Lignin Nanoparticles Deliver Novel Thymine Biomimetic Photo-Adducts with Antimelanoma Activity
Abstract
:1. Introduction
2. Results
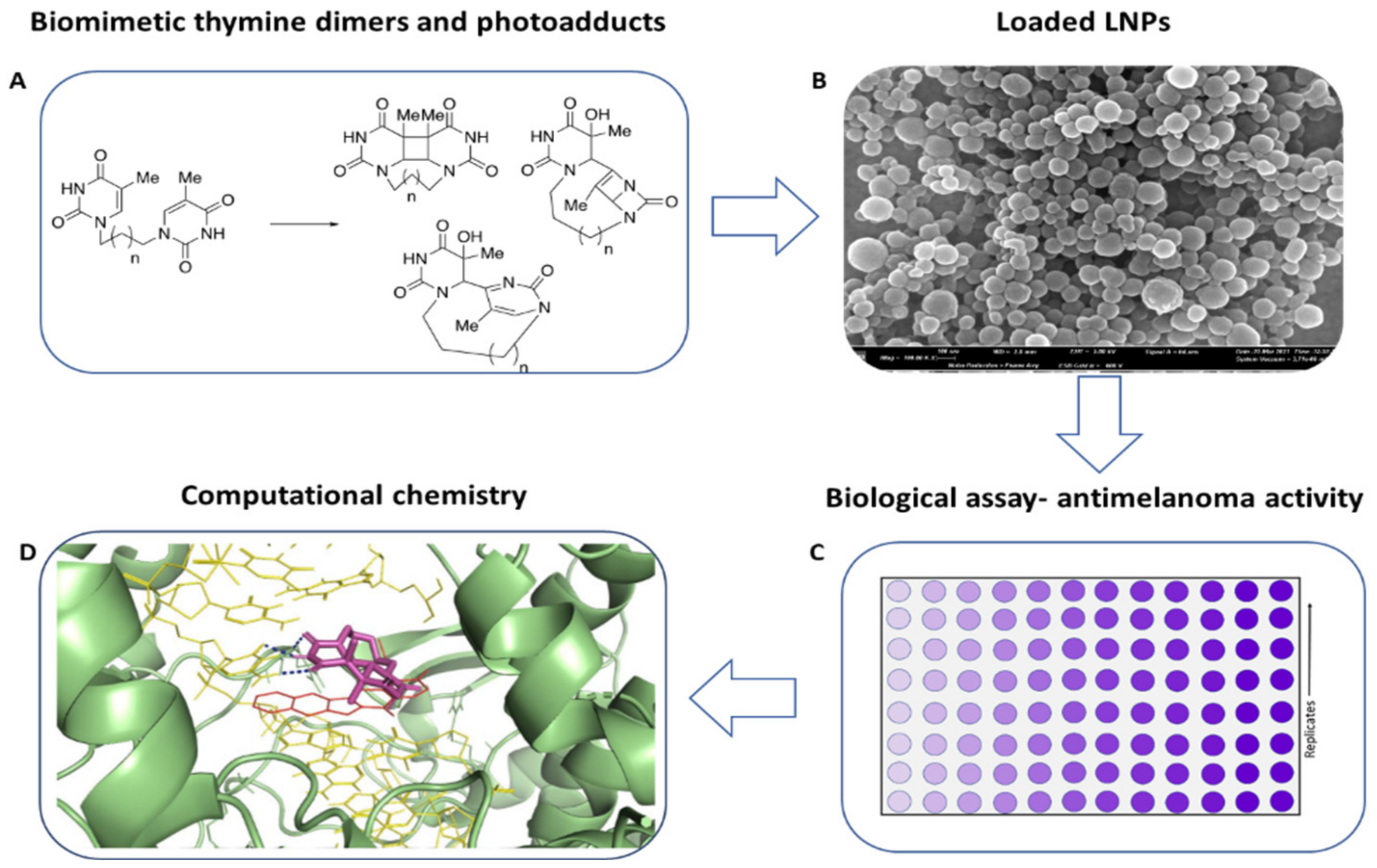
2.1. Summary and Concise Description of the Study
2.2. Synthesis of CPD, (6-4)PP, and DV Photo-Adducts by UV-Irradiation of Biomimetic Thymine Dimers 4a-d
2.3. Lignin Nanoparticles as a Drug Delivery System for Biomimetic Thymine Dimers and Photo-Adducts
2.4. UV Shielding Capacity of Loaded LNPs
2.5. Releasing Properties
2.6. Biological Activity
2.7. In Silico Molecular Docking Analysis
3. Materials and Methods
3.1. Synthesis and Characterization of Thymine Biomimetic Dimer 4a-d
3.2. Synthesis of Photo-Adducts CPDs 6a-d, (6-4)PPs 7a-d, and DVs 8a-d
3.3. Preparation and Loading of Lignin Nanoparticles
3.4. UV Photo-Protective Effect of Loaded LNPs
3.5. Kinetic Release of Lignin Nanoparticles
3.6. Biological Assay
3.7. In Silico Molecular Docking Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shen, Y.; Stanislauskas, M.; Li, G.; Zheng, D.; Liu, L. Epigenetic and genetic dissections of UV-induced global gene dysregulation in skin cells through multi-omics analyses. Sci. Rep. 2017, 7, 42646. [Google Scholar] [CrossRef] [Green Version]
- Ravanat, J.L.; Douki, T.; Cadet, J. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B 2001, 63, 88–102. [Google Scholar] [CrossRef]
- Cadet, J.; Mouret, S.; Ravanat, J.L.; Douki, T. Photoinduced damage to cellular DNA: Direct and photosensitized reactions. Photochem. Photobiol. 2012, 88, 1048–1065. [Google Scholar] [CrossRef]
- Saha, L.K.; Wakasugi, M.; Akter, S.; Akter, S.; Prasad, R.; Wilson, S.H.; Shimizu, N.; Sasanuma, H.; Huang, S.N.; Agama, K.; et al. Topoisomerase I-driven repair of UV-induced damage in NER-deficient cells. Proc. Natl. Acad. Sci. USA 2020, 117, 14412–14420. [Google Scholar] [CrossRef]
- Cadet, J.; Wagner, J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef]
- Napolitano, A.; Panzella, L.; Monfrecola, G.; d’Ischia, M. Pheomelanin-induced oxidative stress: Bright and dark chemistry bridging red hair phenotype and melanoma. Pigment Cell Melanoma Res. 2014, 27, 721–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giglia-Mari, G.; Zotter, A.; Vermeulen, W. DNA damage response. Cold Spring Harb. Perspect. Biol. 2011, 3, a000745. [Google Scholar] [CrossRef] [PubMed]
- Athar, M.; Kim, A.L.; Ahmad, N.; Mukhtar, H.; Gautier, J.; Bickers, D.R. Mechanism of ultraviolet B-induced cell cycle arrest in G2/M phase in immortalized skin keratinocytes with defective p53. Biochem. Biophys. Res. Commun. 2000, 277, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.S.; Harry, E.L.; Liljendahl, T.S.; Segerbäck, D. DNA nucleotide excision repair, where do all the cyclobutane pyrimidine dimers go? Cell Cycle 2013, 12, 1642. [Google Scholar] [CrossRef] [Green Version]
- Kemp, M.G.; Hu, J. PostExcision Events in Human Nucleotide Excision Repair. Photochem. Photobiol. 2016, 93, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Douki, T. Formation of UV-induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018, 17, 1816–1841. [Google Scholar] [CrossRef]
