [CrossRef]
- Abuqayyas, L.; Balthasar, J.P. Application of PBPK modeling to predict monoclonal antibody disposition in plasma and tissues in mouse models of human colorectal cancer. J. Pharm. Pharm. 2012, 39, 683–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, R.K. Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res. 1990, 50, 814s–819s. [Google Scholar]
- Christiansen, J.; Rajasekaran, A.K. Biological impediments to monoclonal antibody-based cancer immunotherapy. Mol. Cancer Ther. 2004, 3, 1493–1501. [Google Scholar] [PubMed]
- Thomas, V.A.; Balthasar, J.P. Understanding Inter-Individual Variability in Monoclonal Antibody Disposition. Antibodies 2019, 8, 56. [Google Scholar] [CrossRef] [Green Version]
- Carlier, M.; Noe, M.; Roberts, J.A.; Stove, V.; Verstraete, A.G.; Lipman, J.; De Waele, J.J. Population pharmacokinetics and dosing simulations of cefuroxime in critically ill patients: Non-standard dosing approaches are required to achieve therapeutic exposures. J. Antimicrob. Chemother. 2014, 69, 2797–2803. [Google Scholar] [CrossRef] [Green Version]
- Darwich, A.S.; Ogungbenro, K.; Hatley, O.J.; Rostami-Hodjegan, A. Role of pharmacokinetic modeling and simulation in precision dosing of anticancer drugs. Transl. Cancer Res. 2017, 6, S1512–S1529. [Google Scholar] [CrossRef] [Green Version]
- Evelina, C.; Guidi, M.; Khoudour, N.; Pascaline, B.-R.; Fabre, E.; Tlemsani, C.; Arrondeau, J.; Francois, G.; Vidal, M.; Schneider, M.P.; et al. Population Pharmacokinetics of Erlotinib in Patients With Non-small Cell Lung Cancer: Its Application for Individualized Dosing Regimens in Older Patients. Clin. Ther. 2020, 42, 1302–1316. [Google Scholar] [CrossRef]
- Polli, J.R.; Engler, F.A.; Balthasar, J.P. Physiologically Based Modeling of the Pharmacokinetics of “Catch-and-Release” Anti-Carcinoembryonic Antigen Monoclonal Antibodies in Colorectal Cancer Xenograft Mouse Models. J. Pharm. Sci. 2019, 108, 674–691. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, Y.; Chang, H.-P.; Chang, H.-Y.; Guo, L.; Shah, D.K. Effect of Size on Solid Tumor Disposition of Protein Therapeutics. Drug Metab. Dispos. 2019, 47, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Barnes, S.L.; Whisenant, J.G.; Xia, L.; Yankeelov, T.E. Techniques and applications of dynamic contrast enhanced magnetic resonance imaging in cancer. In Proceedings of the International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC’14), Chicago, IL, USA, 26–30 August 2014; pp. 4264–4267. [Google Scholar] [CrossRef] [Green Version]
- An, Y.Y.; Kim, S.H.; Kang, B.J.; Lee, A.W. Treatment Response Evaluation of Breast Cancer after Neoadjuvant Chemotherapy and Usefulness of the Imaging Parameters of MRI and PET/CT. J. Korean Med. Sci. 2015, 30, 808–815. [Google Scholar] [CrossRef]
- Tofts, P.S.; Brix, G.; Buckley, D.L.; Evelhoch, J.L.; Henderson, E.; Knopp, M.V.; Larsson, H.B.; Lee, T.Y.; Mayr, N.A.; Parker, G.J.; et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: Standardized quantities and symbols. J. Magn. Reson. Imaging 1999, 10, 223–232. [Google Scholar] [CrossRef]
- Cuenod, C.A.; Balvay, D. Perfusion and vascular permeability: Basic concepts and measurement in DCE-CT and DCE-MRI. Diagn. Interv. Imaging 2013, 94, 1187–1204. [Google Scholar] [CrossRef] [Green Version]
- Wegner, C.S.; Gaustad, J.V.; Andersen, L.M.; Simonsen, T.G.; Rofstad, E.K. Diffusion-weighted and dynamic contrast-enhanced MRI of pancreatic adenocarcinoma xenografts: Associations with tumor differentiation and collagen content. J. Transl. Med. 2016, 14, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eikenes, L.; Bruland, O.S.; Brekken, C.; Davies Cde, L. Collagenase increases the transcapillary pressure gradient and improves the uptake and distribution of monoclonal antibodies in human osteosarcoma xenografts. Cancer Res. 2004, 64, 4768–4773. [Google Scholar] [CrossRef] [Green Version]
