Making Sense of “Nonsense” and More: Challenges and Opportunities in the Genetic Code Expansion, in the World of tRNA Modifications
Abstract
:1. Introduction
2. The Principles of Engineering the Genetic Codes
2.1. Directed Evolution of Orthogonal Translational System of Expanded Codons
2.2. Site-Specific Incorporation of ncAA into Proteins by Engineering of the Codon Anticodon Interface
3. tRNA Modifications Affect the Synthesis of Non-Natural Proteins
3.1. tRNA Processing by Intracellular Machinery May Influence Incorporation Efficiency in GCE Systems
3.2. Modulation of o-tRNA Transport across Cellular Barriers in Eukaryotic Expression Hosts as a Means to Improve GCE
3.3. Modifications of the Target Protein Producer Organism to Allow an Efficient o-tRNA Processing
Improvement of Host Organism’s Cellular Fitness upon Alteration of Its Metabolism by Integrated o-tRNAs
3.4. Upregulation of Metabolic Precursors for o-tRNA Modifications
3.4.1. Methylation as a Target
3.4.2. Thiolation as a Target
3.4.3. Acetylation as a Target
3.4.4. Isopentenylation as a Target
3.4.5. Adenosine-to-Inosine Editing as a Target
3.5. tRNA Modification Tunable Transcripts
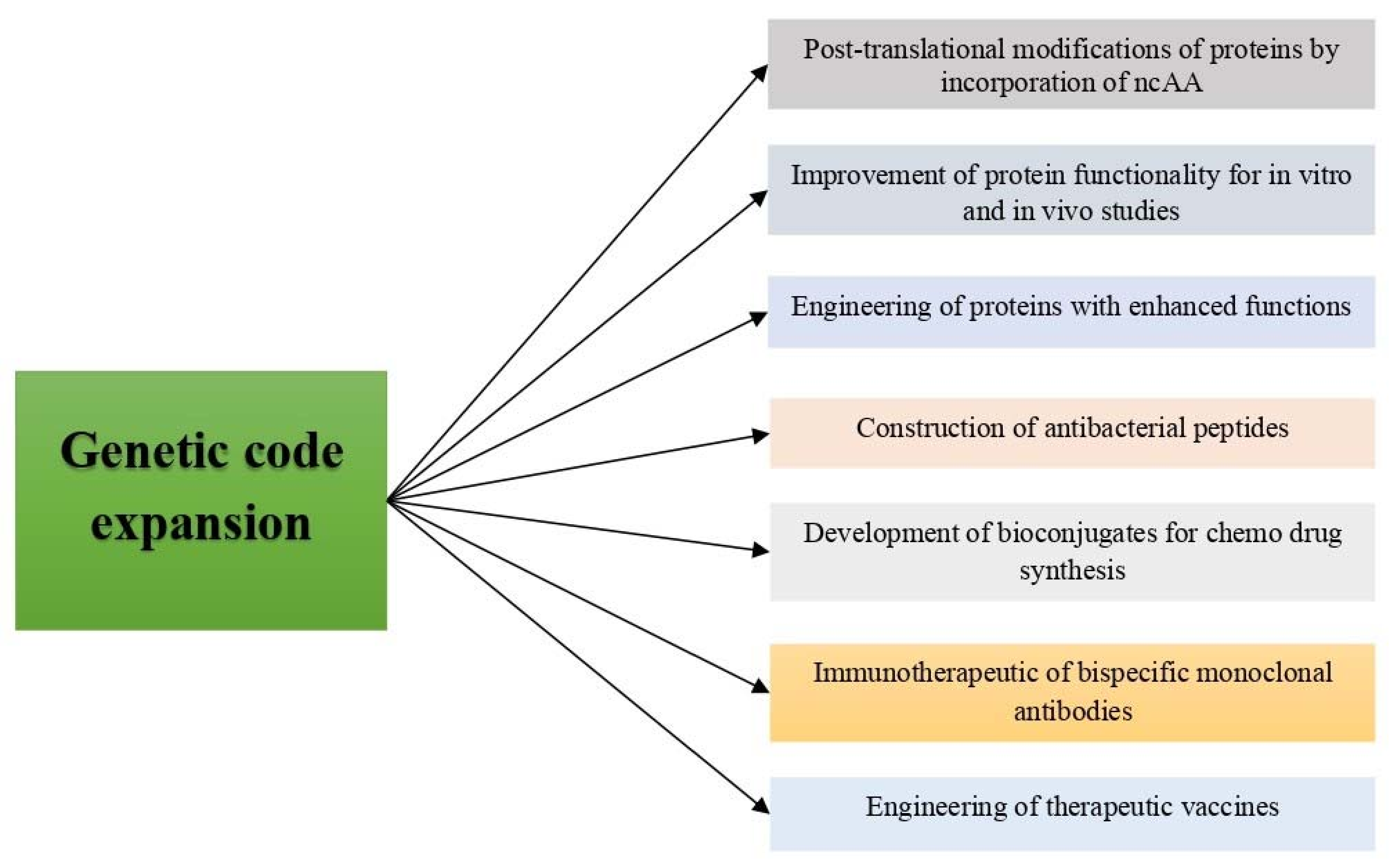
4. Applications of Genetic Code Expansion
4.1. Studies on Post-Translational Modifications (PTMs) of Proteins by Incorporation of ncAA
4.2. Improvement of Protein Stability for In Vitro and In Vivo Studies
4.3. Engineering of Proteins with Enhanced Functions
4.4. Construction of Biosynthesized Therapeutic Peptides
4.5. Development of Bioconjugates for Chemo Drug Synthesis
4.6. Immunotherapeutic of Bispecific Monoclonal Antibodies (BsMAb)
4.7. Engineered Therapeutic Vaccines
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aaRS | aminoacyl-tRNA synthetase |
| ADC | antibody-drug conjugate |
| ASL | anticodon stem-loop |
| BCMA | B-cell maturation antigen |
| EF-Tu | elongation factor translation thermos unstable |
| EGFR | epidermal growth factor receptor |
| FPs | fluorescent proteins |
| GCE | genetic code expansion |
| GRO | genomically recoded organism |
| LXR | liver X receptor |
| MAGE | multiplex automated genome engineering |
| MjRS | methanocaldococcus jannaschii tyrosyl-tRNA synthetase |
| MoTTs | modification tunable transcripts |
| ncAA | noncanonical amino acid |
| NAAs | natural amino acids |
| ORF | open reading frame |
| OTSs | orthogonal translation systems |
| o-tRNA | orthogonal-tRNA |
| PACE | phage-assisted continuous evolution |
| PAcF | p-acetylphenylalanine |
| PTMs | post-translational modifications |
| RBP | retinol-binding protein |
| RF-1 | release factor 1 |
| RiPPs | ribosomally synthesized and post-translationally modified peptides |
| SCI | supplementation-based incorporation |
| SICLOPPS | split intein catalyzed ligation of proteins and peptides |
| URM1 | ubiquitin-related modifier-1 |
References