- Schuch, A.P.; Moreno, N.C.; Schuch, N.J.; Menck, C.; Garcia, C. Sunlight damage to cellular DNA: Focus on oxidatively generated lesions. Free Radic. Biol. Med. 2017, 107, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Choi, J.H.; Gaddameedhi, S.; Kemp, M.G.; Reardon, J.T.; Sancar, A. Nucleotide excision repair in human cells: Fate of the excised oligonucleotide carrying DNA damage in vivo. J. Biol. Chem. 2013, 288, 20918–20926. [Google Scholar] [CrossRef] [Green Version]
- Kciuk, M.; Marciniak, B.; Mojzych, M.; Kontek, R. Focus on UV-Induced DNA Damage and Repair-Disease Relevance and Protective Strategies. Int. J. Mol. Sci. 2020, 21, 7264. [Google Scholar] [CrossRef] [PubMed]
- Lanza, A.; Tornaletti, S.; Rodolfo, C.; Scanavini, M.C.; Pedrini, A.M. Human DNA topoisomerase I-mediated cleavages stimulated by ultraviolet light-induced DNA damage. J. Biol. Chem. 1996, 271, 6978–6986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, D.; Rosenstein, B.S.; Muller, M.T. Ultraviolet-induced DNA damage stimulates topoisomerase I-DNA complex formation in vivo: Possible relationship with DNA repair. Cancer Res. 1998, 58, 976–984. [Google Scholar] [PubMed]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer. 2006, 6, 789–802. [Google Scholar] [CrossRef]
- Pommier, Y.; Barcelo, J.M.; Rao, V.A.; Sordet, O.; Jobson, A.G.; Thibaut, L.; Miao, Z.H.; Seiler, J.A.; Zhang, H.; Marchand, C.; et al. Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2006, 81, 179–229. [Google Scholar] [CrossRef] [Green Version]
- Beiu, C.; Giurcaneanu, C.; Grumezescu, A.M. Nanosystems for Improved Targeted Therapies in Melanoma. J. Clin. Med. 2020, 9, 318. [Google Scholar] [CrossRef] [Green Version]
- Vlasceanu, G.M.; Victor, L.; Maricica, H.; Raluca, T.; Vlad, O.; Gheorghe, I.; Bolocan, A.; Grumezescu, A.M.; Holban, A.M. Nanostructures for cancer therapy: From targeting to selective toxicology. In Nanostructures for Cancer Therapy; Ficai, A., Grumezescu, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 831–847. [Google Scholar] [CrossRef]
- Piccinino, D.; Capecchi, E.; Tomaino, E.; Gabellone, S.; Gigli, V.; Avitabile, D.; Saladino, R. Lignin as Green Antioxidant and UV Shielding Ingredient for Sunscreen Applications. Antioxidants 2021, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Dreis, C.D.; Vince, R. Photoprotection of DNA (in vitro) by acyclothymidine dinucleosides. Bioorg. Med. Chem. Lett. 2013, 23, 620–623. [Google Scholar] [CrossRef]
- Raza, A.; Ericson, M.E.; Nugent, J.S.; Dreis, C.D.; Vince, R. A bio-mimetic approach to DNA photoprotection. J. Investig. Dermatol. 2014, 134, 559–562. [Google Scholar] [CrossRef] [Green Version]
- Nugent, J.S.; Vince, R.; Raza, A. Topical Acyclothymidine Dinucleosides (aTds) Promote Non-UV-Mediated Endogenous Defense Mechanisms in Guinea Pig Skin. J. Investig. Dermatol. 2015, 135, 1687–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moidoveanu, S.C.; David, V. (Eds.) Derivatization Reactions for Analytes with Various Functional Groups. In Sample Preparation in Chromatography; Elsevier: Amsterdam, The Netherlands, 2002; Volume 65, pp. 639–845. [Google Scholar] [CrossRef]
- Hanessian, S.; Saladino, R.; Nunez, J.C. On the binding site of quinolone antibacterials. An attempt to probe the shen model. Bioorg. Med. Chem. Lett. 1996, 6, 2333–2338. [Google Scholar] [CrossRef]
- Varghese, A.J.; Wang, S.Y. Thymine-thymine adduct as a photoproduct of thymine. Science 1968, 160, 186–187. [Google Scholar] [CrossRef] [PubMed]
