Role of GALNT2 on Insulin Sensitivity, Lipid Metabolism and Fat Homeostasis
Abstract
1. Introduction
2. Insulin Resistance and Metabolic Diseases
3. GALNT2 Gene, Protein Function and Targets
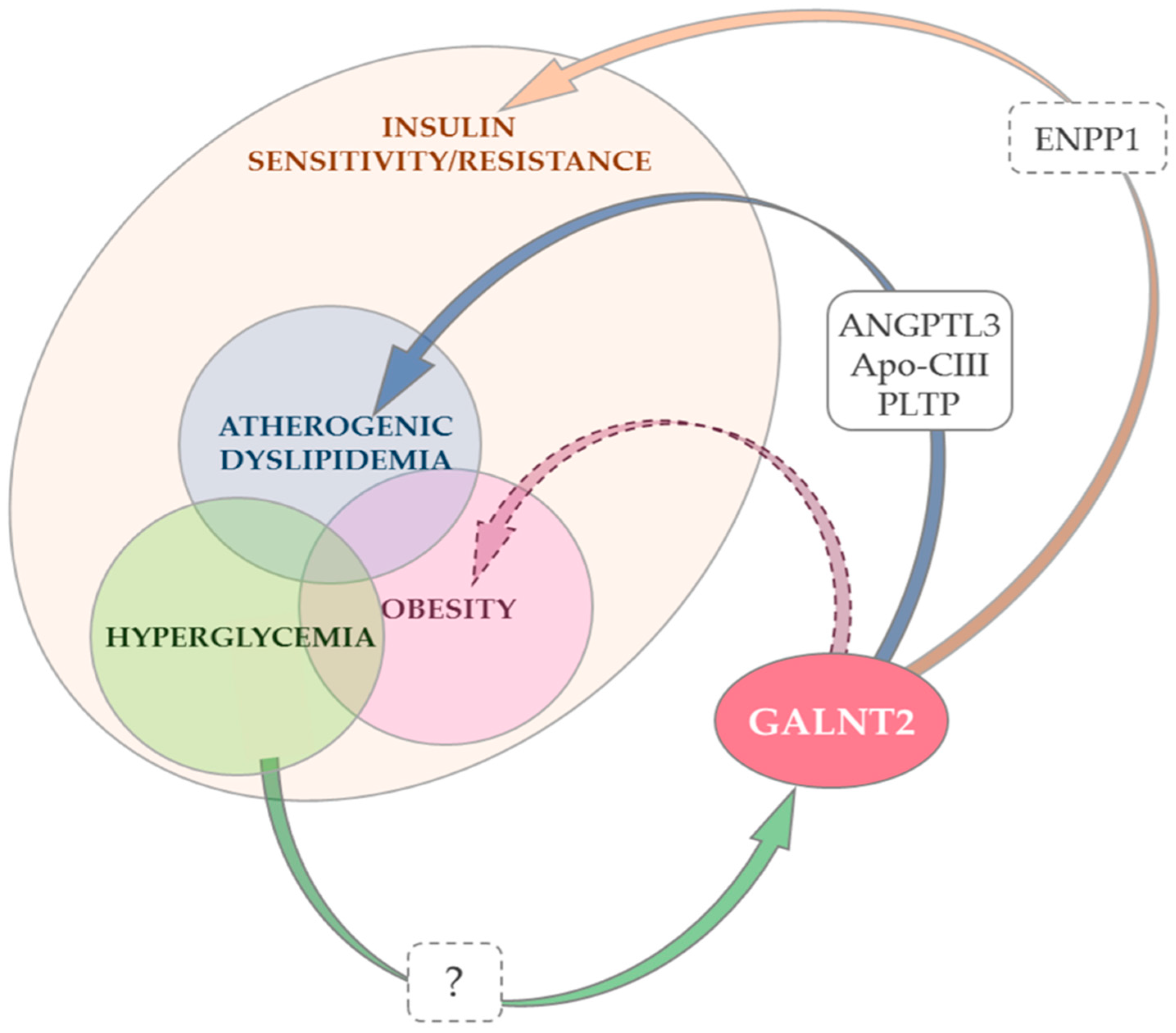
4. GALNT2 and Insulin Sensitivity
5. GALNT2 and Atherogenic Dyslipidemia
6. GALNT2, Type 2 Diabetes and Hyperglycemia
7. GALNT2 and Obesity
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ANGPTL3 | angiopoietin-like protein 3 |
| ApoC-III | apolipoprotein C3 |
| CAD | coronary artery disease |
| CDG2T | congenital disorder of glycosylation |
| ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| GalNAc | N-acetyl-D-galactosamine |
| GalNAc-T2 | polypeptide N-acetyl-galactosaminyl-transferase 2 |
| HDL | high-density lipoprotein |
| HOMA | homeostasis model assessment |
| IRS | insulin receptor substrates |
| LOF | loss of function |
| LPL | lipoprotein lipase |
| PEPCK | phosphoenolpyruvate carboxykinase |
| PIP3 | phosphatidyl-inositol-3,4,5-triphosphate |
| PLTP | phospholipid transfer protein |
| PTPN1 | protein tyrosine phosphatase non-receptor type 1 |
| PWBC | peripheral white blood cells |
| SH2 | Src homology 2 |
| SHIP2 | SH2-containing inositol phosphatase 2 |
| TG | triglycerides |
| TRIB3 | tribbles pseudokinase 3 |
References
- Lira-Navarrete, E.; de Las Rivas, M.; Compañón, I.; Pallarés, M.C.; Kong, Y.; Iglesias-Fernández, J.; Bernardes, G.J.; Peregrina, J.M.; Rovira, C.; Bernadó, P.; et al. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat. Commun. 2015, 6, 6937. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bennett, E.P.; Takio, K.; Sørensen, T.; Bonding, N.; Clausen, H. Purification and cDNA cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J. Biol. Chem. 1995, 270, 24156–24165. [Google Scholar] [CrossRef]
- Bennett, E.P.; Weghuis, D.O.; Merkx, G.; van Kessel, A.G.; Eiberg, H.; Clausen, H. Genomic organization and chromosomal localization of three members of the UDP-N-acetylgalactosamine: Polypeptide N-acetylgalactosaminyltransferase family. Glycobiology 1998, 8, 547–555. [Google Scholar] [CrossRef]
- Pisano, A.; Redmond, J.W.; Williams, K.L.; Gooley, A.A. Glycosylation sites identified by solid-phase Edman degradation: O-linked glycosylation motifs on human glycophorin A. Glycobiology 1993, 3, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.E.; Lund, O.; Nielsen, J.O.; Brunak, S. O-GLYCBASE: A revised database of O-glycosylated proteins. Nucleic Acids Res. 1996, 24, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Birch, H.; Rapacki, K.; Brunak, S.; Hansen, J.E. O-GLYCBASE version 4.0: A revised database of O-glycosylated proteins. Nucleic Acids Res. 1999, 27, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Roman, T.S.; Marvelle, A.F.; Fogarty, M.P.; Vadlamudi, S.; Gonzalez, A.J.; Buchkovich, M.L.; Huyghe, J.R.; Fuchsberger, C.; Jackson, A.U.; Wu, Y.; et al. Multiple Hepatic Regulatory Variants at the GALNT2 GWAS Locus Associated with High-Density Lipoprotein Cholesterol. Am. J. Hum. Genet. 2015, 97, 801–815. [Google Scholar] [CrossRef]
