Effect of 1-Ethyl-3-methylimidazolium Tetrafluoroborate and Acetate Ionic Liquids on Stability and Amyloid Aggregation of Lysozyme
Abstract
:1. Introduction
2. Results
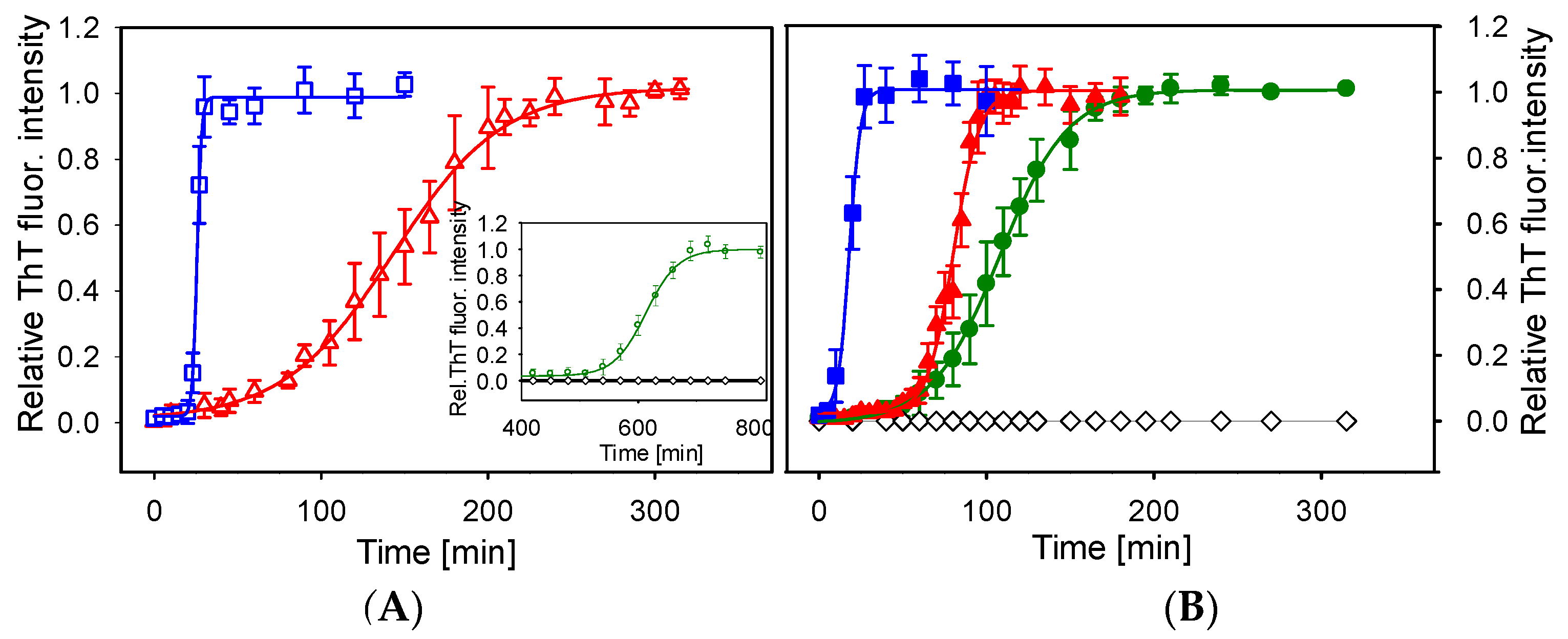
2.1. Kinetics of Lysozyme Amyloid Fibrillization in ILs
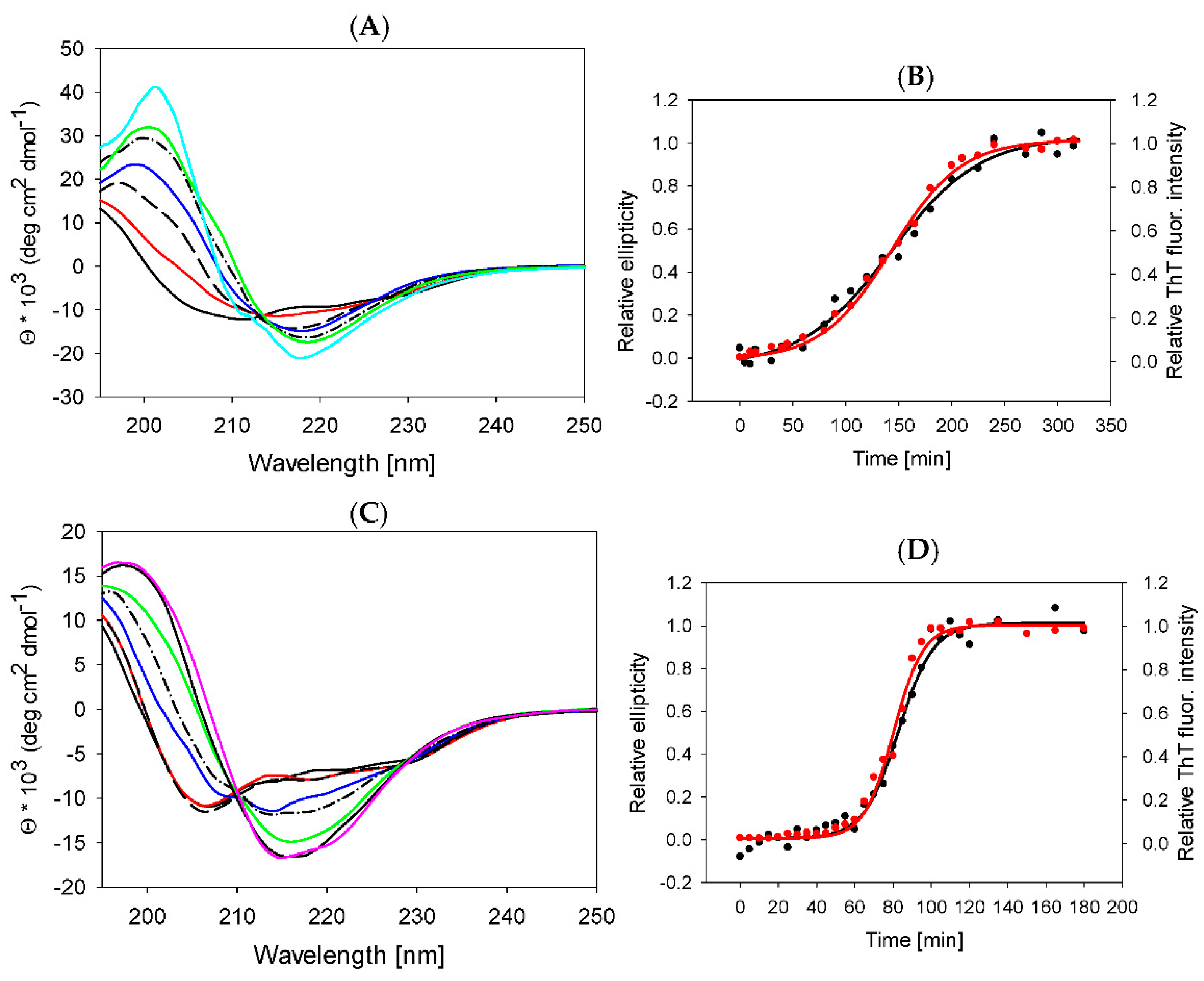
2.2. Thermal Stability of Lysozyme in the Presence of ILs
2.3. Morphology of Lysozyme Amyloid Aggregates
2.4. Lysozyme Fibrils Secondary Structure
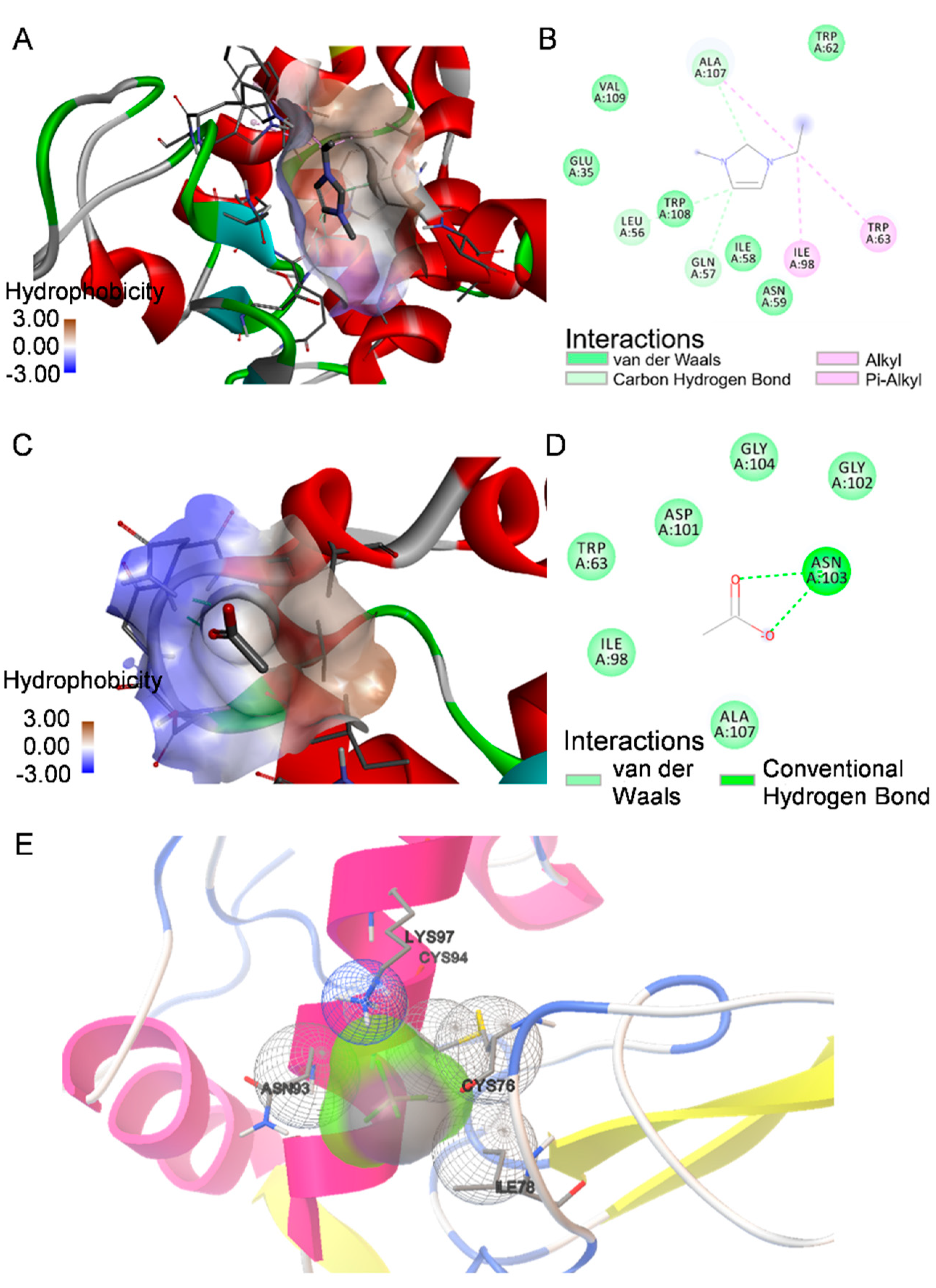
2.5. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Preparation of Lysozyme Amyloid Fibrils
4.3. Thioflavin T Fluorescence Assay
4.4. Differential Scanning Calorimetry (DSC)
4.5. Atomic Force Microscopy (AFM)
4.6. Data Processing of AFM Topographies
4.7. Circular Dichroism (CD) Spectroscopy
4.8. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Small, D.H.; Mok, S.S.; Bornstein, J.C. Alzheimer’s disease and A toxicity: From top to bottom. Nat. Rev. Neurosci. 2001, 2, 595–598. [Google Scholar] [CrossRef]
- Petkova, A.T.; Ishii, Y.; Balbach, J.J.; Antzutkin, O.N.; Leapman, R.D.; Delaglio, F. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA 2002, 99, 16742–16747. [Google Scholar] [CrossRef] [Green Version]
- DeMarco, M.L.; Dagget, V. From conversion to aggregation: Protofibril formation of the prion protein. Proc. Natl. Acad. Sci. USA 2004, 101, 2293–2298. [Google Scholar] [CrossRef] [Green Version]
- Arimon, M.; Díez-Pérez, I.; Kogan, M.J.; Durany, N.; Giralt, E.; Sanz, F.; Fernàndez-Busquets, X. Fine structure study of Abeta1-42 fibrillogenesis with atomic force microscopy. FASEB J. 2005, 19, 1344–1346. [Google Scholar] [CrossRef]
- Guo, S.; Akhremitchev, B.B. Packing density and structural heterogeneity of insulin amyloid fibrils measured by AFM nanoindentation. Biomacromolecules 2006, 7, 1630–1636. [Google Scholar] [CrossRef]
- Cherny, I.; Gazit, E. Amyloids: Not only pathological agents but also ordered nanomaterials. Angew. Chem. Int. Ed. 2008, 47, 4062–4069. [Google Scholar] [CrossRef]
- Assenza, S.; Adamcik, J.; Mezzenga, R.; De Los Rios, P. Universal behavior in the mesoscale properties of amyloid fibrils. Phys Rev Lett. 2014, 113, 268103. [Google Scholar] [CrossRef]
- Serpell, L.C.; Sunde, M.; Blake, C.C. The molecular basis of amyloidosis. Cell. Mol. Life. Sci. 1997, 53, 871–887. [Google Scholar] [CrossRef]
