Impact of ER Stress and ER-Mitochondrial Crosstalk in Huntington’s Disease
Abstract
1. Introduction
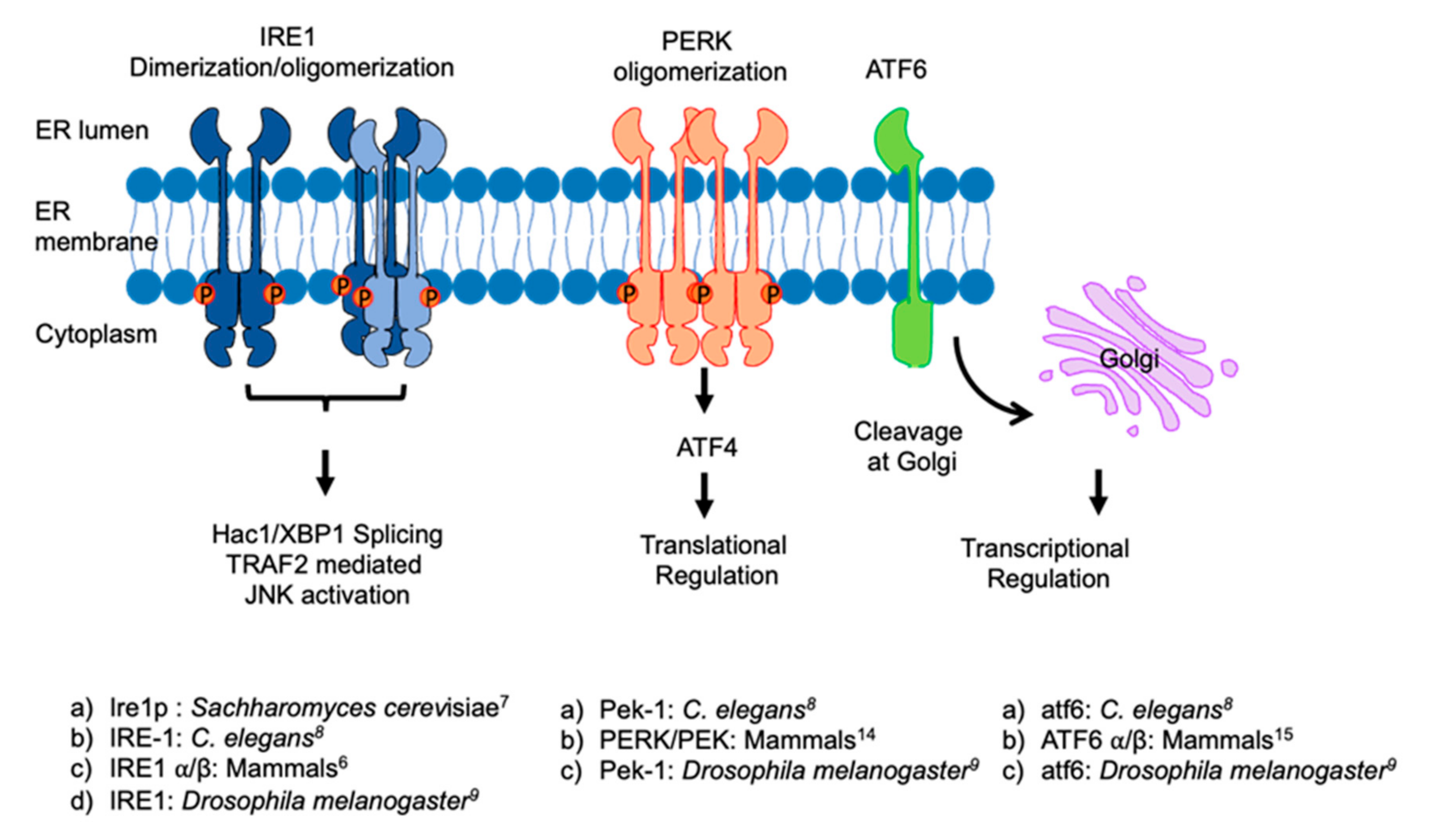
2. ER and ER Stress Response
3. ER Stress Proteins and Their Involvement in HD Pathology
3.1. IRE1
3.2. PERK
3.3. ATF6
3.4. ERAD
4. MAM Proteins and Their Involvement in HD Pathology
4.1. Mitochondrial Dynamics Protein (Drp1, Fis1, Opa1, Mfn1)
4.2. Sigma-1 Receptor S1R
5. Therapeutic Intervention for HD Targeting through ER Stress Pathways
5.1. Targeting UPR Sensors
5.2. Targeting MAM Proteins
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Graveland, G.A.; Williams, R.S.; DiFiglia, M. Evidence for Degenerative and Regenerative Changes in Neostriatal Spiny Neurons in Huntington’s Disease. Science 1985, 227, 770–773. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Crick, S.L.; Ruff, K.M.; Garai, K.; Frieden, C.; Pappu, R.V. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA 2013, 110, 20075–20080. [Google Scholar] [CrossRef]
- Baias, M.; Smith, P.E.S.; Shen, K.; Joachimiak, L.A.; Żerko, S.; Koźmiński, W.; Frydman, J.; Frydman, L. Structure and Dynamics of the Huntingtin Exon-1 N-Terminus: A Solution NMR Perspective. J. Am. Chem. Soc. 2017, 139, 1168–1176. [Google Scholar] [CrossRef]
- Ramdzan, Y.M.; Trubetskov, M.; Ormsby, A.R.; Newcombe, E.; Sui, X.; Tobin, M.J.; Bongiovanni, M.N.; Gras, S.; Dewson, G.; Miller, J.; et al. Huntingtin Inclusions Trigger Cellular Quiescence, Deactivate Apoptosis, and Lead to Delayed Necrosis. Cell Rep. 2017, 19, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, Z.; Thakur, A.K.; Wetzel, R.; Gierasch, L.M. In-cell Aggregation of a Polyglutamine-containing Chimera Is a Multistep Process Initiated by the Flanking Sequence. J. Biol. Chem. 2007, 282, 36736–36743. [Google Scholar] [CrossRef] [PubMed]
- Duennwald, M.L.; Jagadish, S.; Muchowski, P.J.; Lindquist, S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 11045–11050. [Google Scholar] [CrossRef]
- Sakahira, H.; Breuer, P.; Hayer-Hartl, M.K.; Hartl, F.U. Molecular chaperones as modulators of polyglutamine protein aggrega-tion and toxicity. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. S4), 16412–16418. [Google Scholar]
- Kim, Y.E.; Hosp, F.; Frottin, F.; Ge, H.; Mann, M.; Hayer-Hartl, M.; Hartl, F.U. Soluble Oligomers of PolyQ-Expanded Huntingtin Target a Multiplicity of Key Cellular Factors. Mol. Cell 2016, 63, 951–964. [Google Scholar] [CrossRef]
- Leitman, J.; Ulrich Hartl, F.; Lederkremer, G.Z. Soluble forms of polyQ-expanded huntingtin rather than large aggregates cause endoplasmic reticulum stress. Nat. Commun. 2013, 4, 2753. [Google Scholar] [CrossRef]
- Duennwald, M.L.; Lindquist, S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008, 22, 3308–3319. [Google Scholar] [CrossRef]
- Perkins, H.T.; Allan, V.J.; Waigh, T.A. Network organisation and the dynamics of tubules in the endoplasmic reticulum. Sci. Rep. 2021, 11, 16230. [Google Scholar] [CrossRef]
- Shibata, Y.; Shemesh, T.; Prinz, W.A.; Palazzo, A.F.; Kozlov, M.M.; Rapoport, T.A. Mechanisms Determining the Morphology of the Peripheral ER. Cell 2010, 143, 774–788. [Google Scholar] [CrossRef]
- Jan, C.H.; Williams, C.C.; Weissman, J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 2014, 346, 1257521. [Google Scholar] [CrossRef]
- Reid, D.W.; Chen, Q.; Tay, A.S.-L.; Shenolikar, S.; Nicchitta, C.V. The Unfolded Protein Response Triggers Selective mRNA Release from the Endoplasmic Reticulum. Cell 2014, 158, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, M.G.; Higuchi-Sanabria, R.; Garcia, G.; Tsui, C.K.; Dillin, A. Beyond the cell factory: Homeostatic regulation of and by the UPR ER. Sci. Adv. 2020, 6, eabb9614. [Google Scholar] [CrossRef]
- Mori, K. Signalling Pathways in the Unfolded Protein Response: Development from Yeast to Mammals. J. Biochem. 2009, 146, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Karagöz, G.E.; Acosta-Alvear, D.; Nguyen, H.; Lee, C.P.; Chu, F.; Walter, P. An unfolded protein-induced conformational switch activates mammalian IRE1. eLife 2017, 6, e30700. [Google Scholar] [CrossRef] [PubMed]
- Pincus, D.; Chevalier, M.W.; Aragón, T.; van Anken, E.; Vidal, S.E.; El-Samad, H.; Walter, P. BiP Binding to the ER-Stress Sensor Ire1 Tunes the Homeostatic Behavior of the Unfolded Protein Response. PLoS Biol. 2010, 8, e1000415. [Google Scholar] [CrossRef]
- Gardner, B.M.; Walter, P. Unfolded Proteins Are Ire1-Activating Ligands That Directly Induce the Unfolded Protein Response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, D.; Kimata, Y.; Kohno, K.; Iwawaki, T. Activation of mammalian IRE1α upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp. Cell Res. 2009, 315, 2496–2504. [Google Scholar] [CrossRef]
- Kopp, M.C.; Nowak, P.R.; Larburu, N.; Adams, C.J.; Ali, M.M. In vitro FRET analysis of IRE1 and BiP association and dissociation upon endoplasmic reticulum stress. eLife 2018, 7, e30257. [Google Scholar] [CrossRef]
- Sundaram, A.; Appathurai, S.; Plumb, R.; Mariappan, M. Dynamic changes in complexes of IRE1α, PERK, and ATF6α during endoplasmic reticulum stress. Mol. Biol. Cell 2018, 29, 1376–1388. [Google Scholar] [CrossRef]
- Tirasophon, W.; Welihinda, A.A.; Kaufman, R.J. A stress response pathway from the endoplasmic reticulum to the nucleus re-quires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998, 12, 1812–1824. [Google Scholar] [CrossRef] [PubMed]
- Nikawa, J.-I.; Yamashita, S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 1992, 6, 1441–1446. [Google Scholar] [CrossRef]
- Shen, X.; Ellis, R.E.; Lee, K.; Liu, C.Y.; Yang, K.; Solomon, A.; Yoshida, H.; Morimoto, R.; Kurnit, D.M.; Mori, K.; et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 2001, 107, 893–903. [Google Scholar] [CrossRef]
- Hollien, J.; Weissman, J.S. Decay of Endoplasmic Reticulum-Localized mRNAs During the Unfolded Protein Response. Science 2006, 313, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.-H.; Heidtman, K.; Hotamisligil, G.S.; Glimcher, L.H. Dual and opposing roles of the unfolded protein response regulated by IRE1 and XBP1 in proinsulin processing and insulin secretion. Proc. Natl. Acad. Sci. USA 2011, 108, 8885–8890. [Google Scholar] [CrossRef]