- Varghese, A.J. Photochemistry of thymidine in ice. Biochemistry 1970, 9, 4781–4787. [Google Scholar] [CrossRef]
- Wagner, P.J.; Bucheck, D.J. Photodimerization of Thymine and Uracil in Acetonitrile. J. Am. Chem. Soc. 1970, 92, 181–185. [Google Scholar] [CrossRef]
- Taylor, J.S.; Lu, H.F.; Kotyk, J.J. Quantitative conversion of the (6-4) photoproduct of TpdC to its Dewar valence isomer upon exposure to simulated sunlight. Photochem. Photobiol. 1990, 51, 161–167. [Google Scholar] [CrossRef]
- Kohei, T.; Chikahide, M.; Fumio, H.; Shigenori, I. Chemical synthesis and translesion replication of a cis-syn cyclobutane thymine-uracil dimer. Nucleic Acids Res. 2004, 32, 1738–1745. [Google Scholar] [CrossRef] [Green Version]
- Tohnai, N.; Miyata, M.; Yasui, N.; Mochizuki, E.; Kai, K.; Inaki, Y. Photodimerization of Thymine Derivatives in Single Crystal. J. Photopolym. Sci. Technol. 1998, 11, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Cadet, J.; Voituriez, L.; Frank, E.; Hruska, L.S.K.; de Leeuw, F.A.A.M.; Altona, A. Characterization of thymidine ultraviolet photoproducts. Cyclobutane dimers and 5,6-dihydrothymidines. Can. J. Chem. 1985, 63, 2861. [Google Scholar] [CrossRef] [Green Version]
- Gut, I.G.; Wood, P.D.; Redmond, R.W. Interaction of triplet photosensitizers with nucleotides and DNA in aqueous solution at room temperature. J. Am. Chem. Soc. 1996, 118, 2366–2373. [Google Scholar] [CrossRef]
- Ramamurthy, V.; Sivaguru, J. Supramolecular Photochemistry as a Potential Synthetic Tool: Photocycloaddition. Chem. Rev. 2016, 116, 9914–9993. [Google Scholar] [CrossRef] [PubMed]
- Epe, B.; Henzl, H.; Adam, W.; Saha-Moller, C.R. Endonuclease-sensitive DNA modifications induced by acetone and acetophenone as photosensitizers. Nucleic Acids Res. 1993, 21, 863–869. [Google Scholar] [CrossRef] [Green Version]
- Kan, L.S.; Voituriez, L.; Cadet, J. The Dewar valence isomer of the (6-4) photoadduct of thymidylyl-(3’-5’)-thymidine monophosphate: Formation, alkaline lability and conformational properties. J. Photochem. Photobiol. B 1992, 12, 339–357. [Google Scholar] [CrossRef]
- Nagpal, A.; Dhankhar, D.; Cesario, T.C.; Li, R.; Chen, J.; Rentzepis, P.M. Thymine dissociation and dimer formation: A Raman and synchronous fluorescence spectroscopic study. Proc. Natl. Acad. Sci. USA 2021, 118, e2025263118. [Google Scholar] [CrossRef]
- Shigenori, I.; Masato, S.; Hiroyuki, K.; Eiko, O. Synthesis of a Phosphoramidite Coupling Unit of the Pyrimidine (6-4) Pyrimidone Photoproduct and Its Incorporation into Oligodeoxynucleotides. J. Am. Chem. Soc. 1996, 118, 7642–7643. [Google Scholar] [CrossRef]
- Yokoyama, H.; Mizutani, R. Structural biology of DNA (6-4) photoproducts formed by ultraviolet radiation and interactions with their binding proteins. Int. J. Mol. Sci. 2014, 15, 20321–20338. [Google Scholar] [CrossRef] [Green Version]
- Mu, W.; Han, Q.; Luo, Z.; Wang, Y. Production of cis-syn thymine-thymine cyclobutane dimer oligonucleotide in the presence of acetone photosensitizer. Anal. Biochem. 2006, 353, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Gutierrez-Bayona, N.E.; Taylor, J.S. The effect of flanking bases on direct and triplet sensitized cyclobutane pyrimidine dimer formation in DNA depends on the dipyrimidine, wavelength and the photosensitizer. Nucleic Acids Res. 2021, 49, 4266–4280. [Google Scholar] [CrossRef]