- Smolskaya, S.; Andreev, Y.A. Site-Specific Incorporation of Unnatural Amino Acids into Escherichia coli Recombinant Protein: Methodology Development and Recent Achievement. Biomolecules 2019, 9, 255. [Google Scholar] [CrossRef] [Green Version]
- Rodnina, M.V. Translation in Prokaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032664. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.L.J.; Poruri, K.; Martinis, S.A. tRNA Synthetase: tRNA Aminoacylation and Beyond. Wiley Interdiscip. Rev. RNA 2014, 5, 461–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczanowska, M.; Rydén-Aulin, M. Ribosome Biogenesis and the Translation Process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007, 71, 477–494. [Google Scholar] [CrossRef] [Green Version]
- Petry, S.; Brodersen, D.E.; Murphy, F.V.; Dunham, C.M.; Selmer, M.; Tarry, M.J.; Kelley, A.C.; Ramakrishnan, V. Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon. Cell 2005, 123, 1255–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, T.D. Natural Variation in the Genetic Code. Annu. Rev. Genet. 1987, 21, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Lehman, N. Molecular Evolution: Please Release Me, Genetic Code. Curr. Biol. 2001, 11, R63–R66. [Google Scholar] [CrossRef] [Green Version]
- Gesteland, R.F.; Atkins, J.F. Recoding: Dynamic Reprogramming of Translation. Annu. Rev. Biochem. 1996, 65, 741–768. [Google Scholar] [CrossRef] [PubMed]
- Rother, M.; Krzycki, J.A. Selenocysteine, Pyrrolysine, and the Unique Energy Metabolism of Methanogenic Archaea. Archaea 2010, 2010, 453642. [Google Scholar] [CrossRef] [Green Version]
- Schwark, D.G.; Schmitt, M.A.; Fisk, J.D. Dissecting the Contribution of Release Factor Interactions to Amber Stop Codon Reassignment Efficiencies of the Methanocaldococcus Jannaschii Orthogonal Pair. Genes 2018, 9, 546. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Sun, S.B.; Furman, J.L.; Xiao, H.; Schultz, P.G. A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia coli. Biochemistry 2013, 52, 1828–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L. Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts. Acc. Chem. Res. 2017, 50, 2767–2775. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crécy-Lagard, V. Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; de Crécy-Lagard, V.; Marck, C. Deciphering Synonymous Codons in the Three Domains of Life: Co-Evolution with Specific tRNA Modification Enzymes. FEBS Lett. 2010, 584, 252–264. [Google Scholar] [CrossRef]
- Hao, B.; Gong, W.; Ferguson, T.K.; James, C.M.; Krzycki, J.A.; Chan, M.K. A New UAG-Encoded Residue in the Structure of a Methanogen Methyltransferase. Science 2002, 296, 1462–1466. [Google Scholar] [CrossRef]
- Srinivasan, G.; James, C.M.; Krzycki, J.A. Pyrrolysine Encoded by UAG in Archaea: Charging of a UAG-Decoding Specialized tRNA. Science 2002, 296, 1459–1462. [Google Scholar] [CrossRef]
- Nozawa, K.; O’Donoghue, P.; Gundllapalli, S.; Araiso, Y.; Ishitani, R.; Umehara, T.; Söll, D.; Nureki, O. Pyrrolysyl-tRNA Synthetase–tRNA Pyl Structure Reveals the Molecular Basis of Orthogonality. Nature 2009, 457, 1163–1167. [Google Scholar] [CrossRef] [Green Version]