- Rubio, C.; Pincus, D.; Korennykh, A.; Schuck, S.; El-Samad, H.; Walter, P. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J. Cell Biol. 2011, 193, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Hollien, J.; Lin, J.H.; Li, H.; Stevens, N.; Walter, P.; Weissman, J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009, 186, 323–331. [Google Scholar] [CrossRef]
- Tam, A.B.; Koong, A.C.; Niwa, M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep. 2014, 9, 850–858. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef]
- Chan, C.-P.; Kok, K.-H.; Tang, H.-M.V.; Wong, C.-M.; Jin, D.-Y. Internal ribosome entry site-mediated translational regulation of ATF4 splice variant in mammalian unfolded protein response. Biochim. Biophys. Acta (BBA)–Bioenerg. 2013, 1833, 2165–2175. [Google Scholar] [CrossRef]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Y.; Tang, J.; Jiang, J.; Chen, Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim. et Biophys. Sin. 2015, 47, 146–147. [Google Scholar] [CrossRef]
- Haze, K.; Yoshida, H.; Yanagi, H.; Yura, T.; Mori, K. Mammalian Transcription Factor ATF6 Is Synthesized as a Transmembrane Protein and Activated by Proteolysis in Response to Endoplasmic Reticulum Stress. Mol. Biol. Cell 1999, 10, 3787–3799. [Google Scholar] [CrossRef]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Davé, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER Stress Induces Cleavage of Membrane-Bound ATF6 by the Same Proteases that Process SREBPs. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Hong, M.; Luo, S.; Baumeister, P.; Huang, J.-M.; Gogia, R.K.; Li, M.; Lee, A. Underglycosylation of ATF6 as a Novel Sensing Mechanism for Activation of the Unfolded Protein Response. J. Biol. Chem. 2004, 279, 11354–11363. [Google Scholar] [CrossRef]
- Shen, J.; Chen, X.; Hendershot, L.; Prywes, R. ER Stress Regulation of ATF6 Localization by Dissociation of BiP/GRP78 Binding and Unmasking of Golgi Localization Signals. Dev. Cell 2002, 3, 99–111. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, J.; Arenzana, N.; Tirasophon, W.; Kaufman, R.J.; Prywes, R. Activation of ATF6 and an ATF6 DNA Binding Site by the Endoplasmic Reticulum Stress Response. J. Biol. Chem. 2000, 275, 27013–27020. [Google Scholar] [CrossRef]
- Ast, T.; Aviram, N.; Chuartzman, S.G.; Schuldiner, M. A cytosolic degradation pathway, prERAD, monitors pre-inserted secretory pathway proteins. J. Cell Sci. 2014, 127, 3017–3023. [Google Scholar] [CrossRef]
- Habeck, G.; Ebner, F.A.; Shimada-Kreft, H.; Kreft, S.G. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J. Cell Biol. 2015, 209, 621. [Google Scholar] [CrossRef]
- Xia, D.; Tang, W.K.; Ye, Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene 2016, 583, 64–77. [Google Scholar] [CrossRef]
- Naidoo, N.; Ferber, M.; Master, M.; Zhu, Y.; Pack, A.I. Aging Impairs the Unfolded Protein Response to Sleep Deprivation and Leads to Proapoptotic Signaling. J. Neurosci. 2008, 28, 6539–6548. [Google Scholar] [CrossRef]
- Jin, H.; Komita, M.; Aoe, T. Decreased Protein Quality Control Promotes the Cognitive Dysfunction Associated with Aging and Environmental Insults. Front. Neurosci. 2018, 12, 753. [Google Scholar] [CrossRef]
- Brandstaetter, H.; Kruppa, A.J.; Buss, F. Huntingtin is required for ER-to-Golgi transport and for secretory vesicle fusion at the plasma membrane. Dis. Model. Mech. 2014, 7, 1335–1340. [Google Scholar] [CrossRef]
- Trettel, F.; Rigamonti, D.; Hilditch-Maguire, P.; Wheeler, V.C.; Sharp, A.H.; Persichetti, F.; Cattaneo, E.; Macdonald, M.E. Dominant phenotypes produced by the HD mutation in STHdhQ111 striatal cells. Hum. Mol. Genet. 2000, 9, 2799–2809. [Google Scholar] [CrossRef]
- Nishitoh, H.; Matsuzawa, A.; Tobiume, K.; Saegusa, K.; Takeda, K.; Inoue, K.; Hori, S.; Kakizuka, A.; Ichijo, H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002, 16, 1345–1355. [Google Scholar] [CrossRef]
- Reijonen, S.; Putkonen, N.; Nørremølle, A.; Lindholm, D.; Korhonen, L. Inhibition of endoplasmic reticulum stress counteracts neuronal cell death and protein aggregation caused by N-terminal mutant huntingtin proteins. Exp. Cell Res. 2008, 314, 950–960. [Google Scholar] [CrossRef]
- Carnemolla, A.; Fossale, E.; Agostoni, E.; Michelazzi, S.; Calligaris, R.; De Maso, L.; Del Sal, G.; MacDonald, M.E.; Persichetti, F. Rrs1 is involved in endoplasmic reticulum stress response in Huntington disease. J. Biol. Chem. 2009, 284, 18167–18173. [Google Scholar] [CrossRef]
- Hyrskyluoto, A.; Bruelle, C.; Lundh, S.H.; Do, H.T.; Kivinen, J.; Rappou, E.; Reijonen, S.; Waltimo, T.; Petersén, Å.; Lindholm, D.; et al. Ubiquitin-specific protease-14 reduces cellular aggregates and protects against mutant huntingtin-induced cell degeneration: Involvement of the proteasome and ER stress-activated kinase IRE1α. Hum. Mol. Genet. 2014, 23, 5928–5939. [Google Scholar] [CrossRef]
- Lee, H.; Noh, J.-Y.; Oh, Y.; Kim, Y.; Chang, J.-W.; Chung, C.-W.; Lee, S.-T.; Kim, M.; Ryu, H.; Jung, Y.-K. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum. Mol. Genet. 2011, 21, 101–114. [Google Scholar] [CrossRef]
- Vidal, R.L.; Figueroa, A.; Court, F.A.; Thielen, P.; Molina, C.; Wirth, C.; Caballero, B.; Kiffin, R.; Segura-Aguilar, J.; Cuervo, A.M.; et al. Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum. Mol. Genet. 2012, 21, 2245–2262. [Google Scholar] [CrossRef]
- Hill, S.M.; Wrobel, L.; Ashkenazi, A.; Fernandez-Estevez, M.; Tan, K.; Bürli, R.W.; Rubinsztein, D.C. VCP/p97 regulates Beclin-1-dependent autophagy initiation. Nat. Chem. Biol. 2021, 17, 448–455. [Google Scholar] [CrossRef]
- Ghosh, D.K.; Roy, A.; Ranjan, A. The ATPase VCP/p97 functions as a disaggregase against toxic Huntingtin-exon1 aggregates. FEBS Lett. 2018, 592, 2680–2692. [Google Scholar] [CrossRef]
- Becher, M.W.; Kotzuk, J.A.; Sharp, A.H.; Davies, S.W.; Bates, G.P.; Price, D.L.; Ross, C.A. Intranuclear Neuronal Inclusions in Huntington’s Disease and Dentatorubral and Pallidoluysian Atrophy: Correlation between the Density of Inclusions andIT15CAG Triplet Repeat Length. Neurobiol. Dis. 1998, 4, 387–397. [Google Scholar] [CrossRef]
- Ordway, J.M.; Tallaksen-Greene, S.; Gutekunst, C.-A.; Bernstein, E.M.; Cearley, J.A.; Wiener, H.W.; Dure, L.S.; Lindsey, R.; Hersch, S.M.; Jope, R.S.; et al. Ectopically Expressed CAG Repeats Cause Intranuclear Inclusions and a Progressive Late Onset Neurological Phenotype in the Mouse. Cell 1997, 91, 753–763. [Google Scholar] [CrossRef]