- Petkova, A.T.; Leapman, R.D.; Guo, Z.; Yau, W.M.; Mattson, M.P.; Tycko, R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science 2005, 307, 262–265. [Google Scholar] [CrossRef]
- Kodali, R.; Wetzel, R. Polymorphism in the intermediates and products of amyloid assembly. Curr. Opin. Struct. Biol. 2007, 17, 48–57. [Google Scholar] [CrossRef]
- Close, W.; Neumann, M.; Schmidt, A.; Hora, M.; Annamalai, K.; Schmidt, M.; Reif, B.; Schmidt, V.; Grigorieff, N.; Fändrich, M. Physical basis of amyloid fibril polymorphism. Nat. Commun. 2018, 9, 699. [Google Scholar] [CrossRef]
- Gosal, W.S.; Morten, I.J.; Hewitt, E.W.; Smith, D.A.; Thomson, N.H.; Radford, S.E. Competing pathways determine fibril morphology in the self-assembly of β2-microglobulin into amyloid. J. Mol. Biol. 2005, 351, 850–864. [Google Scholar] [CrossRef]
- Goldsbury, C.; Frey, P.; Olivieri, V.; Aebi, U.; Müller, S.A. Multiple assembly pathways underlie amyloid-beta fibril polymorphisms. J. Mol. Biol. 2005, 352, 282–298. [Google Scholar] [CrossRef]
- Foderà, V.; Zaccone, A.; Lattuada, M.; Donald, A.M. Electrostatics Controls the Formation of Amyloid Superstructures in Protein Aggregation. Phys. Rev. Lett. 2013, 111, 108105. [Google Scholar] [CrossRef] [Green Version]
- Auer, S. Nucleation of Polymorphic Amyloid Fibrils. Biophys. J. 2015, 108, 1176–1186. [Google Scholar] [CrossRef] [Green Version]
- Risør, M.W.; Juhl, D.W.; Bjerring, M.; Mathiesen, J.; Enghild, J.J.; Nielsen, N.C.; Otzen, D.E. Critical Influence of Cosolutes and Surfaces on the Assembly of Serpin-Derived Amyloid Fibrils. Biophys. J. 2017, 113, 580–596. [Google Scholar] [CrossRef] [Green Version]
- Welton, T. Room-Temperature Ionic Liquids. Solvents for Synthesis and Catalysis. Chem. Rev. 1999, 99, 2071–2084. [Google Scholar] [CrossRef]
- Marsh, K.N.; Deer, A.; Wu, A.C.-T.; Tran, E.; Klamt, A. Room Temperature Ionic Liquids as Replacements for Conventional Solvents—A Review. Korean J. Chem. Eng. 2002, 19, 357–362. [Google Scholar] [CrossRef]
- Ghandi, K.A. Review of Ionic Liquids, Their Limits and Applications. Green Sustain. Chem. 2014, 4, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef]
- Summers, C.A.; Flowers, R.A. Protein renaturation by the liquid organic salt ethylammonium nitrate. Protein Sci. 2000, 9, 2001–2008. [Google Scholar] [CrossRef] [Green Version]