- Khetarpal, S.A.; Schjoldager, K.T.; Christoffersen, C.; Raghavan, A.; Edmondson, A.C.; Reutter, H.M.; Ahmed, B.; Ouazzani, R.; Peloso, G.M.; Vitali, C.; et al. Loss of Function of GALNT2 Lowers High-Density Lipoproteins in Humans, Nonhuman Primates, and Rodents. Cell Metab. 2016, 24, 234–245. [Google Scholar] [CrossRef]
- Marucci, A.; Mangiacotti, D.; Trischitta, V.; Di Paola, R. GALNT2 mRNA levels are associated with serum triglycerides in humans. Endocrine 2016, 53, 331–334. [Google Scholar] [CrossRef]
- Willer, C.J.; Sanna, S.; Jackson, A.U.; Scuteri, A.; Bonnycastle, L.L.; Clarke, R.; Heath, S.C.; Timpson, N.J.; Najjar, S.S.; Stringham, H.M.; et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008, 40, 161–169. [Google Scholar] [CrossRef]
- Willer, C.J.; Mohlke, K.L. Finding genes and variants for lipid levels after genome-wide association analysis. Curr. Opin. Lipidol. 2012, 23, 98–103. [Google Scholar] [CrossRef]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef]
- Kathiresan, S.; Melander, O.; Guiducci, C.; Surti, A.; Burtt, N.P.; Rieder, M.J.; Cooper, G.M.; Roos, C.; Voight, B.F.; Havulinna, A.S.; et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008, 40, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Marucci, A.; di Mauro, L.; Menzaghi, C.; Prudente, S.; Mangiacotti, D.; Fini, G.; Lotti, G.; Trischitta, V.; Di Paola, R. GALNT2 expression is reduced in patients with Type 2 diabetes: Possible role of hyperglycemia. PLoS ONE 2013, 8, e70159. [Google Scholar] [CrossRef] [PubMed]
- Almon, R.R.; DuBois, D.C.; Lai, W.; Xue, B.; Nie, J.; Jusko, W.J. Gene expression analysis of hepatic roles in cause and development of diabetes in Goto-Kakizaki rats. J. Endocrinol. 2009, 200, 331–346. [Google Scholar] [CrossRef]
- Lee, Y.H.; Nair, S.; Rousseau, E.; Allison, D.B.; Page, G.P.; Tataranni, P.A.; Bogardus, C.; Permana, P.A. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: Increased expression of inflammation-related genes. Diabetologia 2005, 48, 1776–1783. [Google Scholar] [CrossRef]
- Reynolds, E.G.M.; Neeley, C.; Lopdell, T.J.; Keehan, M.; Dittmer, K.; Harland, C.S.; Couldrey, C.; Johnson, T.J.J.; Tiplady, K.; Worth, G.; et al. Non-additive association analysis using proxy phenotypes identifies novel cattle syndromes. Nat. Genet. 2021, 53, 949–954. [Google Scholar] [CrossRef]
- Reaven, G.M. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes 1988, 37, 1595–1607. [Google Scholar] [CrossRef]