- Taylor, J.P.; Tanaka, F.; Robitschek, J.; Sandoval, C.M.; Taye, A.; Markovic-Plese, S.; Fischbeck, K.H. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 2003, 12, 749–757. [Google Scholar] [CrossRef]
- Cummings, C.J.; Reinstein, E.; Sun, Y.; Antalffy, B.; Jiang, Y.-H.; Ciechanover, A.; Orr, H.; Beaudet, A.L.; Zoghbi, H.Y. Mutation of the E6-AP Ubiquitin Ligase Reduces Nuclear Inclusion Frequency While Accelerating Polyglutamine-Induced Pathology in SCA1 Mice. Neuron 1999, 24, 879–892. [Google Scholar] [CrossRef]
- Arrasate, M.; Mitra, S.; Schweitzer, E.S.; Segal, M.R.; Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 2004, 431, 805–810. [Google Scholar] [CrossRef]
- Iwata, A.; Riley, B.E.; Johnston, J.A.; Kopito, R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005, 280, 40282–40292. [Google Scholar] [CrossRef] [PubMed]
- Bae, D.; Jones, R.E.; Hollien, J. Regulation of Blos1 by IRE1 prevents the accumulation of Huntingtin protein aggregates. bio-Rxiv 2021. [Google Scholar] [CrossRef]
- Pu, J.; Schindler, C.; Jia, R.; Jarnik, M.; Backlund, P.; Bonifacino, J.S. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 2015, 33, 176–188. [Google Scholar] [CrossRef]
- Ravikumar, B.; Vacher, C.; Berger, Z.; Davies, J.E.; Luo, S.; Oroz, L.G.; Scaravilli, F.; Easton, D.F.; Duden, R.; O’Kane, C.J.; et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004, 36, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.; Zhang, Y.; Bertolotti, A.; Zeng, H.; Ron, D. Perk Is Essential for Translational Regulation and Cell Survival during the Unfolded Protein Response. Mol. Cell 2000, 5, 897–904. [Google Scholar] [CrossRef]
- Kouroku, Y.; Fujita, E.; Tanida, I.; Ueno, T.; Isoai, A.; Kumagai, H.; Ogawa, S.; Kaufman, R.J.; Kominami, E.; Momoi, T. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007, 14, 230–239. [Google Scholar] [CrossRef]
- Ma, R.-H.; Ni, Z.-J.; Thakur, K.; Zhang, F.; Zhang, Y.-Y.; Zhang, J.-G.; Wei, Z.-J. Natural Compounds Play Therapeutic Roles in Various Human Pathologies via Regulating Endoplasmic Reticulum Pathway. Med. Drug Discov. 2020, 8, 100065. [Google Scholar] [CrossRef]
- Leitman, J.; Barak, B.; Benyair, R.; Shenkman, M.; Ashery, U.; Hartl, F.U.; Lederkremer, G.Z. ER stress-induced eIF2-alpha phosphorylation un-derlies sensitivity of striatal neurons to pathogenic huntingtin. PLoS ONE 2014, 9, e90803. [Google Scholar] [CrossRef]
- Ganz, J.; Shacham, T.; Kramer, M.; Shenkman, M.; Eiger, H.; Weinberg, N.; Iancovici, O.; Roy, S.; Simhaev, L.; Da’Adoosh, B.; et al. A novel specific PERK activator reduces toxicity and extends survival in Huntington’s disease models. Sci. Rep. 2020, 10, 6875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Szabo, E.; Michalak, M.; Opas, M. Endoplasmic reticulum stress during the embryonic development of the central nervous system in the mouse. Int. J. Dev. Neurosci. 2007, 25, 455–463. [Google Scholar] [CrossRef]
- Cho, Y.M.; Jang, Y.S.; Jang, Y.M.; Chung, S.M.; Kim, H.S.; Lee, J.H.; Jeong, S.W.; Kim, I.K.; Kim, J.J.; Kim, K.S.; et al. Induction of unfolded protein response during neuronal in-duction of rat bone marrow stromal cells and mouse embryonic stem cells. Exp. Mol. Med. 2009, 41, 440–452. [Google Scholar] [CrossRef]
- Naughton, M.C.; McMahon, J.M.; Fitzgerald, U. Differential activation of ER stress pathways in myelinating cerebellar tracts. Int. J. Dev. Neurosci. 2015, 47, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Matus, S.; Lisbona, F.; Torres, M.; Leon, C.; Thielen, P.; Hetz, C. The Stress Rheostat: An Interplay Between the Unfolded Protein Response (UPR) and Autophagy in Neurodegeneration. Curr. Mol. Med. 2008, 8, 157–172. [Google Scholar] [CrossRef]
- Fernandez-Fernandez, M.R.; Ferrer, I.; Lucas, J.J. Impaired ATF6α processing, decreased Rheb and neuronal cell cycle re-entry in Huntington’s disease. Neurobiol. Dis. 2011, 41, 23–32. [Google Scholar] [CrossRef] [PubMed]
- López-Hurtado, A.; Burgos, D.F.; González, P.; Dopazo, X.M.; González, V.; Rábano, A.; Mellström, B.; Naranjo, J.R. Inhibition of DREAM-ATF6 interaction delays onset of cognition deficit in a mouse model of Huntington’s disease. Mol. Brain 2018, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, J.R.; Zhang, H.; Villar, D.; González, P.; Dopazo, X.M.; Morón-Oset, J.; Higueras, E.; Oliveros, J.C.; Arrabal, M.D.; Prieto, A.; et al. Activating transcription factor 6 derepression mediates neuroprotection in Huntington disease. J. Clin. Investig. 2016, 126, 627–638. [Google Scholar] [CrossRef]
- Wang, X.; Xu, M.; Frank, J.A.; Ke, Z.-J.; Luo, J. Thiamine deficiency induces endoplasmic reticulum stress and oxidative stress in human neurons derived from induced pluripotent stem cells. Toxicol. Appl. Pharmacol. 2017, 320, 26–31. [Google Scholar] [CrossRef]
- Yang, H.; Liu, C.; Zhong, Y.; Luo, S.; Monteiro, M.J.; Fang, S. Huntingtin interacts with the cue domain of gp78 and inhibits gp78 binding to ubiquitin and p97/VCP. PLoS ONE 2010, 5, e8905. [Google Scholar] [CrossRef]
- Lajoie, P.; Snapp, E.L. Changes in BiP availability reveal hypersensitivity to acute endoplasmic reticulum stress in cells ex-pressing mutant huntingtin. J. Cell Sci. 2011, 124, 3332–3343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ryskamp, D.A.; Korban, S.; Zhemkov, V.; Kraskovskaya, N.; Bezprozvanny, I. Neuronal Sigma-1 Receptors: Signaling Functions and Protective Roles in Neurodegenerative Diseases. Front. Neurosci. 2019, 13, 862. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Jeong, S.-Y.; Karbowski, M.; Smith, C.L.; Youle, R.J. Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis. Mol. Biol. Cell 2004, 15, 5001–5011. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Carelli, V.; Manfredi, G.; Chan, D.C. Proteolytic Cleavage of Opa1 Stimulates Mitochondrial Inner Membrane Fusion and Couples Fusion to Oxidative Phosphorylation. Cell Metab. 2014, 19, 630–641. [Google Scholar] [CrossRef]
- Yu, R.; Jin, S.; Lendahl, U.; Nistér, M.; Zhao, J. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 2019, 38, e99748. [Google Scholar] [CrossRef]
- Hyrskyluoto, A.; Pulli, I.; Törnqvist, K.; Ho, T.H.; Korhonen, L.; Lindholm, D. Sigma-1 receptor agonist PRE084 is protective against mutant huntingtin-induced cell degeneration: Involvement of calpastatin and the NF-κB pathway. Cell Death Dis. 2013, 4, e646. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, R.; Oliván, S.; Rando, A.; Casas, C.; Osta, R.; Navarro, X. Sigma-1R Agonist Improves Motor Function and Motoneuron Survival in ALS Mice. Neurotherapeutics 2012, 9, 814–826. [Google Scholar] [CrossRef] [PubMed]