- Goto, N.; Bazar, G.; Kovacs, Z.; Kunisada, M.; Morita, H.; Kizaki, S.; Sugiyama, H.; Tsenkova, R.; Nishigori, C. Detection of UV-induced cyclobutane pyrimidine dimers by near-infrared spectroscopy and aquaphotomics. Sci. Rep. 2015, 5, 11808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Rabbi, M.; Kim, M.; Ke, C.; Lee, W.; Clark, R.L.; Mieczkowski, P.A.; Marszalek, P.E. UVA generates pyrimidine dimers in DNA directly. Biophys. J. 2009, 96, 1151–1158. [Google Scholar] [CrossRef] [Green Version]
- Mouret, S.; Baudouin, C.; Charveron, M.; Favier, A.; Cadet, J.; Douki, T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. USA 2006, 103, 13765–13770. [Google Scholar] [CrossRef] [Green Version]
- Piccinino, D.; Capecchi, E.; Botta, L.; Bizzarri, B.M.; Bollella, P.; Antiochia, R.; Saladino, R. Layer-by-Layer Preparation of Microcapsules and Nanocapsules of Mixed Polyphenols with High Antioxidant and UV-Shielding Properties. Biomacromolecules 2018, 19, 3883–3893. [Google Scholar] [CrossRef]
- Alqahtani, M.S.; Alqahtani, A.; Kazi, M.; Ahmad, M.Z.; Alahmari, A.; Alsenaidy, M.A.; Syed, S. Wound-healing potential of curcumin loaded lignin nanoparticles. J. Drug. Deliv. Sci. Technol. 2020, 60, 102020. [Google Scholar] [CrossRef]
- Siddiqui, L.; Bag, J.; Seetha; Mittal, D.; Leekha, A.; Mishra, H.; Mishra, M.; Verma, A.K.; Mishra, P.K.; Ekielski, A.; et al. Assessing the potential of lignin nanoparticles as drug carrier: Synthesis, cytotoxicity and genotoxicity studies. Int. J. Biol. Macromol. 2020, 152, 786–802. [Google Scholar] [CrossRef]
- Li, H.; Deng, Y.; Wu, H.; Ren, Y.; Qiu, X.; Zheng, D.; Li, C. Self-assembly of kraft lignin into nanospheres in dioxane-water mixtures. Holzforschung 2016, 70, 725–731. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, D.Y.; Choi, I.G.; Choi, J.W. Investigation of Molecular Size Effect on the Formation of Lignin Nanoparticles by Nanoprecipitation. Appl. Sci. 2020, 10, 4910. [Google Scholar] [CrossRef]
- Sipponen, M.H.; Lange, H.; Crestini, C.; Henn, A.; Österberg, M. Lignin for Nano- and Microscaled Carrier Systems: Applications, Trends, and Challenges. ChemSusChem 2019, 12, 2039–2054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Terrasson, V.; Guénin, E. Lignin Nanoparticles and Their Nanocomposites. Nanomaterials 2021, 11, 1336. [Google Scholar] [CrossRef] [PubMed]
- Piccinino, D.; Capecchi, E.; Delfino, I.; Crucianelli, M.; Conte, N.; Avitabile, D.; Saladino, R. Green and Scalable Preparation of Colloidal Suspension of Lignin Nanoparticles and Its Application in Eco-friendly Sunscreen Formulations. ACS Omega 2021, 6, 21444–21456. [Google Scholar] [CrossRef] [PubMed]
- Parisi, N.; Paz-Alvarez, M.; Matts, P.J.; Lever, R.; Hadgraft, J.; Lane, M.E. Topical delivery of hexamidine. Int. J. Pharm. 2016, 506, 332–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Liu, R.; Hu, L.Q.; Zou, Z.F.; Si, C.L. Lignin Nanoparticle as a Novel Green Carrier for the Efficient Delivery of Resveratrol. ACS Sustain. Chem. Eng. 2017, 5, 8241–8249. [Google Scholar] [CrossRef]
- Alqahtani, M.S.; Alqahtani, A.H.; Al-Thabit, A.; Roni, M.A.; Syed, R. Novel lignin nanoparticles for oral drug delivery. J. Mater. Chem. B. 2019, 7, 4461–4473. [Google Scholar] [CrossRef]