- Lange, C.; Patil, G.; Rudolph, R. Ionic liquids as refolding additives: N′-alkyl and N′-(ω-hydroxyalkyl) N-methylimidazolium chlorides. Protein Sci. 2005, 14, 2693–2701. [Google Scholar] [CrossRef]
- Wasserscheid, P.; Welton, T. (Eds.) Ionic Liquids in Synthesi; Wiley-VCH: Weinheim, Germany, 2003; pp. 9–44. [Google Scholar]
- Anderson, J.L.; Armstrong, D.W.; Wei, G.T. Ionic liquids in analytical chemistry. Anal Chem. 2006, 78, 2892–2902. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.L.; Zhang, J.M.; Jiang, J.; Chen, C.F.; Xi, F. Atom transfer radical copolymerization of n-hexylmaleimide and styrene in an ionic liquid. J. Polym. Sci. Part A 2002, 40, 3360–3366. [Google Scholar] [CrossRef]
- Kubisa, P. Application of ionic liquids as solvents for polymerization processes. Prog. Polym. Sci. 2004, 29, 3–12. [Google Scholar] [CrossRef]
- Chandra, P.; Shinde, S.S.; Biradar, A.V. Tailor made ionic liquids: Catalyst and media for organic transformations. Curr. Org. Chem. 2015, 19, 728–742. [Google Scholar] [CrossRef]
- Reddy, P.N.; Padmaja, P.; Reddy, B.V.S.; Rambabu, G. Ionic liquid/water mixture promoted organic transformations. RSC Adv. 2005, 5, 51035–51054. [Google Scholar] [CrossRef]
- Fuller, J.; Carkin, R.T.; Osteryoung, R.A. The Room Temperature Ionic Liquid 1-Ethyl-3-methylimidazolium Tetrafluoroborate: Electrochemical Couples and Physical Properties. J. Electrochem. Soc. 1997, 144, 3881–3886. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Shadjou, N.; Eskandani, M.; Guardia, M. de la Room-temperature ionic liquid-based electrochemical nanobiosensors. TrAC Trends Anal. Chem. 2012, 41, 58–74. [Google Scholar] [CrossRef]
- van Rantwijk, F.; Sheldon, R.A. Biocatalysis in Ionic Liquids. Chem. Rev. 2007, 107, 2757–2785. [Google Scholar] [CrossRef]
- Naushad, M.; Alothman, Z.A.; Khan, A.B.; Ali, M. Effect of ionic liquid on activity, stability, and structure of enzymes: A review. Int. J. Biol. Macromol. 2012, 51, 555–560. [Google Scholar] [CrossRef]
- Gao, W.W.; Zhang, F.X.; Zhang, G.X.; Zhou, C.H. Key factors affecting the activity and stability of enzymes in ionic liquids and novel applications in biocatalysis. Biochem. Eng. J. 2015, 99, 67–84. [Google Scholar] [CrossRef]