- Wan, W.; Tharp, J.M.; Liu, W.R. Pyrrolysyl-tRNA Synthetase: An Ordinary Enzyme but an Outstanding Genetic Code Expansion Tool. Biochim. Biophys. Acta 2014, 1844, 1059–1070. [Google Scholar] [CrossRef] [Green Version]
- Mukai, T.; Kobayashi, T.; Hino, N.; Yanagisawa, T.; Sakamoto, K.; Yokoyama, S. Adding L-Lysine Derivatives to the Genetic Code of Mammalian Cells with Engineered Pyrrolysyl-tRNA Synthetases. Biochem. Biophys. Res. Commun. 2008, 371, 818–822. [Google Scholar] [CrossRef]
- Bianco, A.; Townsley, F.M.; Greiss, S.; Lang, K.; Chin, J.W. Expanding the Genetic Code of Drosophila Melanogaster. Nat. Chem. Biol. 2012, 8, 748–750. [Google Scholar] [CrossRef]
- Kato, Y. Tight Translational Control Using Site-Specific Unnatural Amino Acid Incorporation with Positive Feedback Gene Circuits. ACS Synth. Biol. 2018, 7, 1956–1963. [Google Scholar] [CrossRef]
- Strømgaard, A.; Jensen, A.A.; Strømgaard, K. Site-Specific Incorporation of Unnatural Amino Acids into Proteins. ChemBioChem 2004, 5, 909–916. [Google Scholar] [CrossRef]
- Vargas-Rodriguez, O.; Sevostyanova, A.; Söll, D.; Crnković, A. Upgrading Aminoacyl-tRNA Synthetases for Genetic Code Expansion. Curr. Opin. Chem. Biol. 2018, 46, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Schultz, P.G. Expanding the Genetic Code. Chem. Commun. 2002, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumas, A.; Lercher, L.; Spicer, C.D.; Davis, B.G. Designing Logical Codon Reassignment—Expanding the Chemistry in Biology. Chem. Sci. 2015, 6, 50–69. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Schultz, P.G. Expanding the Genetic Code. Angew. Chem. Int. Ed. 2004, 44, 34–66. [Google Scholar] [CrossRef]
- Barciszewska, M.Z.; Perrigue, P.M.; Barciszewski, J. tRNA—The Golden Standard in Molecular Biology. Mol. Biosyst. 2016, 12, 12–17. [Google Scholar] [CrossRef]
- Italia, J.S.; Addy, P.S.; Wrobel, C.J.J.; Crawford, L.A.; Lajoie, M.J.; Zheng, Y.; Chatterjee, A. An Orthogonalized Platform for Genetic Code Expansion in Both Bacteria and Eukaryotes. Nat. Chem. Biol. 2017, 13, 446–450. [Google Scholar] [CrossRef]
- Iraha, F.; Oki, K.; Kobayashi, T.; Ohno, S.; Yokogawa, T.; Nishikawa, K.; Yokoyama, S.; Sakamoto, K. Functional Replacement of the Endogenous Tyrosyl-tRNA Synthetase–tRNATyr Pair by the Archaeal Tyrosine Pair in Escherichia coli for Genetic Code Expansion. Nucleic Acids Res. 2010, 38, 3682–3691. [Google Scholar] [CrossRef] [Green Version]