- Maurice, T.; Su, T.-P.; Privat, A. Sigma1 (σ1) receptor agonists and neurosteroids attenuate β25–35-amyloid peptide-induced amnesia in mice through a common mechanism. Neuroscience 1998, 83, 413–428. [Google Scholar] [CrossRef]
- Smirnova, E.; Griparic, L.; Shurland, D.-L.; van der Bliek, A.M. Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells. Mol. Biol. Cell 2001, 12, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Otsuga, D.; Keegan, B.R.; Brisch, E.; Thatcher, J.W.; Hermann, G.; Bleazard, W.; Shaw, J.M. The Dynamin-related GTPase, Dnm1p, Controls Mitochondrial Morphology in Yeast. J. Cell Biol. 1998, 143, 333–349. [Google Scholar] [CrossRef]
- Mozdy, A.D.; McCaffery, J.M.; Shaw, J.M. Dnm1p Gtpase-Mediated Mitochondrial Fission Is a Multi-Step Process Requiring the Novel Integral Membrane Component Fis1p. J. Cell Biol. 2000, 151, 367–380. [Google Scholar] [CrossRef]
- Yu, R.; Liu, T.; Jin, S.-B.; Ning, C.; Lendahl, U.; Nistér, M.; Zhao, J. MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff. Sci. Rep. 2017, 7, 880. [Google Scholar] [CrossRef]
- Song, Z.; Ghochani, M.; McCaffery, J.M.; Frey, T.G.; Chan, D.C. Mitofusins and OPA1 Mediate Sequential Steps in Mitochondrial Membrane Fusion. Mol. Biol. Cell 2009, 20, 3525–3532. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Shi, X.; Boopathy, S.; McDonald, J.; Smith, A.W.; Chao, L.H. Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. eLife 2020, 9, e50973. [Google Scholar] [CrossRef]
- Song, W.; Chen, J.; Petrilli, A.; Liot, G.; Klinglmayr, E.; Zhou, Y.; Poquiz, P.; Tjong, J.; Pouladi, M.A.; Hayden, M.R.; et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 2011, 17, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Giacomello, M.; Hudec, R.; Lopreiato, R.; Ermak, G.; Lim, D.; Malorni, W.; Davies, K.J.A.; Carafoli, E.; Scorrano, L. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol. Med. 2010, 2, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Moody, J.P.; Edgerly, C.K.; Bordiuk, O.L.; Cormier, K.; Smith, K.; Beal, M.F.; Ferrante, R.J. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum. Mol. Genet. 2010, 19, 3919–3935. [Google Scholar] [CrossRef] [PubMed]
- Shirendeb, U.P.; Calkins, M.J.; Manczak, M.; Anekonda, V.; Dufour, B.; McBride, J.L.; Mao, P.; Reddy, P.H. Mutant huntingtin’s interaction with mi-tochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degenera-tion in Huntington’s disease. Hum. Mol. Genet. 2011, 21, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.N.; Yu, Y.V.; Gundemir, S.; Jo, C.; Cui, M.; Tieu, K.; Johnson, G.V.W. Impaired Mitochondrial Dynamics and Nrf2 Signaling Contribute to Compromised Responses to Oxidative Stress in Striatal Cells Expressing Full-Length Mutant Huntingtin. PLoS ONE 2013, 8, e57932. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.U.; Ebert, A.E.; Haileselassie, B.; Mochly-Rosen, D. Drp1/Fis1-mediated mitochondrial fragmentation leads to lysosomal dysfunction in cardiac models of Huntington’s disease. J. Mol. Cell Cardiol. 2019, 127, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Hering, T.; Kojer, K.; Birth, N.; Hallitsch, J.; Taanman, J.W.; Orth, M. Mitochondrial cristae remodelling is associated with disrupted OPA1 oligomerisation in the Huntington’s disease R6/2 fragment model. Exp. Neurol. 2017, 288, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Zhemkov, V.; Ditlev, J.A.; Lee, W.-R.; Wilson, M.; Liou, J.; Rosen, M.K.; Bezprozvanny, I. The role of sigma 1 receptor in organization of endoplasmic reticulum signaling microdomains. eLife 2021, 10, e65192. [Google Scholar] [CrossRef]
- Palmer, C.P.; Mahen, R.; Schnell, E.; Djamgoz, M.B.; Aydar, E. Sigma-1 Receptors Bind Cholesterol and Remodel Lipid Rafts in Breast Cancer Cell Lines. Cancer Res. 2007, 67, 11166–11175. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Sigma-1 Receptor Chaperones at the ER-Mitochondrion Interface Regulate Ca2+ Signaling and Cell Survival. Cell 2007, 131, 596–610. [Google Scholar] [CrossRef]
- Ryskamp, D.; Wu, J.; Geva, M.; Kusko, R.; Grossman, I.; Hayden, M.; Bezprozvanny, I. The sigma-1 receptor mediates the beneficial effects of pridopidine in a mouse model of Huntington disease. Neurobiol. Dis. 2017, 97, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, S.; Van Der Gaag, B.; Cortese, F.A.B. RNAi mechanisms in Huntington’s disease therapy: siRNA versus shRNA. Transl. Neurodegener. 2017, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Datson, N.A.; González-Barriga, A.; Kourkouta, E.; Weij, R.; Van De Giessen, J.; Mulders, S.; Kontkanen, O.; Heikkinen, T.; Lehtimäki, K.; Van Deutekom, J.C.T. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS ONE 2017, 12, e0171127. [Google Scholar] [CrossRef]
- Southwell, A.L.; Kordasiewicz, H.B.; Langbehn, D.; Skotte, N.H.; Parsons, M.P.; Villanueva, E.B.; Caron, N.S.; Østergaard, M.E.; Anderson, L.M.; Xie, Y.; et al. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci. Transl. Med. 2018, 10, eaar3959. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.M.; Miniarikova, J.; Juhas, S.; Vallès, A.; Bohuslavova, B.; Juhasova, J.; Skalnikova, H.K.; Vodicka, P.; Valekova, I.; Brouwers, C.; et al. AAV5-miHTT Gene Therapy Demonstrates Broad Distribution and Strong Human Mutant Huntingtin Lowering in a Huntington’s Disease Minipig Model. Mol. Ther. 2018, 26, 2163–2177. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Chang, R.; Yang, H.; Zhao, T.; Hong, Y.; Kong, H.E.; Sun, X.; Qin, Z.; Jin, P.; Li, S.; et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Investig. 2017, 127, 2719–2724. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, M.; Juzwa, W.; Krzyzosiak, W.J.; Olejniczak, M. Precise Excision of the CAG Tract from the Huntingtin Gene by Cas9 Nickases. Front. Neurosci. 2018, 12, 75. [Google Scholar] [CrossRef]
- Precious, S.V.; Zietlow, R.; Dunnett, S.B.; Kelly, C.M.; Rosser, A.E. Is there a place for human fetal-derived stem cells for cell replacement therapy in Huntington’s disease? Neurochem. Int. 2017, 106, 114–121. [Google Scholar] [CrossRef]