- Hompland, T.; Gulliksrud, K.; Ellingsen, C.; Rofstad, E.K. Assessment of the interstitial fluid pressure of tumors by dynamic contrast-enhanced magnetic resonance imaging with contrast agents of different molecular weights. Acta Oncol. 2013, 52, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Gaustad, J.V.; Simonsen, T.G.; Smistad, R.; Wegner, C.S.; Andersen, L.M.; Rofstad, E.K. Early effects of low dose bevacizumab treatment assessed by magnetic resonance imaging. BMC Cancer 2015, 15, 900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kickingereder, P.; Wiestler, B.; Graf, M.; Heiland, S.; Schlemmer, H.P.; Wick, W.; Wick, A.; Bendszus, M.; Radbruch, A. Evaluation of dynamic contrast-enhanced MRI derived microvascular permeability in recurrent glioblastoma treated with bevacizumab. J. Neurooncol. 2015, 121, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Joo, B.; Park, M.S.; Shin, S.J.; Ahn, J.B.; Kim, M.J. Dynamic Contrast-Enhanced Magnetic Resonance Imaging as a Surrogate Biomarker for Bevacizumab in Colorectal Cancer Liver Metastasis: A Single-Arm, Exploratory Trial. Cancer Res. Treat. 2016, 48, 1210–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urva, S.R.; Yang, V.C.; Balthasar, J.P. Physiologically based pharmacokinetic model for T84.66: A monoclonal anti-CEA antibody. J. Pharm. Sci. 2010, 99, 1582–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, V.A.; Balthasar, J.P. Sorafenib Decreases Tumor Exposure to an Anti-carcinoembryonic Antigen Monoclonal Antibody in a Mouse Model of Colorectal Cancer. AAPS J. 2016, 18, 923–932. [Google Scholar] [CrossRef]
- Shuhendler, A.J.; Prasad, P.; Cai, P.; Hui, K.K.; Henderson, J.T.; Rauth, A.M.; Wu, X.Y. Matrigel alters the pathophysiology of orthotopic human breast adenocarcinoma xenografts with implications for nanomedicine evaluation. Nanomedicine 2013, 9, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, E.Q.; Balthasar, J.P. Monoclonal antibody pharmacokinetics and pharmacodynamics. Clin. Pharmacol. Ther. 2008, 84, 548–558. [Google Scholar] [CrossRef]
- Bartelink, I.H.; Jones, E.F.; Shahidi-Latham, S.K.; Lee, P.R.E.; Zheng, Y.; Vicini, P.; van ′t Veer, L.; Wolf, D.; Iagaru, A.; Kroetz, D.L.; et al. Tumor Drug Penetration Measurements Could Be the Neglected Piece of the Personalized Cancer Treatment Puzzle. Clin. Pharmacol. Ther. 2018. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2018, 20, 273–286. [Google Scholar] [CrossRef]
- Hwang, T.J.; Carpenter, D.; Lauffenburger, J.C.; Wang, B.; Franklin, J.M.; Kesselheim, A.S. Failure of Investigational Drugs in Late-Stage Clinical Development and Publication of Trial Results. JAMA Intern. Med. 2016, 176, 1826–1833. [Google Scholar] [CrossRef]
- Hudis, C.A. Trastuzumab—Mechanism of Action and Use in Clinical Practice. N. Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Lievre, A.; Bachet, J.B.; Le Corre, D.; Boige, V.; Landi, B.; Emile, J.F.; Cote, J.F.; Tomasic, G.; Penna, C.; Ducreux, M.; et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006, 66, 3992–3995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karapetis, C.S.; Khambata-Ford, S.; Jonker, D.J.; O’Callaghan, C.J.; Tu, D.; Tebbutt, N.C.; Simes, R.J.; Chalchal, H.; Shapiro, J.D.; Robitaille, S.; et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008, 359, 1757–1765. [Google Scholar] [CrossRef] [Green Version]