- Ferrannini, E.; Buzzigoli, G.; Bonadonna, R.; Giorico, M.A.; Oleggini, M.; Graziadei, L.; Pedrinelli, R.; Brandi, L.; Bevilacqua, S. Insulin resistance in essential hypertension. N. Engl. J. Med. 1987, 317, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Eckel, R.H.; Alberti, K.G.; Grundy, S.M.; Zimmet, P.Z. The metabolic syndrome. Lancet 2010, 375, 181–183. [Google Scholar] [CrossRef]
- Hedblad, B.; Nilsson, P.; Engström, G.; Berglund, G.; Janzon, L. Insulin resistance in non-diabetic subjects is associated with increased incidence of myocardial infarction and death. Diabet. Med. 2002, 19, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.W.; D’Agostino, R.B.; Parise, H.; Sullivan, L.; Meigs, J.B. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation 2005, 112, 3066–3072. [Google Scholar] [CrossRef] [PubMed]
- Wannamethee, S.G.; Shaper, A.G.; Lennon, L.; Morris, R.W. Metabolic syndrome vs Framingham Risk Score for prediction of coronary heart disease, stroke, and type 2 diabetes mellitus. Arch. Intern. Med. 2005, 165, 2644–2650. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.E.; do Carmo, J.M.; da Silva, A.A.; Wang, Z.; Hall, M.E. Obesity, kidney dysfunction and hypertension: Mechanistic links. Nat. Rev. Nephrol. 2019, 15, 367–385. [Google Scholar] [CrossRef]
- WHO. Noncommunicable Diseases, Progress Monitor 2017; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Kahn, C.R. Banting Lecture. Insulin action, diabetogenes, and the cause of type II diabetes. Diabetes 1994, 43, 1066–1084. [Google Scholar] [CrossRef]
- Martin, B.C.; Warram, J.H.; Krolewski, A.S.; Bergman, R.N.; Soeldner, J.S.; Kahn, C.R. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: Results of a 25-year follow-up study. Lancet 1992, 340, 925–929. [Google Scholar] [CrossRef]
- Weyer, C.; Bogardus, C.; Mott, D.M.; Pratley, R.E. The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J. Clin. Investig. 1999, 104, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef]
- Smith, U.; Kahn, B.B. Adipose tissue regulates insulin sensitivity: Role of adipogenesis, de novo lipogenesis and novel lipids. J. Intern. Med. 2016, 280, 465–475. [Google Scholar] [CrossRef]
- Reaven, G.M. Insulin resistance: The link between obesity and cardiovascular disease. Med. Clin. N. Am. 2011, 95, 875–892. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, J.M.; Stern, J.H.; Scherer, P.E. The cell biology of fat expansion. J. Cell Biol. 2015, 208, 501–512. [Google Scholar] [CrossRef]
- Vatier, C.; Vantyghem, M.C.; Storey, C.; Jéru, I.; Christin-Maitre, S.; Fève, B.; Lascols, O.; Beltrand, J.; Carel, J.C.; Vigouroux, C.; et al. Monogenic forms of lipodystrophic syndromes: Diagnosis, detection, and practical management considerations from clinical cases. Curr. Med. Res. Opin. 2019, 35, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Van Maldergem, L.; Magré, J.; Khallouf, T.E.; Gedde-Dahl, T.; Delépine, M.; Trygstad, O.; Seemanova, E.; Stephenson, T.; Albott, C.S.; Bonnici, F.; et al. Genotype-phenotype relationships in Berardinelli-Seip congenital lipodystrophy. J. Med. Genet. 2002, 39, 722–733. [Google Scholar] [CrossRef]