- Sipponen, M.H.; Lange, H.; Ago, M.; Crestini, C. Understanding Lignin Aggregation Processes. A Case Study: Budesonide Entrapment and Stimuli Controlled Release from Lignin Nanoparticles. ACS Sustain. Chem. Eng. 2018, 6, 9342–9351. [Google Scholar] [CrossRef]
- Wang, M.; Yang, D.; Xu, Q.; Li, P.; Yi, C.; Qian, Y.; Qiu, X. Highly efficient evaporation method to prepare pH-responsive lignin-hollow-nanosphere with controllable size and its application in oral drug delivery. Ind. Crops Prod. 2021, 162, 113230. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, X.; Shi, Y.; Gao, B.; Wu, J.; Kirk, T.B.; Xu, J.; Xue, W. Green synthesis of lignin nanoparticle in aqueous hydrotropic solution toward broadening the window for its processing and application. Chem. Eng. J. 2018, 346, 217–225. [Google Scholar] [CrossRef]
- Andeme Ela, R.C.; Tajiri, M.; Newberry, N.K.; Heiden, P.A.; Ong, R.G. Double-Shell Lignin Nanocapsules Are a Stable Vehicle for Fungicide Encapsulation and Release. ACS Sustain. Chem. Eng. 2020, 8, 17299–17306. [Google Scholar] [CrossRef]
- Tao, J.; Chow, S.F.; Zheng, Y. Application of flash nanoprecipitation to fabricate poorly water-soluble drug nanoparticles. Acta Pharm. Sin. B. 2019, 9, 4–18. [Google Scholar] [CrossRef]
- D’Addio, S.M.; Prud’homme, R.K. Controlling drug nanoparticle formation by rapid precipitation. Adv. Drug Deliv. Rev. 2011, 63, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.K.; Ekielski, A. The Self-Assembly of Lignin and Its Application in Nanoparticle Synthesis: A Short Review. Nanomaterials 2019, 9, 243. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Feng, X.; Yang, D.; Yi, C.; Qiu, X. Pi-Pi stacking of the aromatic groups in lignosulfonates. BioResources 2012, 7, 1145–1156. [Google Scholar] [CrossRef]
- Davies, R.J.; Malone, J.F.; Gan, Y.; Cardin, C.J.; Lee, M.P.; Neidle, S. High-resolution crystal structure of the intramolecular d(TpA) thymine-adenine photoadduct and its mechanistic implications. Nucleic Acids Res. 2007, 35, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Di Pasquale, N.; Marchisio, D.L.; Barresi, A.A.; Carbone, P. Solvent Structuring and Its Effect on the Polymer Structure and Processability: The Case of Water-Acetone Poly-ε-caprolactone Mixtures. J. Phys. Chem. B 2014, 118, 13258–13267. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Deng, Y.; Liu, B.; Ren, Y.; Liang, J.; Qian, Y.; Qiu, X.; Li, C.; Zheng, D. Preparation of Nanocapsules via the Self-Assembly of Kraft Lignin: A Totally Green Process with Renewable Resources. ACS Sustain. Chem. Eng. 2016, 4, 1946–1953. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, X.; Qian, Y.; Xiong, W.; Yang, D. pH-responsive lignin-based complex micelles: Preparation, characterization and application in oral drug delivery. Chem. Eng. J. 2017, 327, 1176–1183. [Google Scholar] [CrossRef]
- Zhou, Y.; Han, Y.; Li, G.; Yang, S.; Xiong, F.; Chu, F. Preparation of targeted lignin-based hollow nanoparticles for the delivery of doxorubicin. Nanomaterials 2019, 9, 188. [Google Scholar] [CrossRef] [Green Version]
- Yiamsawas, D.; Kangwansupamonkon, W.; Kiatkamjornwong, S. Lignin-based nanogels for the release of payloads in alkaline conditions. Eur. Polym. J. 2021, 145, 110241. [Google Scholar] [CrossRef]
