A Structural Perspective on the RNA Editing of Plant Respiratory Complexes
Abstract
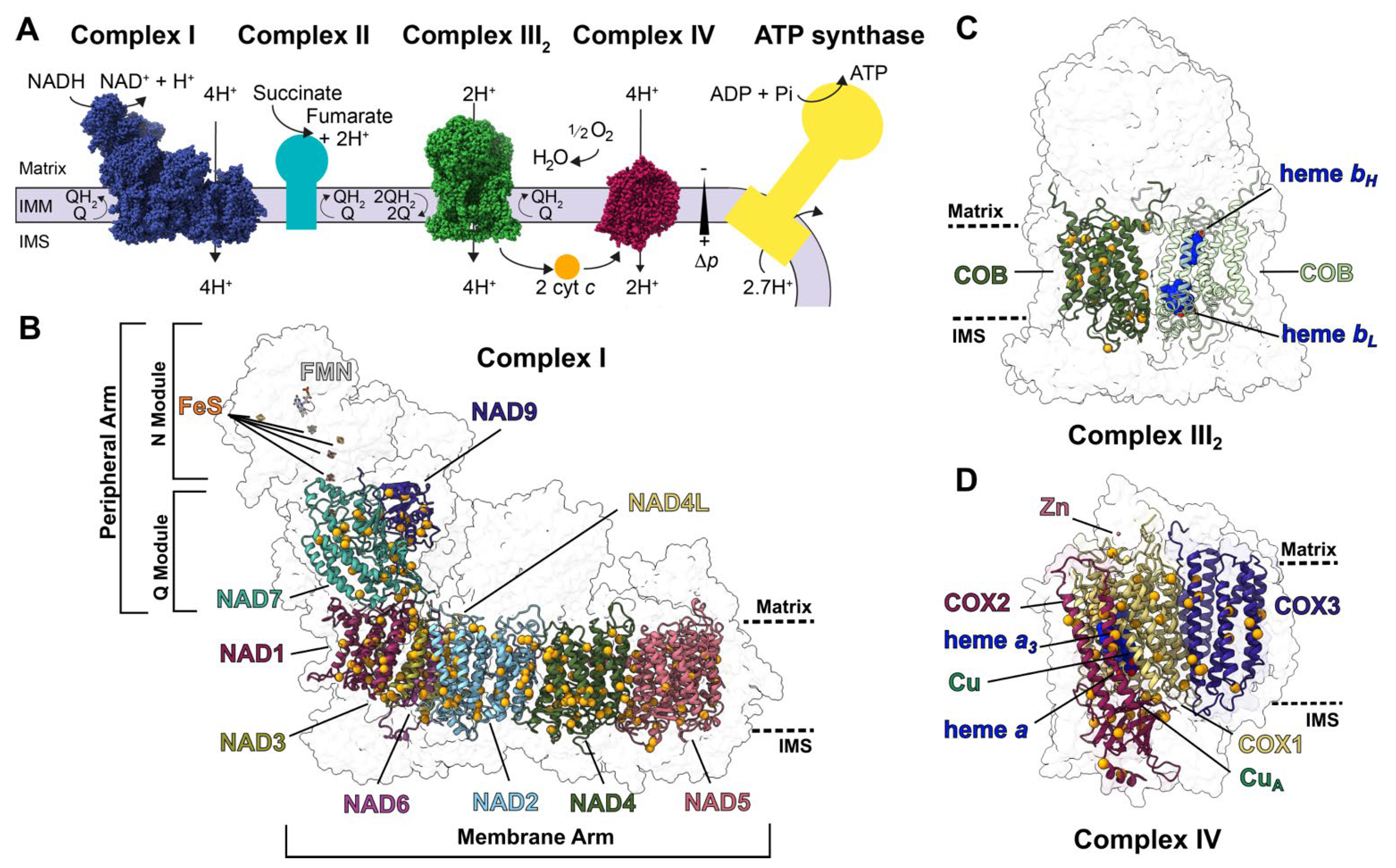
:1. Introduction
2. Approach
2.1. Structural and Sequence Characterization of Editing Sites
2.2. Structural and Functional Consequences of PPR Mutants
2.2.1. Mutations in Single-Site PPR Proteins
2.2.2. Mutations in Multi-Site PPR Proteins
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Covello, P.S.; Gray, M.W. RNA editing in plant mitochondria. Nature 1989, 341, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.; LaMattina, L.; Bonnard, G.; Weil, J.-H.; Grienenberger, J.-M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 1989, 341, 660–662. [Google Scholar] [CrossRef]
- Hiesel, R.; Wissinger, B.; Schuster, W.; Brennicke, A. RNA editing in plant mitochondria. Science 1989, 246, 1632–1634. [Google Scholar] [CrossRef] [PubMed]
- Hoch, B.; Maier, R.M.; Appel, K.; Igloi, G.L.; Kössel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 1991, 353, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Härtel, B.; Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef]
- Edera, A.A.; Gandini, C.L.; Sanchez-Puerta, M.V. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 2018, 97, 215–231. [Google Scholar] [CrossRef]
- Barkan, A.; Small, A.I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Giege, P.; Brennicke, A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 1999, 96, 15324–15329. [Google Scholar] [CrossRef] [Green Version]
- Maldonado, M.; Padavannil, A.; Zhou, L.; Guo, F.; A Letts, J. Atomic structure of a mitochondrial complex I intermediate from vascular plants. eLife 2020, 9, e56664. [Google Scholar] [CrossRef] [PubMed]
- Soufari, H.; Parrot, C.; Kuhn, L.; Waltz, F.; Hashem, Y. Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM. Nat. Commun. 2020, 11, 5195. [Google Scholar] [CrossRef]
- Maldonado, M.; Guo, F.; Letts, J.A. Atomic structures of respiratory complex III2, complex IV, and supercomplex III2-IV from vascular plants. eLife 2021, 10, e62047. [Google Scholar] [CrossRef]
- Klusch, N.; Senkler, J.; Yildiz, Ö.; Kühlbrandt, W.; Braun, H.-P. A ferredoxin bridge connects the two arms of plant mitochondrial complex I. Plant Cell 2021, 33, 2072–2091. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, D.G.; Ferguson, S.J. Bioenergetics 4; Academic Press: London, UK, 2013. [Google Scholar]
- O’Leary, B.M.; Asao, S.; Millar, A.H.; Atkin, O.K. Core principles which explain variation in respiration across biological scales. New Phytol. 2019, 222, 670–686. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, O.; De Clercq, I.; Van Breusegem, F.; Whelan, J. Interaction between hormonal and mitochondrial signalling during growth, development and in plant defence responses. Plant Cell Environ. 2016, 39, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Van Aken, O.; Schwarzländer, M.; Belt, K.; Millar, A.H. The Roles of Mitochondrial Reactive Oxygen Species in Cellular Signaling and Stress Response in Plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sazanov, L.A. A giant molecular proton pump: Structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 2015, 16, 375–388. [Google Scholar] [CrossRef]