- Gomes, K.B.; Pardini, V.C.; Ferreira, A.C.; Fernandes, A.P. Phenotypic heterogeneity in biochemical parameters correlates with mutations in AGPAT2 or Seipin genes among Berardinelli-Seip congenital lipodystrophy patients. J. Inherit. Metab. Dis. 2005, 28, 1123–1131. [Google Scholar] [CrossRef]
- Knebel, B.; Müller-Wieland, D.; Kotzka, J. Lipodystrophies-Disorders of the Fatty Tissue. Int. J. Mol. Sci. 2020, 21, 8778. [Google Scholar] [CrossRef]
- Cohen, P. The twentieth century struggle to decipher insulin signalling. Nat. Rev. Mol. Cell Biol. 2006, 7, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef]
- Saltiel, A.R. Insulin signaling in health and disease. J. Clin. Investig. 2021, 131, e142241. [Google Scholar] [CrossRef] [PubMed]
- Roberts, F.; Zhu, D.; Farquharson, C.; Macrae, V.E. ENPP1 in the Regulation of Mineralization and Beyond. Trends Biochem. Sci. 2019, 44, 616–628. [Google Scholar] [CrossRef]
- Maddux, B.A.; Sbraccia, P.; Kumakura, S.; Sasson, S.; Youngren, J.; Fisher, A.; Spencer, S.; Grupe, A.; Henzel, W.; Stewart, T.A. Membrane glycoprotein PC-1 and insulin resistance in non-insulin-dependent diabetes mellitus. Nature 1995, 373, 448–451. [Google Scholar] [CrossRef]
- Maddux, B.A.; Goldfine, I.D. Membrane glycoprotein PC-1 inhibition of insulin receptor function occurs via direct interaction with the receptor alpha-subunit. Diabetes 2000, 49, 13–19. [Google Scholar] [CrossRef][Green Version]
- Maddux, B.; Chang, Y.; Accili, D.; McGuinness, O.; Youngren, J.; Goldfine, I. Overexpression of the insulin receptor inhibitor PC-1/ENPP1 induces insulin resistance and hyperglycemia. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E746–E749. [Google Scholar] [CrossRef]
- Di Paola, R.; Caporarello, N.; Marucci, A.; Dimatteo, C.; Iadicicco, C.; Del Guerra, S.; Prudente, S.; Sudano, D.; Miele, C.; Parrino, C.; et al. ENPP1 affects insulin action and secretion: Evidences from in vitro studies. PLoS ONE 2011, 6, e19462. [Google Scholar] [CrossRef] [PubMed]
- Dimatteo, C.; Marucci, A.; Palazzo, A.; Cisternino, C.; Marsano, R.M.; Trischitta, V.; Di Paola, R. Role of somatomedin-B-like domains on ENPP1 inhibition of insulin signaling. Biochim. Biophys. Acta 2013, 1833, 552–558. [Google Scholar] [CrossRef][Green Version]
- Prudente, S.; Sesti, G.; Pandolfi, A.; Andreozzi, F.; Consoli, A.; Trischitta, V. The mammalian tribbles homolog TRIB3, glucose homeostasis, and cardiovascular diseases. Endocr. Rev. 2012, 33, 526–546. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R. Putting the brakes on insulin signaling. N. Engl. J. Med. 2003, 349, 2560–2562. [Google Scholar] [CrossRef]