- Patrick, M.H. Studies on thymine-derived UV photoproducts in DNA-I. Formation and biological role of pyrimidine adducts in DNA. Photochem. Photobiol. 1977, 25, 357–372. [Google Scholar] [CrossRef]
- Strozyk, E.; Kulms, D. The role of AKT/mTOR pathway in stress response to UV-irradiation: Implication in skin carcinogenesis by regulation of apoptosis, autophagy and senescence. Int. J. Mol. Sci. 2013, 14, 15260–15285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegedűs, C.; Boros, G.; Fidrus, E.; Kis, G.N.; Antal, M.; Juhász, T.; Janka, E.A.; Jankó, L.; Paragh, G.; Emri, G.; et al. PARP1 Inhibition Augments UVB-Mediated Mitochondrial Changes—Implications for UV-Induced DNA Repair and Photocarcinogenesis. Cancers 2020, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Emmert, S.; Kobayashi, N.; Khan, S.G.; Kraemer, K.H. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc. Natl. Acad. Sci. USA 2000, 97, 2151–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, Y.N.; Shiomi, N.; Koike, M.; Ikawa, M.; Okabe, M.; Hirota, S.; Kitamura, Y.; Kitagawa, M.; Matsunaga, T.; Nikaido, O.; et al. Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol. Cell. Biol. 2020, 19, 2366–2372. [Google Scholar] [CrossRef] [Green Version]
- Hiramoto, T.; Matsunaga, T.; Ichihashi, M.; Nikaido, O.; Fujiwara, Y.; Mishima, Y. Repair of 254 nm ultraviolet-induced (6-4) photoproducts: Monoclonal antibody recognition and differential defects in xeroderma pigmentosum complementation groups A, D, and variant. J. Investig. Dermatol. 1989, 93, 703–706. [Google Scholar] [CrossRef] [Green Version]
- Köberle, B.; Roginskaya, V.; Wood, R.D. XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair 2006, 5, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Nakane, H.; Takeuchi, S.; Yuba, S.; Saijo, M.; Nakatsu, Y.; Murai, H.; Nakatsuru, Y.; Ishikawa, T.; Hirota, S.; Kitamura, Y. High incidence of ultraviolet-B-or chemical-carcinogen-induced skin tumors in mice lacking the xeroderma pigmentosum group A gene. Nature 1995, 377, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Jhappan, C.; Noonan, F.; Merlino, G. Ultraviolet radiation and cutaneous malignant melanoma. Oncogene 2003, 22, 3099–3112. [Google Scholar] [CrossRef] [Green Version]
- Botta, L.; Filippi, S.; Bizzarri, B.M.; Zippilli, C.; Meschini, R.; Pogni, R.; Baratto, M.C.; Villanova, L.; Saladino, R. Synthesis and Evaluation of Artemisinin-Based Hybrid and Dimer Derivatives as Antimelanoma Agents. ACS Omega 2020, 5, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Kotla, N.G.; Singh, S.; Maddiboyina, B.; Sunnapu, O.; Webster, T.J. A novel dissolution media for testing drug release from a nanostructured polysaccharide-based colon specific drug delivery system: An approach to alternative colon media. Int. J. Nanomed. 2016, 11, 1089–1095. [Google Scholar] [CrossRef] [Green Version]
- Badmus, J.A.; Ekpo, O.E.; Hussein, A.A.; Meyer, M.; Hiss, D.C. Cytotoxic and cell cycle arrest properties of two steroidal alkaloids isolated from Holarrhena floribunda (G. Don) T. Durand & Schinz leaves. BMC Complement Altern. Med. 2019, 19, 112. [Google Scholar] [CrossRef] [Green Version]