- Scior, A.; Buntru, A.; Arnsburg, K.; Ast, A.; Iburg, M.; Juenemann, K.; Pigazzini, M.L.; Mlody, B.; Puchkov, D.; Priller, J.; et al. Complete suppression of Htt fibrilization and dis-aggregation of Htt fibrils by a trimeric chaperone complex. EMBO J. 2018, 37, 282–299. [Google Scholar] [CrossRef] [PubMed]
- Emerich, D. Cellular delivery of human CNTF prevents motor and cognitive dysfunction in a rodent model of Huntington’s disease. Cell Transplant. 1997, 6, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Suelves, N.; Kirkham-McCarthy, L.; Lahue, R.S.; Ginés, S. A selective inhibitor of histone deacetylase 3 prevents cognitive deficits and suppresses striatal CAG repeat expansions in Huntington’s disease mice. Sci. Rep. 2017, 7, 6082. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kim, W.; Jang, J.; Isacson, O.; Seo, H. Gene therapy by proteasome activator, PA28γ, improves motor coordination and proteasome function in Huntington’s disease YAC128 mice. Neuroscience 2016, 324, 20–28. [Google Scholar] [CrossRef] [PubMed]
- McGarry, A.; Leinonen, M.; Kieburtz, K.; Geva, M.; Olanow, C.W.; Hayden, M. Effects of Pridopidine on Functional Capacity in Early-Stage Participants from the PRIDE-HD Study. J. Huntingt. Dis. 2020, 9, 371–380. [Google Scholar] [CrossRef] [PubMed]
- BioSpace. Prilenia Enrolls First Patients into Its PROOF-HD Phase 3 Clinical Trial for Huntington’s Disease in the United States. BioSpace. 27 October 2020. Available online: https://www.biospace.com/article/prilenia-enrolls-first-patients-into-its-proof-hd-phase-3-clinical-trial-for-huntington-s-disease-in-the-united-states/ (accessed on 18 September 2021).
- Grande, V.; Ornaghi, F.; Comerio, L.; Restelli, E.; Masone, A.; Corbelli, A.; Tolomeo, D.; Capone, V.; Axten, J.M.; Laping, N.J.; et al. PERK inhibition delays neurodegeneration and improves motor function in a mouse model of Marinesco-Sjögren syndrome. Hum. Mol. Genet. 2018, 27, 2477–2489. [Google Scholar] [CrossRef] [PubMed]
- Mercado, G.; Castillo, V.; Soto, P.; López, N.; Axten, J.M.; Sardi, S.P.; Hoozemans, J.J.; Hetz, C. Targeting PERK signaling with the small molecule GSK2606414 prevents neurodegeneration in a model of Parkinson’s disease. Neurobiol. Dis. 2018, 112, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Radford, H.; Moreno, J.A.; Verity, N.; Halliday, M.; Mallucci, G.R. PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol. 2015, 130, 633–642. [Google Scholar] [CrossRef]
- Wang, F.; Song, W.; Brancati, G.; Segatori, L. Inhibition of Endoplasmic Reticulum-associated Degradation Rescues Native Folding in Loss of Function Protein Misfolding Diseases. J. Biol. Chem. 2011, 286, 43454–43464. [Google Scholar] [CrossRef]
- Wang, Q.; Shinkre, B.A.; Lee, J.-G.; Weniger, M.A.; Liu, Y.; Chen, W.; Wiestner, A.; Trenkle, W.C.; Ye, Y. The ERAD Inhibitor Eeyarestatin I Is a Bifunctional Compound with a Membrane-Binding Domain and a p97/VCP Inhibitory Group. PLoS ONE 2010, 5, e15479. [Google Scholar] [CrossRef]
- Guo, X.; Sun, X.; Hu, D.; Wang, Y.-J.; Fujioka, H.; Vyas, R.; Chakrapani, S.; Joshi, A.U.; Luo, Y.; Mochly-Rosen, D.; et al. VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease. Nat. Commun. 2016, 7, 12646. [Google Scholar] [CrossRef] [PubMed]
- Matus, S.; Nassif, M.; Glimcher, L.H.; Hetz, C. XBP-1 deficiency in the nervous system reveals a homeostatic switch to activate autophagy. Autophagy 2009, 5, 1226–1228. [Google Scholar] [CrossRef]
- Zuleta, A.; Vidal, R.L.; Armentano, D.; Parsons, G.; Hetz, C. AAV-mediated delivery of the transcription factor XBP1s into the striatum reduces mutant Huntingtin aggregation in a mouse model of Huntington’s disease. Biochem. Biophys. Res. Commun. 2012, 420, 558–563. [Google Scholar] [CrossRef]
- Ferrante, R.J.; Kubilus, J.K.; Lee, J.; Ryu, H.; Beesen, A.; Zucker, B.; Smith, K.; Kowall, N.W.; Ratan, R.R.; Luthi-Carter, R.; et al. Histone Deacetylase Inhibition by Sodium Butyrate Chemotherapy Ameliorates the Neurodegenerative Phenotype in Huntington’s Disease Mice. J. Neurosci. 2003, 23, 9418–9427. [Google Scholar] [CrossRef] [PubMed]
- Keene, C.D.; Rodrigues, C.M.P.; Eich, T.; Chhabra, M.S.; Steer, C.J.; Low, W.C. Tauroursodeoxycholic Acid, a Bile Acid, Is Neuroprotective in a Transgenic Animal Model of Huntington’s Disease. Proc. Natl. Acad. Sci. USA 2002, 99, 10671–10676. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, L.G.G.C.; Lindsten, K.; Masucci, M.G.; Dantuma, N.P. Aggregate Formation Inhibits Proteasomal Degradation of Polyglutamine Proteins. Hum. Mol. Genet. 2002, 11, 2689–2700. [Google Scholar] [CrossRef] [PubMed]
- Bence, N.F. Impairment of the Ubiquitin-Proteasome System by Protein Aggregation. Science 2001, 292, 1552–1555. [Google Scholar] [CrossRef]
- Jana, N.R.; Zemskov, E.A.; Wang, G.H.; Nukina, N. Altered Proteasomal Function due to the Expression of Polyglutamine-Expanded Truncated N-Terminal Huntingtin Induces Apoptosis by Caspase Activation through Mitochondrial Cytochrome c Release. Hum. Mol. Genet. 2001, 10, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.-W.; Hong, J.-Y.; Zhang, S.-X.; Jiang, L.-L.; Hu, H.-Y. PolyQ-Expanded Proteins Impair Cellular Proteostasis of Ataxin-3 through Sequestering the Co-Chaperone HSJ1 into Aggregates. Sci. Rep. 2021, 11, 7815. [Google Scholar] [CrossRef]
- Houghton, C.A.; Fassett, R.G.; Coombes, J.S. Sulforaphane and Other Nutrigenomic Nrf2 Activators: Can the Clinician’s Expectation Be Matched by the Reality? Oxid. Med. Cell. Longev. 2016, 2016, 7857186. [Google Scholar] [CrossRef]
- Koduah, P.; Paul, F.; Dörr, J.-M. Vitamin D in the Prevention, Prediction and Treatment of Neurodegenerative and Neuroinflammatory Diseases. EPMA J. 2017, 8, 313–325. [Google Scholar] [CrossRef]
- Chel, V.G.; Ooms, M.E.; van der Bent, J.; Veldkamp, F.; Roos, R.A.; Achterberg, W.P.; Lips, P. High Prevalence of Vitamin D Deficiency and Insufficiency in Patients with Manifest Huntington Disease: An Explorative Study. Dermatoendocrinology 2013, 5, 348–351. [Google Scholar] [CrossRef][Green Version]