- Heimer, P.; Tietze, A.A.; Bohm, M.; Giernoth, R.; Kuchenbuch, A.; Stark, A.; Leipold, E.; Heinemann, S.H.; Kandt, C.; Imhof, D. Application of Room-Temperature Aprotic and Protic Ionic Liquids for xidative Folding of Cysteine-Rich Peptides. Chembiochem. 2014, 15, 2754–2765. [Google Scholar] [CrossRef]
- Wakayama, R.; Uchiyama, S.; Hall, D. Ionic liquids and protein folding—Old tricks for new solvents. Biophys. Rev. 2019, 11, 209–225. [Google Scholar] [CrossRef]
- Fujita, K.; MacFarlane, D.R.; Forsyth, M. Protein solubilising and stabilising ionic liquids. Chem. Commun. 2005, 38, 4804–4806. [Google Scholar] [CrossRef]
- Kumar, A.; Venkatesu, P. Does the stability of proteins in ionic liquids obey the Hofmeister series? J. Biol. Macrmol. 2014, 63, 244–253. [Google Scholar] [CrossRef]
- Patel, R.; Kumari, M.; Khan, A.B. Recent advances in the applications of ionic liquids in protein stability and activity: A review. Appl. Biochem. Biotechnol. 2014, 172, 3701–3720. [Google Scholar] [CrossRef]
- Kumar, A.; Venkatesu, P. Innovative aspects of protein stability in ionic liquid mixtures. Biophys. Rev. 2018, 10, 841–846. [Google Scholar] [CrossRef]
- Hwang, H.; Choi, H.; Kim, H.K.; Jo, D.H.; Kim, T.D. Ionic liquids promote amyloid formation from alpha-synuclein. Anal. Biochem. 2009, 386, 293–295. [Google Scholar] [CrossRef]
- Bae, S.Y.; Kim, S.; Hwang, H.; Kim, H.K.; Yoon, H.C.; Kim, J.H.; Lee, S.; Kim, T.D. Amyloid formation and disaggregation of α-synuclein and its tandem repeat (α-TR). Biochem. Biophys. Res. Commun. 2010, 400, 531–536. [Google Scholar] [CrossRef]
- Takekiyo, T.; Yamada, N.; Nakazawa, C.T.; Amo, Y.; Asano, A.; Yoshimura, Y. Formation of α-synuclein aggregates in aqueous ethylammonium nitrate solutions. Biopolymers 2020, 111, e23352. [Google Scholar] [CrossRef]
- Debeljuh, N.; Barrow, C.C.; Byrne, N. The impact of ionic liquids on amyloid fibrilization of Aβ16-22: Tuning the rate of fibrilization using a reverse Hofmeister strategy. Phys. Chem. Chem. Phys. 2011, 13, 16534–16536. [Google Scholar] [CrossRef] [Green Version]
- Takekiyo, T.; Yamada, N.; Amo, T.; Yoshimura, Y. Aggregation selectivity of amyloid β1-11 peptide in aqueous ionic liquid solutions. Peptide Sci. 2020, 112, e24138. [Google Scholar] [CrossRef]
- Takekiyo, A.; Yamaguchi, E.; Abe, H.; Yoshimura, Y. Suppression Effect on the Formation of Insulin Amyloid by the Use of Ionic Liquids. ACS Sustainable Chem. Eng. 2016, 4, 422–428. [Google Scholar] [CrossRef]