- Vasconcelos, Z.S.; Ralph, A.; Calcagno, D.Q.; Dos Santos Barbosa, G.; Nascimento Pedrosa, T.; Antony, L.P.; de Arruda Cardoso Smith, M.; de Lucas Chazin, E.; Vasconcelos, T.; Montenegro, R.C.; et al. Anticancer potential of benzothiazolic derivative (E)-2-((2-(benzo[d]thiazol-2-yl)hydrazono)methyl)-4-nitrophenol against melanoma cells. Toxicol. In Vitro 2018, 50, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Staker, B.L.; Feese, M.D.; Cushman, M.; Pommier, Y.; Zembower, D.; Stewart, L.; Burgin, A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005, 48, 2336–2345. [Google Scholar] [CrossRef]
- Pal, S.; Kumar, V.; Kundu, B.; Bhattacharya, D.; Preethy, N.; Reddy, M.P.; Talukdar, A. Ligand-based Pharmacophore Modeling, Virtual Screening and Molecular Docking Studies for Discovery of Potential Topoisomerase I Inhibitors. Comput. Struct. Biotechnol. 2019, 17, 291–310. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Botta, L.; Filippi, S.; Zippilli, C.; Cesarini, S.; Bizzarri, B.M.; Cirigliano, A.; Rinaldi, T.; Paiardini, A.; Fiorucci, D.; Saladino, R.; et al. Artemisinin Derivatives with Antimelanoma Activity Show Inhibitory Effect against Human DNA Topoisomerase 1. ACS Med. Chem. Lett. 2020, 11, 1035–1040. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/EWater Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, R.; Matsuno, S.; Sakagami, H.; Okada, Y.; Shirataki, Y. Search of new cytotoxic crude materials against human oral squamous cell carcinoma using 1H NMR-based metabolomics. Anticancer Res. 2014, 34, 4117–4120. [Google Scholar]






| Entry | Conditions a | Product(s) | Spacer b | Conversion (%) | Yield (%) |
|---|---|---|---|---|---|
| 1 | A | 4a (5a) | 3 | 14 | 22 (78) |
| 2 | B | 4a (5a) | 3 | >99 | 85 (8.7) |
| 3 | C | 4b (5b) | 4 | >99 | 89.6 (10.4) |
| 4 | D | 4c (5c) | 5 | >99 | 83.5 (16.5) |
| 5 | E | 4d (5d) | 6 | >99 | 68 (32) |
| Entry | Substrate | Condition | Conversion (%) | TS:CS ratio a | Product(s) (%) | Yield (%) |
|---|---|---|---|---|---|---|
| 1 | 4a | A | >99% | 4.0:1.0 | 6a (7a) [8a] | 20 (12) [15] |
| 2 | 4a | B | >99% | 4.3:0.9 | 6a (7a) [8a] | 26 (16) [19] |
| 3 | 4a | C | >99% | 4.2:0.9 | 6a (7a) [8a] | 31 (17) [28] |
| 4 | 4b | C | >99% | 3.1:1.0 | 6b (7a) [8a] | 33 (26) [21] |
| 5 | 4c | C | >99% | 4.0:1.0 | 6c (7a) [8a] | 38 (16) [14] |
| 6 | 4d | C | >99% | 3.5:1.0 | 6d [8a] | 39 [23] |
| Entry | II Solvent (mL) b | 1:5 c | 1:10 | 1:20 |
|---|---|---|---|---|
| 1 | ×1 | Precipitate | Precipitate | Precipitate |
| 2 | ×2 | Stable colloid | Stable colloid | Low stable colloid |
| 3 | ×4 | Stable colloid | Stable colloid | Low stable colloid |
| 4 | ×6 | Low stable colloid | Low stable colloid | Low stable colloid |
| 5 | ×8 | Absence of LNPs | Aggregated LNPs | Aggregated LNPs |
| 6 | ×10 | Absence of LNPs | Absence of LNPs | Absence of LNPs |
| 7 | ×12 | Absence of LNPs | Absence of LNPs | Absence of LNPs |
| 8 | ×14 | Absence of LNPs | Absence of LNPs | Absence of LNPs |
| Entry | Compound(s) | Compd/KL Ratio | LE% | LC% | Compd/KL Ratio | LE% | LC% |
|---|---|---|---|---|---|---|---|
| 1 | 4a | 1:5 | 49.94 | 5.59 | 1:10 | 49.26 | 4.93 |
| 2 | 4b | 1:5 | 51.91 | 10.38 | 1:10 | 52.40 | 4.24 |
| 3 | 4c | 1:5 | 69.70 | 12.52 | 1:10 | 62.59 | 6.97 |
| 4 | 4d | 1:5 | 71.85 | 14.37 | 1:10 | 71.41 | 7.14 |
| 5 | 6a | 1:5 | 37.40 | 6.93 | 1:10 | 41.12 | 6.81 |
| 6 | 6b | 1:5 | 50.94 | 9.59 | 1:10 | 50.74 | 9.33 |