- Mark, K.A.; Dumas, K.J.; Bhaumik, D.; Schilling, B.; Davis, S.; Oron, T.R.; Sorensen, D.J.; Lucanic, M.; Brem, R.B.; Melov, S.; et al. Vitamin D Promotes Protein Homeostasis and Longevity via the Stress Response Pathway Genes Skn-1, Ire-1, and Xbp-1. Cell Rep. 2016, 17, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Koshenov, Z.; Oflaz, F.E.; Hirtl, M.; Pilic, J.; Bachkoenig, O.A.; Gottschalk, B.; Madreiter-Sokolowski, C.T.; Rost, R.; Malli, R.; Graier, W.F. Sigma-1 Receptor Promotes Mitochondrial Bioenergetics by Orchestrating ER Ca Leak during Early ER Stress. Metabolites 2021, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Miralles, M.; Geva, M.; Tan, J.Y.; Yusof, N.A.B.M.; Cha, Y.; Kusko, R.; Tan, L.J.; Xu, X.; Grossman, I.; Orbach, A.; et al. Early Pridopidine Treatment Improves Behavioral and Transcriptional Deficits in YAC128 Huntington Disease Mice. JCI Insight 2017, 2. [Google Scholar] [CrossRef]
- Kusko, R.; Dreymann, J.; Ross, J.; Cha, Y.; Escalante-Chong, R.; Garcia-Miralles, M.; Tan, L.J.; Burczynski, M.E.; Zeskind, B.; Laifenfeld, D.; et al. Large-Scale Transcriptomic Analysis Reveals That Pridopidine Reverses Aberrant Gene Expression and Activates Neuroprotective Pathways in the YAC128 HD Mouse. Mol. Neurodegener. 2018, 13. [Google Scholar] [CrossRef]
- Squitieri, F.; Di Pardo, A.; Favellato, M.; Amico, E.; Maglione, V.; Frati, L. Pridopidine, a Dopamine Stabilizer, Improves Motor Performance and Shows Neuroprotective Effects in Huntington Disease R6/2 Mouse Model. J. Cell. Mol. Med. 2015, 19, 2540–2548. [Google Scholar] [CrossRef]
- Reilmann, R.; McGarry, A.; Grachev, I.D.; Savola, J.M.; Borowsky, B.; Eyal, E.; Gross, N.; Langbehn, D.; Schubert, R.; Wickenberg, A.T.; et al. Safety and Efficacy of Pridopidine in Patients with Huntington’s Disease (PRIDE-HD): A Phase 2, Randomised, Placebo-Controlled, Multicentre, Dose-Ranging Study. Lancet Neurol. 2019, 18, 165–176. [Google Scholar] [CrossRef]

| Sensor Proteins | Functions of Proteins during HD | References |
|---|---|---|
| IRE1 | Inhibition of autophagy via IRE-1-TRAF2 may impair autophagic clearance of mHtt aggregates leading to neuronal degeneration. | Lee et al., 2011 |
| Overexpression of the ubiquitin-specific protease-14 (Usp14) reduces cellular aggregates in mutant Htt-expressing cells. Overexpression of Usp14 in turn was able to inhibit phosphorylation of IRE1α in mutant Htt-overexpressing cells and to protect against cell degeneration and caspase-3 activation. | Hyrskyluoto et al., 2014 | |
| PERK | ERK/eIF2α phosphorylation is involved in polyQ72-induced LC3 conversion, which is a prerequisite for autophagy in the expression of Huntington’s disease. It acted as a cellular defence that inhibited ER-stress-mediated cell death. | Kouroku et al., 2007 |
| This study proposed that use of salubrinal can reduce ER stress and counteract cell death caused by the mutant huntingtin proteins. Here increased phosphorylation of eukaryotic translation initiation factor2 subunit-α (eIF2α) by the salubrinal has shown to be neuroprotective, thereby influencing the PERK pathway. | Reijonen et al., 2008 | |
| ERAD substrate accumulation and the related ER stress are not dependent on the presence and concentration of large visible aggregates but arise when Htt is in a monomeric or oligomeric state. Eventually all the three UPR sensors are activated including PERK. | Leitman et al., 2013 | |
| It reveals that cultured striatal cells and murine brain striatum have remarkably low levels of phosphorylation of translation initiation factor eIF2α. eIF2α phosphorylation was elevated in a striatal cell line stably expressing pathogenic huntingtin and in brain sections of Huntington’s disease model mice. | Leitman et al., 2014 | |
| PromISR-6 is a potent GBZ analogue, has the capability to prolong the phosphorylation time of eIF2α and protein translation inhibition, reducing mutant Htt aggregates and increasing survival in an HD cellular model, apparently by activating autophagy | Sundaram et al., 2019 | |
| Ganz et al. developed a potent small molecule PERK activator MK-28, which showed improvement in cellular and mice models of HD. It rescued cells from undergoing ER-stress induced apoptosis. Also, it significantly improved motor and executive functions and delayed death onset in the HD induced mice model. | Ganz et al., 2020 | |
| The level of eIF2α phosphorylation is significantly lower in the striatal cells. Interestingly, the presence of pathogenic mtHtt increased the eIF2 alpha phosphorylation in STHdh Q111/111 cells and in the murine striatum | Ma et al., 2020 | |
| ATF6 | ATF6α has also been demonstrated to mediate Rheb expression, and both proteins cooperate to maintain the survival of postmitotic neurons. The ATF6α/Rheb pathway is altered in Huntington’s disease as the decrease in ATF6αprocessing is accompanied by a decrease in the accumulation of Rheb. These alterations correlate with the aberrant accumulation of cell cycle re-entry markers in post-mitotic neurons which is accompanied by death of a subset of neurons. It also leads to, compromised execution of the ER-stress response which may contribute to the pathogenesis of HD. | Fernandez et al., 2011 |
| They have studied the effect of TD-induced neurodegeneration and seen that ER stress markers, such as GRP78, XBP-1, CHOP, ATF-6, phosphorylated eIF2α, and cleaved caspase-12 have increased rapidly. | Wang X et al., 2020 |
| MAM Proteins | Functions of Proteins during HD | Reference |
|---|---|---|
| Drp 1 | Aberrant Drp1-mediated mitochondrial fragmentation within the striatum of HD mutant mice, forces mitochondria to place far away from the ER disrupting the ER-mitochondria association and therefore causing changes in Ca2+ efflux and an excessive production of mitochondria superoxide species. | Cherubini et al., 2020 |
| PERK | There is a strong ER stress induction and UPR activation in striatal neurons expressing mHtt. PERK pathway is strongly downregulated in striatal neurons compared to other cell types and brain regions in WT mice and is upregulated in HD. PERK activation is an effective and selective strategy to reduce ER stress, re-establish ER homeostasis and increase the survival of cells and mice expressing mHtt. | Ganz et al., 2020 |
| RyR 2 | RyR channels were oxidized, PKA phosphorylated, and leaky in brain, heart, and diaphragm both in patients with HD and in a murine model of HD. Defective RyR function has been reported in HD, leading to elevated intracellular Ca2+ levels and reduced endoplasmic reticular Ca2+ stores in R6/2 striatal and cortical neurons . | Dridi et al., 2020 |
| ATAD3A | In HD, ATAD3A forms oligomers which bridge Drp1-mediated mitochondrial fragmentation and mtDNA instability, leading to impaired mitochondrial biogenesis and neurodegeneration. | Zhao et al., 2019 |