- Yoshida, K.; Zenin, T.; Fujiyoshi, A.; Sanada, Y.; Yamaguchi, T.; Murata, K.; Takata, S.I.; Hiroi, K.; Takekyio, T.; Yoshimura, Y. The effect of alkyl ammonium ionic liquids on thermal denaturation aggregation of β-lactoglobulin. J. Mol. liq. 2019, 293, 111477. [Google Scholar] [CrossRef]
- Islam, M.M.; Barik, S.; Sarkar, M. Probing the Interactions of 1-Alkyl-3-methylimidazolium Tetrafluoroborate (Alkyl = Octyl, Hexyl, Butyl, and Ethyl) Ionic Liquids with Bovine Serum Albumin: An Alkyl Chain Length- Dependent Study. J. Phys. Chem. B 2019, 123, 1512–1526. [Google Scholar] [CrossRef]
- Byrne, N.; Angell, C.A. Formation and dissolution of hen egg white lysozyme amyloid fibrils in protic ionic liquids. Chem. Commun. 2009, 9, 1046–1048. [Google Scholar] [CrossRef]
- Kalhor, H.R.; Kamizi, M.; Akbari, J.; Heydari, A. Inhibition of amyloid formation by ionic liquids: Ionic liquids affecting intermediate oligomers. Biomacromolecules 2009, 10, 2468–2475. [Google Scholar] [CrossRef]
- Basu, A.; Bhattacharya, S.C.; Kumar, G.S. Influence of the ionic liquid 1-butyl-3-methylimidazolium bromide on amyloid fibrillogenesis in lysozyme: Evidence from photophysical and imaging studies. Int. J. Biol. Macromol. 2018, 107, 2643–2649. [Google Scholar] [CrossRef]
- Silva, N.H.C.S.; Pinto, R.J.B.; Martins, M.A.; Marrucho, I.M.; Ferreira, R.; Correia, I.; Freire, C.S.R.; Marrucho, I.M. Ionic liquids as promoters of fast lysozyme fibrillation. J. Mol. Liq. 2018, 272, 456–467. [Google Scholar] [CrossRef]
- Pillai, V.V.S.; Benedetto, A. Ionic liquids in protein amyloidogenesis: A brief screenshot of the state-of-the-art. Biophys. Rev. 2018, 10, 847–852. [Google Scholar] [CrossRef]
- Marek, J.; Demjenova, E.; Tomori, Z.; Janacek, J.; Zolotova, I.; Valle, F.; Favre, M.; Dietler, G. Interactive measurement and characterization of DNA molecules by analysis of AFM images. Cytometry 2005, 63, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Barrett, W.A.; Mortensen, E.N. Interactive live-wire boundary extraction. Med. Image Anal. 1997, 1, 331–341. [Google Scholar] [CrossRef]
- Khurana, R.; Ionescu-Zanetti, C.; Pope, M.; Li, J.; Nielson, L.; Ramirez-Alvarado, M.; Regan, L.; Fink, A.L.; Carter, S.A. A general model for amyloid fibril assembly based on morphological studies using atomic force microscopy. Biophys. J. 2003, 85, 1135–1144. [Google Scholar] [CrossRef] [Green Version]