- Shin, T.H.; Sung, E.S.; Kim, Y.J.; Kim, K.S.; Kim, S.H.; Kim, S.K.; Lee, Y.D.; Kim, Y.S. Enhancement of the tumor penetration of monoclonal antibody by fusion of a neuropilin-targeting peptide improves the antitumor efficacy. Mol. Cancer Ther. 2014, 13, 651–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, I.; van Rensburg, R.; Strauss, R.; Li, Z.; Wang, H.; Persson, J.; Yumul, R.; Feng, Q.; Song, H.; Bartek, J.; et al. Epithelial junction opener JO-1 improves monoclonal antibody therapy of cancer. Cancer Res. 2011, 71, 7080–7090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viala, M.; Vinches, M.; Alexandre, M.; Mollevi, C.; Durigova, A.; Hayaoui, N.; Homicsko, K.; Cuenant, A.; Gongora, C.; Gianni, L.; et al. Strategies for clinical development of monoclonal antibodies beyond first-in-human trials: Tested doses and rationale for dose selection. Br. J. Cancer 2018, 118, 679–697. [Google Scholar] [CrossRef] [Green Version]
- Thurber, G.M.; Weissleder, R. Quantitating antibody uptake in vivo: Conditional dependence on antigen expression levels. Mol. Imaging Biol. 2011, 13, 623–632. [Google Scholar] [CrossRef] [Green Version]
- Bordeau, B.M.; Balthasar, J.P. Strategies to enhance monoclonal antibody uptake and distribution in solid tumors. Cancer Biol. Med. 2021, 18, 649–664. [Google Scholar] [CrossRef]
- Bordeau, B.M.; Yang, Y.; Balthasar, J.P. Transient Competitive Inhibition Bypasses the Binding Site Barrier to Improve Tumor Penetration of Trastuzumab and Enhance T-DM1 Efficacy. Cancer Res. 2021, 81, 4145–4154. [Google Scholar] [CrossRef]
- Cilliers, C.; Menezes, B.; Nessler, I.; Linderman, J.; Thurber, G.M. Improved Tumor Penetration and Single-Cell Targeting of Antibody-Drug Conjugates Increases Anticancer Efficacy and Host Survival. Cancer Res. 2018, 78, 758–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singha, N.C.; Nekoroski, T.; Zhao, C.; Symons, R.; Jiang, P.; Frost, G.I.; Huang, Z.; Shepard, H.M. Tumor-Associated Hyaluronan Limits Efficacy of Monoclonal Antibody Therapy. Mol. Cancer Ther. 2015, 14, 523–532. [Google Scholar] [CrossRef] [Green Version]
- Khawli, L.A.; Miller, G.K.; Epstein, A.L. Effect of seven new vasoactive immunoconjugates on the enhancement of monoclonal antibody uptake in tumors. Cancer 1994, 73, 824–831. [Google Scholar] [CrossRef]
- Lu, G.; Fakurnejad, S.; Martin, B.A.; van den Berg, N.S.; van Keulen, S.; Nishio, N.; Zhu, A.J.; Chirita, S.U.; Zhou, Q.; Gao, R.W.; et al. Predicting Therapeutic Antibody Delivery into Human Head and Neck Cancers. Clin. Cancer Res. 2020, 26, 2582–2594. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Huang, J.; Liang, J.; Ma, M.; Ye, K.; Shi, C.; Luo, L. The Diagnostic Performance of DCE-MRI in Evaluating the Pathological Response to Neoadjuvant Chemotherapy in Breast Cancer: A Meta-Analysis. Front. Oncol. 2020, 10, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Wang, K.; Sun, X.; Wang, K.; Sun, Y.; Zhang, G.; Shen, B. Parameters of dynamic contrast-enhanced MRI as imaging markers for angiogenesis and proliferation in human breast cancer. Med. Sci. Monit. 2015, 21, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Rahbar, H.; Hippe, D.S.; Rendi, M.H.; Parker, E.U.; Shekar, N.; Hirano, M.; Cheung, K.J.; Partridge, S.C. Dynamic contrast-enhanced breast MRI features correlate with invasive breast cancer angiogenesis. NPJ Breast. Cancer 2021, 7, 42. [Google Scholar] [CrossRef]
- Cilliers, C.; Nessler, I.; Christodolu, N.; Thurber, G.M. Tracking Antibody Distribution with Near-Infrared Fluorescent Dyes: Impact of Dye Structure and Degree of Labeling on Plasma Clearance. Mol. Pharm. 2017, 14, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Nishio, N.; van den Berg, N.S.; Martin, B.A.; Fakurnejad, S.; van Keulen, S.; Colevas, A.D.; Thurber, G.M.; Rosenthal, E.L. Co-administered antibody improves penetration of antibody–dye conjugate into human cancers with implications for antibody–drug conjugates. Nat. Commun. 2020, 11, 5667. [Google Scholar] [CrossRef]