- Goldfine, I.D.; Maddux, B.A.; Youngren, J.F.; Reaven, G.; Accili, D.; Trischitta, V.; Vigneri, R.; Frittitta, L. The role of membrane glycoprotein plasma cell antigen 1/ectonucleotide pyrophosphatase phosphodiesterase 1 in the pathogenesis of insulin resistance and related abnormalities. Endocr. Rev. 2008, 29, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Aspinwall, C.A.; Qian, W.J.; Roper, M.G.; Kulkarni, R.N.; Kahn, C.R.; Kennedy, R.T. Roles of insulin receptor substrate-1, phosphatidylinositol 3-kinase, and release of intracellular Ca2+ stores in insulin-stimulated insulin secretion in beta-cells. J. Biol. Chem. 2000, 275, 22331–22338. [Google Scholar] [CrossRef]
- Kulkarni, R.N.; Brüning, J.C.; Winnay, J.N.; Postic, C.; Magnuson, M.A.; Kahn, C.R. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 1999, 96, 329–339. [Google Scholar] [CrossRef]
- Lyssenko, V.; Almgren, P.; Anevski, D.; Perfekt, R.; Lahti, K.; Nissén, M.; Isomaa, B.; Forsen, B.; Homström, N.; Saloranta, C.; et al. Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes 2005, 54, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Ghani, M.A.; Williams, K.; DeFronzo, R.A.; Stern, M. What is the best predictor of future type 2 diabetes? Diabetes Care 2007, 30, 1544–1548. [Google Scholar] [CrossRef]
- Bennett, E.P.; Mandel, U.; Clausen, H.; Gerken, T.A.; Fritz, T.A.; Tabak, L.A. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. [Google Scholar] [CrossRef] [PubMed]
- Schwientek, T.; Bennett, E.P.; Flores, C.; Thacker, J.; Hollmann, M.; Reis, C.A.; Behrens, J.; Mandel, U.; Keck, B.; Schäfer, M.A.; et al. Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J. Biol. Chem. 2002, 277, 22623–22638. [Google Scholar] [CrossRef]
- Kaneko, M.N.S.; Narimatsu, H.; Saitou, N. The evolutionary history of glycosyltransferase genes. Trends Glycosci. Glycotechnol. 2000, 13, 147–155. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Joshi, H.J.; Kong, Y.; Goth, C.K.; King, S.L.; Wandall, H.H.; Bennett, E.P.; Vakhrushev, S.Y.; Clausen, H. Deconstruction of O-glycosylation—Gal NA c-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 2015, 16, 1713–1722. [Google Scholar] [CrossRef]
- Narimatsu, Y.; Joshi, H.J.; Schjoldager, K.T.; Hintze, J.; Halim, A.; Steentoft, C.; Nason, R.; Mandel, U.; Bennett, E.P.; Clausen, H.; et al. Exploring regulation of protein O-glycosylation in isogenic human HEK293 cells by differential O-glycoproteomics. Mol. Cell. Proteom. 2019, 18, 1396–1409. [Google Scholar] [CrossRef] [PubMed]