| Fis 1 | Heart is highly susceptible to the effects of mHTT. Drp1 hyperactivation through its interaction with Fis1 plays a role in the pathogenesis of polyglutamine induced cardiac mitochondrial dysfunction. Inhibitor for Drp1/Fis1 interaction, reduced pathological mitochondrial fission in mouse and human cell models of HD. | Joshi et al., 2018 |
| CHOP | mHtt expressing striatal cells also exhibited enhanced ER stress in response to serum deprivation, indicated by upregulation of CHOP. The upregulation of CHOP can also downregulate Bcl2 and upregulate pro-apoptotic Bax and Bak. Increase in CHOP expression leads to oxidative stress response in the cells. | Zhou et al., 2018 |
| DISC1 | Disturbance of DISC1-PDE4 interactions and behavioral changes through aggregation of DISC1 in HD.HTT forms a ternary protein complex with the scaffolding protein DISC1 and cAMP-degrading phosphodiesterase 4 (PDE4) to regulate PDE4 activity. Aggregation of DISC1 is significantly accelerated by the presence of mutant HTT, and they recapitulate the PolyQ pathology of HD. Cross-seeding of mutant HTT and DISC1 and the resultant changes in PDE4 activity may underlie the pathology of a specific subset of mental manifestations of HD. | Tanaka et al., 2017 |
| OPA 1 | Integrity of the mitochondrial cristae is compromised in striatum and cortex of the HD mice and that this is most likely caused by impaired OPA1 oligomerisation. Mutant huntingtin reduces OPA1 gene expression and promotes OPA1 cleavage. | Hering et al., 2016 |
| VDAC 1 | The expression of mHtt may promote VDAC closing, a situation known to be accompanied by changes of VDAC selectivity toward cations. This, in turn, may influence metabolite exchange between mitochondria and cytoplasm. Decreased conductance of the open state and decreased voltage-dependence reflect VDAC functional changes occurring in cells with expression of mHtt. | Karachitos et al., 2016 |
| PINK1 | PINK1 overexpression alleviated mitochondrial spheroid formation. Mitophagy is altered in the presence of mHtt and that increasing PINK1/Parkin mitochondrial quality control pathway may improve mitochondrial integrity and neuroprotection in HD. | Khalil et al., 2015 |
| mTOR | Htt promotes signaling by mTORC1 [mechanistic target of rapamycin (mTOR) complex 1] and that this signaling is potentiated by poly-Q–expanded Htt. Htt promotes amino acid mediated mTORC1 signaling that stimulated the interaction of Htt and the guanosine triphosphatase (GTPase) Rheb (a protein that stimulates mTOR activity), and that Htt forms a ternary complex with Rheb and mTOR. | Pryor et al., 2014 |
| Sigma- 1 Receptor | Sig-1R protein levels were decreased in mutant huntingtin-expressing cells. Partial co-localization of Sig-1R with aggregates in mutant huntingtin protein-expressing cells were observed, suggesting that the Sig-1R may be redistributed or delocalized in these cells. | Hyrskyluoto et al., 2013 |
| IRE1 | Regulates mutant huntingtin (HTT) clearance and most studies have shown that IRE1 can control autophagy levels by recruiting the adaptor protein TRAF2 and activating MAPK8 (mitogen-activated protein kinase 8)/JNK | Fouillet et al, 2012; Paolo Remondelli and Maurizio Renna 2017 |
| IP3R | Mutant huntingtin bind to the carboxy-terminal region of the type 1 inositol 1,4,5-trisphosphate receptor (IP3R1) and causes sensitization of IP3R1 to activation by IP3 in planar lipid bilayers and in neuronal cells. InsP3R1 association of ER stress chaperone protein GRP78 is impaired in Huntington’s disease R6/2 model mice, resulting in misregulation of InsP3R1 gating. Impairment of IP3R1 function promotes mitochondria-dependent cell death in the model mice by producing negative effects on mitochondrial Ca2+, the membrane potential and depletion in mitochondrial ATP production. | Higo et al., 2012 |
| Mfn 1 | Levels of Mfn 1 was decreased in mutant Htt neurons, confirming the presence of abnormal mitochondrial dynamics in the cortex of HD patients, and may contribute to neuronal damage in HD patients | Shirendeb et al., 2011 |
| Fis 1 | FIS1 was highly associated with late-stage HD rather than the pre-symptomatic stage. Fis1 recruits Drp1 under cell stress in numerous neurodegenerative disease models. In HD, the balance between fusion and fission is aberrantly shifted toward fission, which is associated with increased levels of Fis1 mRNA and decreased mitofusins in striatal and cortical regions, leading to mitochondrial fragmentation. | Shirendeb et al., 2011; Xiang et al., 2020 |
| Mfn 2 | Mfn2 can directly interact with the N-terminus of mutant Htt, suggesting that Htt can interfere directly with the function of Mfn2 in the promotion of mitochondrial elongation and in the regulation of the apoptotic function of Bax. Mutant Htt, also interfere with the extramitochondrial functions of Mfn2, leading to alterations in the shape of the ER, in the ER levels of Ca2 þ , and last but not least in the tethering of mitochondria to the ER. | Wand et al., 2009; de Brito OM and Scorrano L (2008) |
| ITPR 1/3 | The polyQ-Htt can bind ITPR1 with high affinity, sensitizing the receptor activity by IP3. Blocking the Htt–ITPR1 interaction in vivo was shown to regulate the abnormal calcium signaling in response to glutamate, protecting the neurons from death, and improving motor coordination | Tang et al., 2009 |
| PP2A | Several holoenzyme complexes of PP2A have been isolated from a variety of tissues and have been extensively characterized. PP2A consists of a regulatory subunit termed as PR65. The structure of PR65 is unusual, since it is entirely composed of 15 tandem repeats of a 39-amino-acid sequence, termed as HEAT (huntingtin elongation A subunit TOR, where TOR is target of rapamycin) motif. HEAT sequence is present in huntingtin protein, an elongation factor required for protein synthesis and the TOR kinase. These domains play a role in a variety of interactions between proteins. | Hiroki Takano and James F Gusella 2002 |
| Grp 75 | The interaction between calcium channel IP3R and VDAC1 in the outer mitochondrial membrane is strengthened by Grp75. Compared with Wild type mice, a significant decrease in Grp75 levels was observed in R6/1mice at 12 and 20 weeks of age. | Cherubini et al., 2020 |
| HD Conditions | Regulation | Reference |