- Engler, F.A.; Polli, J.R.; Li, T.; An, B.; Otteneder, M.; Qu, J.; Balthasar, J.P. “Catch-and-Release” Anti-Carcinoembryonic Antigen Monoclonal Antibody Leads to Greater Plasma and Tumor Exposure in a Mouse Model of Colorectal Cancer. J. Pharmacol. Exp. Ther. 2018, 366, 205–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebhart, G.; Lamberts, L.E.; Wimana, Z.; Garcia, C.; Emonts, P.; Ameye, L.; Stroobants, S.; Huizing, M.; Aftimos, P.; Tol, J.; et al. Molecular imaging as a tool to investigate heterogeneity of advanced HER2-positive breast cancer and to predict patient outcome under trastuzumab emtansine (T-DM1): The ZEPHIR trial. Ann. Oncol. 2016, 27, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Menke-van der Houven van Oordt, C.W.; Gootjes, E.C.; Huisman, M.C.; Vugts, D.J.; Roth, C.; Luik, A.M.; Mulder, E.R.; Schuit, R.C.; Boellaard, R.; Hoekstra, O.S.; et al. 89Zr-cetuximab PET imaging in patients with advanced colorectal cancer. Oncotarget 2015, 6, 30384–30393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribba, B.; Boetsch, C.; Nayak, T.; Grimm, H.P.; Charo, J.; Evers, S.; Klein, C.; Tessier, J.; Charoin, J.E.; Phipps, A.; et al. Prediction of the Optimal Dosing Regimen Using a Mathematical Model of Tumor Uptake for Immunocytokine-Based Cancer Immunotherapy. Clin. Cancer Res. 2018, 24, 3325–3333. [Google Scholar] [CrossRef] [Green Version]
- Sampath, L.; Kwon, S.; Ke, S.; Wang, W.; Schiff, R.; Mawad, M.E.; Sevick-Muraca, E.M. Dual-labeled trastuzumab-based imaging agent for the detection of human epidermal growth factor receptor 2 overexpression in breast cancer. J. Nucl. Med. 2007, 48, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Balthasar, J.P. Physiologically-based pharmacokinetic (PBPK) model to predict IgG tissue kinetics in wild-type and FcRn-knockout mice. J. Pharm. Pharm. 2007, 34, 687–709. [Google Scholar] [CrossRef] [PubMed]
- Abuqayyas, L.; Balthasar, J.P. Pharmacokinetic mAb-mAb interaction: Anti-VEGF mAb decreases the distribution of anti-CEA mAb into colorectal tumor xenografts. AAPS J. 2012, 14, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Sourbron, S.P.; Buckley, D.L. On the scope and interpretation of the Tofts models for DCE-MRI. Magn. Reson. Med. 2011, 66, 735–745. [Google Scholar] [CrossRef]
- Chen, H.; Li, F.; Zhao, X.; Yuan, C.; Rutt, B.; Kerwin, W.S. Extended graphical model for analysis of dynamic contrast-enhanced MRI. Magn. Reson. Med. 2011, 66, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Patlak, C.S.; Blasberg, R.G.; Fenstermacher, J.D. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J. Cereb. Blood. Flow Metab. 1983, 3, 1–7. [Google Scholar] [CrossRef]
- Barnes, S.R.; Ng, T.S.; Santa-Maria, N.; Montagne, A.; Zlokovic, B.V.; Jacobs, R.E. ROCKETSHIP: A flexible and modular software tool for the planning, processing and analysis of dynamic MRI studies. BMC Med. Imaging 2015, 15, 19. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felder, S.; LaVin, J.; Ullrich, A.; Schlessinger, J. Kinetics of binding, endocytosis, and recycling of EGF receptor mutants. J. Cell Biol. 1992, 117, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Burke, P.M.; Wiley, H.S. Human mammary epithelial cells rapidly exchange empty EGFR between surface and intracellular pools. J. Cell Physiol. 1999, 180, 448–460. [Google Scholar] [CrossRef]