- Xia, D.; Esser, L.; Tang, W.-K.; Zhou, F.; Zhou, Y.; Yu, L.; Yu, C.-A. Structural analysis of cytochrome bc1 complexes: Implications to the mechanism of function. Biochim. Biophys. Acta 2013, 1827, 1278–1294. [Google Scholar] [CrossRef] [Green Version]
- Rich, P.R. Mitochondrial cytochrome c oxidase: Catalysis, coupling and controversies. Biochem. Soc. Trans. 2017, 45, 813–829. [Google Scholar] [CrossRef]
- Toda, T.; Fujii, S.; Noguchi, K.; Kazama, T.; Toriyama, K. Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. Plant J. 2012, 72, 450–460. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Hou, M.; Sun, F.; Shen, Y.; Xiu, Z.; Wang, X.; Chen, Z.; Sun, S.S.M.; Small, I.; et al. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 2014, 79, 797–809. [Google Scholar] [CrossRef]
- Uchida, M.; Ohtani, S.; Ichinose, M.; Sugita, C.; Sugita, M. The PPR-DYW proteins are required for RNA editing of rps14, cox1 and nad5 transcripts in Physcomitrella patens mitochondria. FEBS Lett. 2011, 585, 2367–2371. [Google Scholar] [CrossRef] [Green Version]
- Fan, K.; Peng, Y.; Ren, Z.; Li, D.; Zhen, S.; Hey, S.; Cui, Y.; Fu, J.; Gu, R.; Wang, J.; et al. Maize Defective Kernel605 Encodes a Canonical DYW-Type PPR Protein that Edits a Conserved Site of nad1 and Is Essential for Seed Nutritional Quality. Plant Cell Physiol. 2020, 61, 1954–1966. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M. MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis. Plant Physiol. 2010, 152, 939–947. [Google Scholar] [CrossRef] [Green Version]
- Von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Fiedorczuk, K.; Letts, J.A.; Degliesposti, G.; Kaszuba, K.; Skehel, M.; Sazanov, L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature 2016, 538, 406–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stroud, D.; Surgenor, E.; Formosa, L.; Reljic, B.; Frazier, A.; Dibley, M.; Osellame, L.D.; Stait, T.; Beilharz, T.; Thorburn, D.; et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 2016, 538, 123–126. [Google Scholar] [CrossRef] [Green Version]
- Ligas, J.; Pineau, E.; Bock, R.; Huynen, M.A.; Meyer, E.H. The assembly pathway of complex I in Arabidopsis thaliana. Plant J. 2019, 97, 447–459. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, X.-Y.; Yang, Y.-Z.; Huang, J.; Sun, F.; Lin, J.; Gu, Z.-Q.; Sayyed, A.; Xu, C.; Tan, B.-C. Empty Pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PLoS Genet. 2019, 15, e1008305. [Google Scholar] [CrossRef] [Green Version]
- Coyne, H.J., III; Ciofi-Baffoni, S.; Banci, L.; Bertini, I.; Zhang, L.; George, G.N.; Winge, D.R. The characterization and role of zinc binding in yeast Cox4. J. Biol. Chem. 2007, 282, 8926–8934. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, S.; Ichinose, M.; Tasaki, E.; Aoki, Y.; Komura, Y.; Sugita, M. Targeted gene disruption identifies three PPR-DYW proteins involved in RNA editing for five editing sites of the moss mitochondrial transcripts. Plant Cell Physiol. 2010, 51, 1942–1949. [Google Scholar] [CrossRef] [Green Version]