|---|---|---|
| Primary neurons of mice showed proteasome dysfunction- and ER stress-induced neuronal cell death, which gives important neuropathological alterations in HD | Overexpression of IRE1 mediated TRAF-2ASK1 and JNK signalling activation | Nishitoh et al., 2002 |
| STHdhQ7/7 striatal cells were treated with tunicamycin, a well-known inducer of ER stress | Bip, CHOP, Herpud1 upregulation,Overexpression of Rrs1, ER stress related marker | Carnemolla et al., 2009 |
| Expanded PolyQ induces cell death in neuronal PC6.3 cells | Downregulation of NF kB via IRE1-TRAF2 ER stress pathway | Reijonen et al., 2008 |
| Accumulation of mHtt in HD mouse model and HD patient striatal tissues was seen | Upregulation of p-IRE1 (ER stress marker) and p62 (Autophagy marker) | Lee et al., 2011 |
| Studying HD pathogenesis through a Fly model (Htt-Q128 fly, a HD model expressing Htt-Q128) | Downregulation of IRE1 rescued rough eye phenotype | Lee et al., 2011 |
| Full-length mHtt transgenic mouse strain YAC128 was used in the study | Silencing XBP-1 expression reduces neuronal loss in the striatum and improves motor performance. It in turn increases expression Foxo1 and autophagy levels | Vidal et al., 2012 |
| Soluble Oligomeric PolyQ expanded HD was expressed in HEK 293 cells in the presence of Htt96Q (containing exon 1 with an expanded tract of 96 glutamines) and in striatal cells | Inhibit ERAD and induces high expression 3 branches of ER stress (IRE1, PERK, and ATF6) | Leitman et al., 2013 |
| Huntingtin-exon 1 aggregates in vitro and in HeLa cells were used in the study | The study identified disaggregase activity of valosin-containing protein (VCP/p97) that can detangle the accumulations of toxic proteins, such as Huntingtin-exon 1 aggregates which can reduce ER stress | Ghosh et al., 2018 |
| Stable MC3T3-E1 cell lines overexpressing exon 1 of either the wild type Huntingtin protein (wtHTT), which contains a string of 23 Gln residues, or a disease-causing mutant Huntingtin (mHTT), which contains 145 Gln’s were used for the study | Activated IRE1 degrades Blos1 mRNA it enhances endosomal microautophagy pathway, protects cells from toxicity of mHtt | Bae et al., 2021 |
| Presence of mHtt in STHdhQ111/111 and murine striatum | Phosphorylation of eIF2alpha is low in striatal cells, and presence of mHtt increase the expression of eIF2alpha | Ma R-H et al., 2020; Leitman et al., 2020 |
| Cellular HD model (a murine striatal cell line) with knock-in of a full-length PolyQ-expanded mHtt (STHdhQ111/111), was used in the study. HD mouse model, the R6/2 with 160 CAG repeats were found to undergo recovery after PERK activation | MK-28 is an activator of PERK. MK-28 significantly improved motor and executive functions and delayed death onset in R6/2 mice, showing no toxicity. PERK activation is beneficial and can help treat aggressive model of HD | Ganz et al., 2020 |
| R6/1 and R6/2 animals (B6CBA-Tg(HDexon1)61Gpb) with expansion of 115 and 150 glutamines respectively. Brain samples from HD patients and age-matched controls were also used | ATF6 alpha is impaired in both animal models of the disease as well as in the brains of HD patients. These alterations are accompanied by a decrease in Rheb. The decreased accumulation of both proteins correlates with the re-entry of neurons into the cell cycle | Fernandez et al., 2010 |
| R6/1 mice model with close to 150 glutamine repeats were used | DREAM inhibition markedly enhance ATF6 expression in the hippocampus and that it might contribute to a delay in memory decline in HD mice. | Hurtado et al., 2018 |
| Thiamine (vitamin B1) deficiency (TD) induced neurodegeneration study. Human neurons differentiated from induced pluripotent stem (iPS) cells was used | The study showed upregulation in different proteins such as GRP78, XBP-1, CHOP and ATF-6 | Wang et al., 2019 |
| Post-mortem neostriatal tissue specimens from 35 adult-onset HD patients was used to carry out the study on both mitochondrial loss and altered mitochondrial morphogenesis with increased mitochondrial fission and reduced fusion in HD. | Mitochondrial dysfunction plays a critical role in the pathogenesis of HD. Increased expression of DRP1 protein and Decreased expression of MFN1 can lead to neuronal loss and disease progression in HD patients | Kim et al., 2010 |
| PC6.3 neuronal cells were used to study that compounds influencing Sig-1R may constitute promising targets for future drug developments in HD. | Sig-1R agonist, PRE084 increases cell survival and counteracts the deleterious effects caused by N-terminal mutant huntingtin proteins in neuronal PC6.3 cells. Sig-1R expression is increased which enhance the levels of cellular antioxidants by activating the NF-kB pathway that is compromised by the expression of mutant huntingtin proteins | Hyrskyluoto et al., 2013 |
| In vitro and In vivo mouse model studies were conducted to see the beneficial effects of S1R during HD | Increase in the levels of S1R through pridopidine in the striatal region exerts long-term beneficial effects on HD. Pridopidine improved motor performance and prolonged survival of R6/2 HD mice and exerted neuroprotective effects in a mouse striatal knock-in cellular model of HD | Ryskamp et al., 2017 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maity, S.; Komal, P.; Kumar, V.; Saxena, A.; Tungekar, A.; Chandrasekar, V. Impact of ER Stress and ER-Mitochondrial Crosstalk in Huntington’s Disease. Int. J. Mol. Sci. 2022, 23, 780. https://doi.org/10.3390/ijms23020780
Maity S, Komal P, Kumar V, Saxena A, Tungekar A, Chandrasekar V. Impact of ER Stress and ER-Mitochondrial Crosstalk in Huntington’s Disease. International Journal of Molecular Sciences. 2022; 23(2):780. https://doi.org/10.3390/ijms23020780
Chicago/Turabian StyleMaity, Shuvadeep, Pragya Komal, Vaishali Kumar, Anshika Saxena, Ayesha Tungekar, and Vaani Chandrasekar. 2022. "Impact of ER Stress and ER-Mitochondrial Crosstalk in Huntington’s Disease" International Journal of Molecular Sciences 23, no. 2: 780. https://doi.org/10.3390/ijms23020780
APA StyleMaity, S., Komal, P., Kumar, V., Saxena, A., Tungekar, A., & Chandrasekar, V. (2022). Impact of ER Stress and ER-Mitochondrial Crosstalk in Huntington’s Disease. International Journal of Molecular Sciences, 23(2), 780. https://doi.org/10.3390/ijms23020780