- McDonagh, C.F.; Huhalov, A.; Harms, B.D.; Adams, S.; Paragas, V.; Oyama, S.; Zhang, B.; Luus, L.; Overland, R.; Nguyen, S.; et al. Antitumor activity of a novel bispecific antibody that targets the ErbB2/ErbB3 oncogenic unit and inhibits heregulin-induced activation of ErbB3. Mol. Cancer Ther. 2012, 11, 582–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusnak, D.W.; Alligood, K.J.; Mullin, R.J.; Spehar, G.M.; Arenas-Elliott, C.; Martin, A.M.; Degenhardt, Y.; Rudolph, S.K.; Haws, T.F., Jr.; Hudson-Curtis, B.L.; et al. Assessment of epidermal growth factor receptor (EGFR, ErbB1) and HER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell Prolif. 2007, 40, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Modjtahedi, H.; Khelwatty, S.A.; Kirk, R.S.; Seddon, A.M.; Essapen, S.; Del Vecchio, C.A.; Wong, A.J.; Eccles, S. Immunohistochemical discrimination of wild-type EGFR from EGFRvIII in fixed tumour specimens using anti-EGFR mAbs ICR9 and ICR10. Br. J. Cancer 2012, 106, 883–888. [Google Scholar] [CrossRef] [Green Version]
- Korc, M.; Meltzer, P.; Trent, J. Enhanced expression of epidermal growth factor receptor correlates with alterations of chromosome 7 in human pancreatic cancer. Proc. Natl. Acad. Sci. USA 1986, 83, 5141–5144. [Google Scholar] [CrossRef] [Green Version]
- D’Argenio, D.Z. ADAPT 5 User’s Guide. In Pharmacokinetic/Pharmacodynamic Systems Analysis Software; Biomedical Simulations Resource: Los Angeles, CA, USA, 2009. [Google Scholar]




| Antibody | Tumor Model | Tumor/Plasma Ratio | Cetuximab/8C2 |
|---|---|---|---|
| 8C2 | LS174T | 0.26 | 1.58 |
| Cetuximab | LS174T | 0.41 | - |
| 8C2 | LS174T/Sorafenib | 0.20 | 1.35 |
| Cetuximab | LS174T/Sorafenib | 0.27 | - |
| 8C2 | NCI-N87 | 0.19 | 2.53 |
| Cetuximab | NCI-N87 | 0.48 | - |
| 8C2 | Panc-1 | 0.08 | 1.50 |
| Cetuximab | Panc-1 | 0.12 | - |
| Parameter | Value | Units | Definition |
|---|---|---|---|
| QTU | 1 × 10−4 | L/min | Tumor blood flow [7] |
| LTU | 4 × 10−6 | L/min | Tumor lymph flow [7] |
| ClupTU | 8.18 × 10−9 | L/min | Initial tumor uptake clearance [14] |
| σTUV | 0.734 | - | Tumor vascular reflection coefficient [7] |
| σTUL | 0.2 | - | Lymph reflection coefficient [7] |
| ClTU | 8.96 × 10−9 | L/min | Initial clearance from endothelial space [14] |
| KDFcRn | 7.5 × 10−7 | M–1 | FcRn-mAb KD [7] |
| CFcRn | 1.64 × 10−5 | M | Tumor FcRn concentration [14] |
| FR | 0.715 | - | Fraction of FcRn bound antibody recycled [7] |
| kgrowth | 8.08 × 10−5 | min–1 | Tumor growth rate [7] |
| VITU | 1.38 × 10−4 | L | Initial Interstitial Volume [14] |
| VETU | 1.25 × 10−6 | L | Initial Endothelial Volume [14] |
| VVTU | 1.75 × 10−5 | L | Initial Vasculature Volume [14] |
| KDEGFR | 1.5 × 10−10 | M–1 | Cetuximab-EGFR KD [4] |
| Kint | 1.38 × 10−3 | min–1 | EGFR Internalization Rate [65,66] |
| ClTMD | Kint × VITU | L/min | Cetuximab Bound EGFR Clearance |
| CEGFR (NCI-N87) | 1.14 × 10−7 | M | EGFR Tumor Concentration [67,68,69] |
| CEGFR (Panc-1) | 9.24 × 10−8 | M | EGFR Tumor Concentration [70] |
| CEGFR (LS174T) | 3.53 × 10−8 | M | EGFR Tumor Concentration [67,68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordeau, B.M.; Polli, J.R.; Schweser, F.; Grimm, H.P.; Richter, W.F.; Balthasar, J.P. Dynamic Contrast-Enhanced Magnetic Resonance Imaging for the Prediction of Monoclonal Antibody Tumor Disposition. Int. J. Mol. Sci. 2022, 23, 679. https://doi.org/10.3390/ijms23020679
Bordeau BM, Polli JR, Schweser F, Grimm HP, Richter WF, Balthasar JP. Dynamic Contrast-Enhanced Magnetic Resonance Imaging for the Prediction of Monoclonal Antibody Tumor Disposition. International Journal of Molecular Sciences. 2022; 23(2):679. https://doi.org/10.3390/ijms23020679
Chicago/Turabian StyleBordeau, Brandon M., Joseph Ryan Polli, Ferdinand Schweser, Hans Peter Grimm, Wolfgang F. Richter, and Joseph P. Balthasar. 2022. "Dynamic Contrast-Enhanced Magnetic Resonance Imaging for the Prediction of Monoclonal Antibody Tumor Disposition" International Journal of Molecular Sciences 23, no. 2: 679. https://doi.org/10.3390/ijms23020679