| 7 | 6c | 1:5 | 51.12 | 11.46 | 1:10 | 47.86 | 10.48 |
| 8 | 6d | 1:5 | 72.61 | 13.22 | 1:10 | 71.98 | 11.54 |
| 9 | 8a | 1:5 | 35.34 | 6.80 | 1:10 | 33.21 | 6.89 |
| 10 | 8b | 1:5 | 52.12 | 9.65 | 1:10 | 50.74 | 9.45 |
| 11 | 8c | 1:5 | 53.23 | 11.58 | 1:10 | 52.86 | 11.67 |
| 12 | 8d | 1:5 | 75.34 | 13.55 | 1:10 | 74.98 | 74.37 |
| Type | Entry | Compd(s) | Time (h) | FB789 (CC50 µM) | SK-Mel28 (IC50 µM) | SI (SKMel28) | RPMI7951 (IC50 µM) | SI (RPMI7951) |
|---|---|---|---|---|---|---|---|---|
| Biomimetic Dimers | 1 | 4a | 24 | 1450 ± 56.18 | 304.3 ± 2.4 | 4.7 | 543.1 ± 2.7 | 2.7 |
| 2 | 4b | 24 | 839.2 ± 10.9 | 169.4 ± 4.6 | 4.9 | 12,446 ± 16 | 0.07 | |
| 3 | 4c | 24 | 1662 ± 36.3 | 292.9 ± 2.5 | 5.7 | 356.7 ± 5.1 | 4.6 | |
| 4 | 4d | 24 | 392.5 ± 5.2 | 169.4 ± 4.6 | 2.3 | 342.8 ± 7.6 | 1.14 | |
| CPDs | 5 | 6a | 24 | 374.9 ± 28.15 | 85.23 ± 0.9 | 4.39 | 138.1 ± 2.1 | 2.7 |
| 6 | 6b | 24 | 109.1 ± 5.4 | 17.89 ± 1.9 | 6.09 | 77.76 ± 1.8 | 1.4 | |
| 7 | 6c | 24 | 234.3 ± 7.3 | 282.4 ± 4.4 | 0.83 | 195.1 ± 3.8 | 1.2 | |
| 8 | 6d | 24 | 376.8 ± 14.8 | 87.91 ± 1.9 | 4.3 | 234.9 ± 1.6 | 1.6 | |
| Dewar Valence | 9 | 8a | 24 | 4935 ± 58.7 | 146.9 ± 2.2 | 33.6 | 94.27 ± 1.9 | 52.3 |
| 10 | 8b | 24 | 12,442 ± 16.05 | 53.07 ± 1.7 | 234.4 | 120.6 ± 2.9 | 103.2 | |
| 11 | 8c | 24 | 250.4 ± 3.4 | 138.8 ± 2.1 | 1.8 | 77.89 ± 1.05 | 3.2 | |
| 12 | 8d | 24 | 33.47 ± 1.4 | 93.08 ± 1.9 | 0.36 | 114.9 ± 6.8 | 0.29 | |
| 13 | LNPs/8a | 2 | 3454 ± 1.9 | 129.1 ± 1.9 | 26.8 | 77.4 ± 1.8 | 44.6 | |
| 4 | 4112 ± 1.7 | 145.4 ± 1.1 | 28.3 | 88.2 ± 0.9 | 46.6 | |||
| 24 | 5012.1 ± 2.1 | 160.8 ± 2.4 | 31.2 | 41.1 ± 2.1 | 47.92 | |||
| 14 | LNPs/8b | 2 | 11,783 ± 1.1 | 59.8 ± 2.1 | 197.0 | 129.5 ± 1.7 | 91.0 | |
| 4 | 13,601 ± 3.1 | 68.7 ± 0.8 | 198.0 | 139.3 ± 1.1 | 97.6 | |||
| 24 | 14,888.1 ± 1.4 | 69.8 ± 0.9 | 213.3 | 148.8 ± 1.4 | 100.1 |
| Entries | Compd(s) | Binding Affinity (Kcal/mol) a | Hydrogen Bonds b | Salt Bridges b | Hydrophobic Interactions b | Π-Cation Interactions b | Π-Stacking Interactions b |
|---|---|---|---|---|---|---|---|
| 1 | 8a | −7.9 | DC112, DA113, ARG364, ASP533, THR718 | - | - | DA113 | - |
| 2 | 8b | −8.8 | DC111, DC112, ARG364, ASP533, THR718 | ASP533 | - | - | - |
| 3 | 4a | −7.1 | DG12, DC112, ARG364, THR718 | - | DT110, LYS532, THR718 | - | DG12 |
| 4 | 4b | −7.3 | DC112, DA113, ARG364 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabellone, S.; Piccinino, D.; Filippi, S.; Castrignanò, T.; Zippilli, C.; Del Buono, D.; Saladino, R. Lignin Nanoparticles Deliver Novel Thymine Biomimetic Photo-Adducts with Antimelanoma Activity. Int. J. Mol. Sci. 2022, 23, 915. https://doi.org/10.3390/ijms23020915
Gabellone S, Piccinino D, Filippi S, Castrignanò T, Zippilli C, Del Buono D, Saladino R. Lignin Nanoparticles Deliver Novel Thymine Biomimetic Photo-Adducts with Antimelanoma Activity. International Journal of Molecular Sciences. 2022; 23(2):915. https://doi.org/10.3390/ijms23020915
Chicago/Turabian StyleGabellone, Sofia, Davide Piccinino, Silvia Filippi, Tiziana Castrignanò, Claudio Zippilli, Davide Del Buono, and Raffaele Saladino. 2022. "Lignin Nanoparticles Deliver Novel Thymine Biomimetic Photo-Adducts with Antimelanoma Activity" International Journal of Molecular Sciences 23, no. 2: 915. https://doi.org/10.3390/ijms23020915