- Niu, L.J. Investigating Self-Assembled Protein Nanotubes Using Atomic Force Microscopy. Chapter 3.2. Fibrillization Processes of Lysozyme. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2009; pp. 76–89. [Google Scholar]
- Bye, J.W.; Falconer, R.J. Thermal stability of lysozyme as a function of ion concentration: A reappraisal of the relationship between the Hofmeister series and protein stability. Protein Sci. 2013, 22, 1563–1570. [Google Scholar] [CrossRef] [Green Version]
- Brudar, S.; Hribar-Lee, B. The Role of Buffers in Wild-Type HEWL Amyloid Fibril Formation Mechanism. Biomolecules 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, N.H.C.S.; Vilela, C.; Pinto, R.J.B.; Martins, M.A.; Marrucho, I.M.; Freire, C.S.R. Tuning lysozyme nanofibers dimensions using deep eutectic solvents for improved reinforcement ability. Int. J. Biol. Macromol. 2018, 115, 518–527. [Google Scholar] [CrossRef]
- Sulatskaya, A.I.; Rodina, N.P.; Povarova, O.I.; Kuznetsova, I.M.; Turoverov, K.K. Different conditions of fibrillogenesis cause polymorphism of lysozyme amyloid fibrils. J. Mol. Struct. 2017, 1140, 52–58. [Google Scholar] [CrossRef]
- Poniková, S.; Antošová, A.; Demjén, E.; Sedláková, D.; Marek, J.; Varhač, R.; Gažová, Z.; Sedlák, E. Lysozyme stability and amyloid fibrillization dependence on Hofmeister anions in acidic pH. J. Biol. Inorg. Chem. 2015, 20, 921–933. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and Reference Databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Singh, O.; Lee, P.Y.; Matysiak, S.; Bermudez, H. Dual mechanism of ionic liquid-induced protein unfolding. Phys. Chem. Chem. Phys. 2020, 22, 19779–19786. [Google Scholar] [CrossRef] [PubMed]
- Ow, S.Y.; Dunstan, D.E. The effect of concentration, temperature and stirring on hen egg white lysozyme amyloid formation. Soft Matter 2013, 9, 9692–9701. [Google Scholar] [CrossRef] [PubMed]
- Klein-Seetharaman, J.; Oikawa, M.; Grimshaw, S.; Duchardt, E.; Ueda, T.; Imoto, T.; Smith, L.; Dobson, C.; Schwalbe, H. Long-range interactions within a nonnative protein. Science 2002, 295, 1719–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Li, S.; Xue, Z.; Hao, M.; Mu, T. Quantifying the hydrogen-bonding interaction between cation andanion of pure [EMIM][Ac] and evidencing the ion pairs existence in its extremely diluted water solution: Via 13C, 1H, 15N and 2D NMR. J. Mol. Struct. 