- Shimada, S.; Shinzawa-Itoh, K.; Baba, J.; Aoe, S.; Shimada, A.; Yamashita, E.; Kang, J.; Tateno, M.; Yoshikawa, S.; Tsukihara, T. Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode. EMBO J. 2017, 36, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Tian, Z.; Lu, L.; Chen, X.; Chen, X.; Zhang, W.; Song, R. Editing of Mitochondrial Transcripts nad3 and cox2 by Dek10 Is Essential for Mitochondrial Function and Maize Plant Development. Genetics 2017, 205, 1489–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Conservation | Structural Type | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subunit | # Edits | High | Inter-Mediate | Low/None | In Membrane | To Hydrophobic | Proline Removal | Close to Functional | At Interface | Unaccounted |
| Complex I | ||||||||||
| NAD1 | 21 | 29% | 43% | 29% | 86% | 48% | 10% | 0% | 33% | 0% |
| NAD2 | 36 | 0% | 11% | 89% | 100% | 61% | 19% | 14% | 31% | 6% |
| NAD3 | 19 | 16% | 47% | 37% | 89% | 47% | 32% | 11% | 74% | 0% |
| NAD4 | 38 | 16% | 24% | 61% | 100% | 50% | 18% | 11% | 34% | 0% |
| NAD4L | 14 | 0% | 36% | 64% | 100% | 79% | 21% | 21% | 86% | 0% |
| NAD5 | 32 | 16% | 16% | 69% | 91% | 63% | 9% | 3% | 28% | 3% |
| NAD6 | 11 | 0% | 18% | 82% | 100% | 45% | 36% | 18% | 73% | 0% |
| Total MA | 171 | 12% | 25% | 63% | 95% | 56% | 19% | 10% | 43% | 2% |
| NAD9 | 13 | 38% | 31% | 31% | 0% | 69% | 0% | 0% | 38% | 0% |
| NAD7 | 22 | 50% | 36% | 14% | 0% | 82% | 5% | 32% | 23% | 0% |
| Total PA | 25 | 46% | 34% | 20% | 0% | 77% | 3% | 20% | 29% | 0% |
| Total CI | 206 | 17% | 27% | 56% | 79% | 60% | 16% | 12% | 41% | 1% |
| Complex III2 | ||||||||||
| COB | 21 | 29% | 29% | 43% | 90% | 43% | 24% | 33% | 14% | 10% |
| Complex IV | ||||||||||
| COX1 | 15 | 53% | 33% | 13% | 93% | 53% | 7% | 53% | 13% | 0% |
| COX2 | 21 | 33% | 38% | 29% | 24% | 71% | 10% | 38% | 29% | 5% |
| COX3 | 12 | 67% | 33% | 0% | 100% | 67% | 42% | 17% | 50% | 0% |
| Total CIV | 48 | 48% | 35% | 17% | 65% | 65% | 17% | 38% | 29% | 2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maldonado, M.; Abe, K.M.; Letts, J.A. A Structural Perspective on the RNA Editing of Plant Respiratory Complexes. Int. J. Mol. Sci. 2022, 23, 684. https://doi.org/10.3390/ijms23020684
Maldonado M, Abe KM, Letts JA. A Structural Perspective on the RNA Editing of Plant Respiratory Complexes. International Journal of Molecular Sciences. 2022; 23(2):684. https://doi.org/10.3390/ijms23020684
Chicago/Turabian StyleMaldonado, Maria, Kaitlyn Madison Abe, and James Anthony Letts. 2022. "A Structural Perspective on the RNA Editing of Plant Respiratory Complexes" International Journal of Molecular Sciences 23, no. 2: 684. https://doi.org/10.3390/ijms23020684
APA StyleMaldonado, M., Abe, K. M., & Letts, J. A. (2022). A Structural Perspective on the RNA Editing of Plant Respiratory Complexes. International Journal of Molecular Sciences, 23(2), 684. https://doi.org/10.3390/ijms23020684