- Bagdonaite, I.; Pallesen, E.M.H.; Ye, Z.; Vakhrushev, S.Y.; Marinova, I.N.; Nielsen, M.I.; Kramer, S.H.; Pedersen, S.F.; Joshi, H.J.; Bennett, E.P.; et al. O-glycan initiation directs distinct biological pathways and controls epithelial differentiation. EMBO Rep. 2020, 21, 1–17. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Lackman, J.J.; Goth, C.K.; Halim, A.; Vakhrushev, S.Y.; Clausen, H.; Petäjä-Repo, U.E. Site-specific O-glycosylation of N-terminal serine residues by polypeptide GalNAc-transferase 2 modulates human δ-opioid receptor turnover at the plasma membrane. Cell. Signal. 2018, 42, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Goth, C.K.; Tuhkanen, H.E.; Khan, H.; Lackman, J.J.; Wang, S.; Narimatsu, Y.; Hansen, L.H.; Overall, C.M.; Clausen, H.; Schjoldager, K.T.; et al. Site-specific O-glycosylation by polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-transferase T2) co-regulates β1-adrenergic receptor N-terminal cleavage. J. Biol. Chem. 2017, 292, 4714–4726. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.C.; Huang, M.J.; Liu, C.H.; Yang, T.L.; Huang, M.C. GALNT2 enhances migration and invasion of oral squamous cell carcinoma by regulating EGFR glycosylation and activity. Oral Oncol. 2014, 50, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.M.; Liu, C.H.; Hu, R.H.; Huang, M.J.; Lee, J.; Chen, C.H.; Huang, J.; Lai, H.S.; Lee, P.H.; Hsu, W.M.; et al. Mucin glycosylating enzyme GALNT2 regulates the malignant character of hepatocellular carcinoma by modifying the EGF receptor. Cancer Res. 2011, 71, 7270–7279. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.L.; Chou, C.H.; Jeng, Y.M.; Lu, M.Y.; Yang, Y.L.; Jou, S.T.; Lin, D.T.; Chang, H.H.; Lin, K.H.; Hsu, W.M.; et al. GALNT2 suppresses malignant phenotypes through IGF-1 receptor and predicts favorable prognosis in neuroblastoma. Oncotarget 2014, 5, 12247–12259. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-T.; Yeh, C.-C.; Liu, S.-Y.; Huang, M.-C.; Lai, I.R. The O-glycosylating enzyme GALNT2 suppresses the malignancy of gastric adenocarcinoma by reducing EGFR activities. Am. J. Cancer Res. 2018, 8, 1739–1751. [Google Scholar]
- Schjoldager, K.T.B.G.; Clausen, H. Site-specific protein O-glycosylation modulates proprotein processing-Deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta-Gen. Subj. 2012, 1820, 2079–2094. [Google Scholar] [CrossRef]
- Goth, C.K.; Halim, A.; Khetarpal, S.A.; Rader, D.J.; Clausen, H.; Schjoldager, K.T.B.G. A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc. Natl. Acad. Sci. USA 2015, 112, 14623–14628. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Zhang, M.; Wang, Y. GALNT2 regulates ANGPTL3 cleavage in cells and in vivo of mice. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Schjoldager, K.T.B.G.; Vester-Christensen, M.B.; Bennett, E.P.; Levery, S.B.; Schwientek, T.; Yin, W.; Blixt, O.; Clausen, H. O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: Possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J. Biol. Chem. 2010, 285, 36293–36303. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.B.; Wang, J.Y.; Sun, Z.L. ANGPTL3 is part of the machinery causing dyslipidemia majorily via LPL inhibition in mastitis mice. Exp. Mol. Pathol. 2017, 103, 242–248. [Google Scholar] [CrossRef]