2015, 1079, 120–129. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, D.; Jucker, M. The amyloid state of proteins in human diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, A.W.P.; Debelouchina, G.T.; Bayro, M.J.; Clare, D.K.; Caporini, M.A.; Bajaj, V.S.; Jaroniec, C.P.; Wang, L.; Ladizhansky, V.; Müller, S.A.; et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc. Natl. Acad. Sci. USA 2013, 110, 5468–5473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satish, L.; Rana, S.; Arakha, M.; Rout, L.; Ekka, B.; Jha, S.; Dash, P.; Sahoo, H. Impact of imidazolium-based ionic liquids on the structure and stability of lysozyme. Spectrosc. Lett. 2016, 49, 383–390. [Google Scholar] [CrossRef]
- Bisht, M.; Kumar, A.; Venkatesu, P. Analysis of the driving force that rule the stability of lysozyme in alkylammonium-based ionic liquids. Int. J. Biol. Macromol. 2015, 81, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Singh, U.K.; Beg, I.; Alanazi, A.M.; Khan, A.A.; Patel, R. Effect of cations and anions of ionic liquids on the stability and activity of lysozyme: Concentration and temperature effect. J. Mol. Liq. 2018, 272, 253–263. [Google Scholar] [CrossRef]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Matas, J.; Shao, Z.; Kittler, J. Estimation of curvature and tangent direction by median filtered differencing. Lect. Notes Comput. Sci. 1995, 974, 83–88. [Google Scholar]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Barca, G.M.J.; Bertoni, C.; Carrington, L.; Datta, D.; De Silva, N.; Deustua, J.E.; Fedorov, D.G.; Gour, J.R.; Gunina, A.O.; Guidez, E.; et al. Recent developments in the general atomic and molecular electronic structure system. J. Chem. Phys. 2020, 152, 154102. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, R.; Mahasenan, K.; Pavlovicz, R.; Li, C.; Tjarks, W. Carborane clusters in computational drug design: A comparative docking evaluation using AutoDock, FlexX, Glide, and Surflex. J. Chem. Inf. Model 2009, 49, 1581–1589. [Google Scholar] [CrossRef] [Green Version]


| tlag (min) | thalf (min) | kagg (min−1) | R | |
|---|---|---|---|---|
| EMIM-ac (% v/v) | ||||
| 0.5 | 558.60 ± 4.40 | 613.50 ± 2.20 | 0.036 ± 0.002 | 0.998 |
| 1 | 77.20 ± 4.10 | 143.10 ± 1.70 | 0.031 ± 0.003 | 0.996 |
| 5 | 19.80 ± 0.30 | 21.90 ± 0.20 | 0.925 ± 0.100 | 0.993 |
| EMIM-BF4 (% v/v) | ||||
| 0.5 | 67.00 ± 1.80 | 107.80 ± 1.40 | 0.049 ± 0.003 | 0.998 |
| 1 | 64.80 ± 1.50 | 80.30 ± 0.70 | 0.130 ± 0.010 | 0.997 |
| 5 | 11.30 ± 1.90 | 18.10 ± 0.80 | 0.290 ± 0.050 | 0.996 |
| Td (°C) | ΔHcal (kJ/mol) | ΔHvH (kJ/mol) | ΔHcal/ΔHvH | R (%) | |
|---|---|---|---|---|---|
| Lys (2 mg/mL) | 66.57 ± 0.02 | 432.20 ± 1.50 | 448.50 ± 1.90 | 1.04 | 98 |
| EMIM-ac (% v/v) | |||||
| 0.5 | 66.44 ± 0.02 | 399.90 ± 2.70 | 512.60 ± 3.20 | 1.28 | 95 |
| 1 | 62.56 ± 0.04 | 363.20 ± 2.50 | 445.90 ± 3.90 | 1.23 | 93 |
| 5 | 60.05 ± 0.04 | 270.10 ± 2.20 | 449.20 ± 6.40 | 1.66 | 72 |
| EMIM-BF4 (% v/v) | |||||
| 0.5 | 62.92 ± 0.03 | 362.20 ± 2.20 | 413.60 ± 3.20 | 1.14 | 96 |
| 1 | 60.38 ± 0.03 | 307.70 ± 3.00 | 412.90 ± 1.40 | 1.34 | 95 |
| 5 | 56.79 ± 0.03 | 177.40 ± 1.80 | 489.40 ± 3.90 | 2.42 | 89 |
| Native Lysozyme | Lysozymein ILs | 1% EMIM-ac | 1% EMIM-BF4 | |
|---|---|---|---|---|
| α-helix (%) | 34 | native | 31 | 31 |
| fibrilar | 5 | 9 | ||
| β-sheet (%) | 16 | native | 19 | 21 |
| fibrilar | 45 | 42 | ||
| β-turn (%) | 21 | native | 21 | 18 |
| fibrilar | 22 | 25 | ||
| unordered (%) | 29 | native | 29 | 30 |
| fibrilar | 28 | 24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedunova, D.; Antosova, A.; Marek, J.; Vanik, V.; Demjen, E.; Bednarikova, Z.; Gazova, Z. Effect of 1-Ethyl-3-methylimidazolium Tetrafluoroborate and Acetate Ionic Liquids on Stability and Amyloid Aggregation of Lysozyme. Int. J. Mol. Sci. 2022, 23, 783. https://doi.org/10.3390/ijms23020783
Fedunova D, Antosova A, Marek J, Vanik V, Demjen E, Bednarikova Z, Gazova Z. Effect of 1-Ethyl-3-methylimidazolium Tetrafluoroborate and Acetate Ionic Liquids on Stability and Amyloid Aggregation of Lysozyme. International Journal of Molecular Sciences. 2022; 23(2):783. https://doi.org/10.3390/ijms23020783
Chicago/Turabian StyleFedunova, Diana, Andrea Antosova, Jozef Marek, Vladimir Vanik, Erna Demjen, Zuzana Bednarikova, and Zuzana Gazova. 2022. "Effect of 1-Ethyl-3-methylimidazolium Tetrafluoroborate and Acetate Ionic Liquids on Stability and Amyloid Aggregation of Lysozyme" International Journal of Molecular Sciences 23, no. 2: 783. https://doi.org/10.3390/ijms23020783