- Garner, B.; Merry, A.H.; Royle, L.; Harvey, D.J.; Rudd, P.M.; Thillet, J. Structural elucidation of the N- and O-glycans of human apolipoprotein(a): Role of o-glycans in conferring protease resistance. J. Biol. Chem. 2001, 276, 22200–22208. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Angiopoietin-like 3 in lipoprotein metabolism. Nat. Rev. Endocrinol. 2017, 13, 731–739. [Google Scholar] [CrossRef]
- Xu, Y.X.; Redon, V.; Yu, H.; Querbes, W.; Pirruccello, J.; Liebow, A.; Deik, A.; Trindade, K.; Wang, X.; Musunuru, K.; et al. Role of angiopoietin-like 3 (ANGPTL3) in regulating plasma level of low-density lipoprotein cholesterol. Atherosclerosis 2018, 268, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Soffer, G.; Sztalryd, C.; Horenstein, R.B.; Holleran, S.; Matveyenko, A.; Thomas, T.; Nandakumar, R.; Ngai, C.; Karmally, W.; Ginsberg, H.N.; et al. Effects of APOC3 Heterozygous Deficiency on Plasma Lipid and Lipoprotein Metabolism. Arter. Thromb. Vasc. Biol. 2019, 39, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Borén, J.; Packard, C.J.; Taskinen, M.R. The Roles of ApoC-III on the Metabolism of Triglyceride-Rich Lipoproteins in Humans. Front. Endocrinol. 2020, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Hegele, R.A.; Ban, M.R.; Hsueh, N.; Kennedy, B.A.; Cao, H.; Zou, G.Y.; Anand, S.; Yusuf, S.; Huff, M.W.; Wang, J. A polygenic basis for four classical Fredrickson hyperlipoproteinemia phenotypes that are characterized by hypertriglyceridemia. Hum. Mol. Genet. 2009, 18, 4189–4194. [Google Scholar] [CrossRef]
- Rao, R.; Albers, J.J.; Wolfbauer, G.; Pownall, H.J. Molecular and macromolecular specificity of human plasma phospholipid transfer protein. Biochemistry 1997, 36, 3645–3653. [Google Scholar] [CrossRef] [PubMed]
- Tall, A.R.; Krumholz, S.; Olivecrona, T.; Deckelbaum, R.J. Plasma phospholipid transfer protein enhances transfer and exchange of phospholipids between very low density lipoproteins and high density lipoproteins during lipolysis. J. Lipid Res. 1985, 26, 842–851. [Google Scholar] [CrossRef]
- Vassalli, P. The pathophysiology of tumor necrosis factors. Annu. Rev. Immunol. 1992, 10, 411–452. [Google Scholar] [CrossRef] [PubMed]
- Marucci, A.; Cozzolino, F.; Dimatteo, C.; Monti, M.; Pucci, P.; Trischitta, V.; Di Paola, R. Role of GALNT2 in the modulation of ENPP1 expression, and insulin signaling and action: GALNT2: A novel modulator of insulin signaling. Biochim. Biophys. Acta 2013, 1833, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Marucci, A.; Antonucci, A.; De Bonis, C.; Mangiacotti, D.; Scarale, M.G.; Trischitta, V.; Di Paola, R. GALNT2 as a novel modulator of adipogenesis and adipocyte insulin signaling. Int. J. Obes. 2019, 43, 2448–2457. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guan, L.; Liu, H.; Liu, Q.; Fan, P.; Bai, H. GALNT2 Gene Variant rs4846914 Is Associated with Insulin and Insulin Resistance Depending on BMI in PCOS Patients: A Case-Control Study. Reprod. Sci. 2021, 28, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Zilmer, M.; Edmondson, A.C.; Khetarpal, S.A.; Alesi, V.; Zaki, M.S.; Rostasy, K.; Madsen, C.G.; Lepri, F.R.; Sinibaldi, L.; Cusmai, R.; et al. Novel congenital disorder of O-linked glycosylation caused by GALNT2 loss of function. Brain 2020, 143, 1114–1126. [Google Scholar] [CrossRef]
- Grundy, S.M. Hypertriglyceridemia, atherogenic dyslipidemia, and the metabolic syndrome. Am. J. Cardiol. 1998, 81, 18B–25B. [Google Scholar] [CrossRef]
- Di Paola, R.; Marucci, A.; Trischitta, V. GALNT2 effect on HDL-cholesterol and triglycerides levels in humans: Evidence of pleiotropy? Nutr. Metab. Cardiovasc. Dis. 2017, 27, 281–282. [Google Scholar] [CrossRef]
- Ghose, S.; Ghosh, S.; Tanwar, V.S.; Tolani, P.; Kutum, R.; Sharma, A.; Bhardwaj, N.; Shamsudheen, K.V.; Verma, A.; Jayarajan, R.; et al. Investigating Coronary Artery Disease methylome through targeted bisulfite sequencing. Gene 2019, 721, 144107. [Google Scholar] [CrossRef]
- Rossetti, L.; Giaccari, A.; DeFronzo, R.A. Glucose toxicity. Diabetes Care 1990, 13, 610–630. [Google Scholar] [CrossRef]
- Antonucci, A.; Marucci, A.; Scarale, M.G.; De Bonis, C.; Mangiacotti, D.; Trischitta, V.; Di Paola, R. Morphological and molecular characterization of GALNT2-mediated adipogenesis. Int. J. Obes. 2021, 45, 1362–1366. [Google Scholar] [CrossRef]
- Tietjen, I.; Hovingh, G.K.; Singaraja, R.R.; Radomski, C.; Barhdadi, A.; McEwen, J.; Chan, E.; Mattice, M.; Legendre, A.; Franchini, P.L.; et al. Segregation of LIPG, CETP, and GALNT2 mutations in Caucasian families with extremely high HDL cholesterol. PLoS ONE 2012, 7, e37437. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonucci, A.; Marucci, A.; Trischitta, V.; Di Paola, R. Role of GALNT2 on Insulin Sensitivity, Lipid Metabolism and Fat Homeostasis. Int. J. Mol. Sci. 2022, 23, 929. https://doi.org/10.3390/ijms23020929
Antonucci A, Marucci A, Trischitta V, Di Paola R. Role of GALNT2 on Insulin Sensitivity, Lipid Metabolism and Fat Homeostasis. International Journal of Molecular Sciences. 2022; 23(2):929. https://doi.org/10.3390/ijms23020929
Chicago/Turabian StyleAntonucci, Alessandra, Antonella Marucci, Vincenzo Trischitta, and Rosa Di Paola. 2022. "Role of GALNT2 on Insulin Sensitivity, Lipid Metabolism and Fat Homeostasis" International Journal of Molecular Sciences 23, no. 2: 929. https://doi.org/10.3390/ijms23020929
APA StyleAntonucci, A., Marucci, A., Trischitta, V., & Di Paola, R. (2022). Role of GALNT2 on Insulin Sensitivity, Lipid Metabolism and Fat Homeostasis. International Journal of Molecular Sciences, 23(2), 929. https://doi.org/10.3390/ijms23020929