- Bryson, D.I.; Fan, C.; Guo, L.-T.; Miller, C.; Söll, D.; Liu, D.R. Continuous Directed Evolution of Aminoacyl-tRNA Synthetases. Nat. Chem. Biol. 2017, 13, 1253–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiram, M.; Haimovich, A.D.; Fan, C.; Wang, Y.-S.; Aerni, H.-R.; Ntai, I.; Moonan, D.W.; Ma, N.J.; Rovner, A.J.; Hong, S.H.; et al. Evolution of Translation Machinery in Recoded Bacteria Enables Multi-Site Incorporation of Nonstandard Amino Acids. Nat. Biotechnol. 2015, 33, 1272–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, H.; Wang, K.; Davis, L.; Garcia-Alai, M.; Chin, J.W. Encoding Multiple Unnatural Amino Acids via Evolution of a Quadruplet-Decoding Ribosome. Nature 2010, 464, 441–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dedkova, L.M.; Hecht, S.M. Expanding the Scope of Protein Synthesis Using Modified Ribosomes. J. Am. Chem. Soc. 2019, 141, 6430–6447. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Brock, A.; Herberich, B.; Schultz, P.G. Expanding the Genetic Code of Escherichia coli. Science 2001, 292, 498–500. [Google Scholar] [CrossRef] [Green Version]
- Crnković, A.; Vargas-Rodriguez, O.; Merkuryev, A.; Söll, D. Effects of Heterologous tRNA Modifications on the Production of Proteins Containing Noncanonical Amino Acids. Bioengineering 2018, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Niu, W.; Schultz, P.G.; Guo, J. An Expanded Genetic Code in Mammalian Cells with a Functional Quadruplet Codon. ACS Chem. Biol. 2013, 8, 1640–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, C.; Xiong, H.; Reynolds, N.M.; Söll, D. Rationally Evolving tRNAPyl for Efficient Incorporation of Noncanonical Amino Acids. Nucleic Acids Res. 2015, 43, e156. [Google Scholar] [CrossRef] [Green Version]
- Polycarpo, C.R.; Herring, S.; Bérubé, A.; Wood, J.L.; Söll, D.; Ambrogelly, A. Pyrrolysine Analogues as Substrates for Pyrrolysyl-tRNA Synthetase. FEBS Lett. 2006, 580, 6695–6700. [Google Scholar] [CrossRef] [Green Version]
- Taki, M.; Tokuda, Y.; Ohtsuki, T.; Sisido, M. Design of Carrier tRNAs and Selection of Four-Base Codons for Efficient Incorporation of Various Nonnatural Amino Acids into Proteins in Spodoptera Frugiperda 21 (Sf21) Insect Cell-Free Translation System. J. Biosci. Bioeng. 2006, 102, 511–517. [Google Scholar] [CrossRef]
- Takimoto, J.K.; Dellas, N.; Noel, J.P.; Wang, L. Stereochemical Basis for Engineered Pyrrolysyl-tRNA Synthetase and the Efficient In Vivo Incorporation of Structurally Divergent Non-Native Amino Acids. ACS Chem. Biol. 2011, 6, 733–743. [Google Scholar] [CrossRef]
- Wang, Y.-S.; Wu, B.; Wang, Z.; Huang, Y.; Wan, W.; Russell, W.K.; Pai, P.-J.; Moe, Y.N.; Russell, D.H.; Liu, W.R. A Genetically Encoded Photocaged Nε-Methyl-l-Lysine. Mol. Biosyst. 2010, 6, 1557–1560. [Google Scholar] [CrossRef] [PubMed]
- Gautier, A.; Nguyen, D.P.; Lusic, H.; An, W.; Deiters, A.; Chin, J.W. Genetically Encoded Photocontrol of Protein Localization in Mammalian Cells. J. Am. Chem. Soc. 2010, 132, 4086–4088. [Google Scholar] [CrossRef] [PubMed]
- Greiss, S.; Chin, J.W. Expanding the Genetic Code of an Animal. J. Am. Chem. Soc. 2011, 133, 14196–14199. [Google Scholar] [CrossRef] [PubMed]
- Odoi, K.A.; Huang, Y.; Rezenom, Y.H.; Liu, W.R. Nonsense and Sense Suppression Abilities of Original and Derivative Methanosarcina Mazei Pyrrolysyl-tRNA Synthetase-tRNA(Pyl) Pairs in the Escherichia coli BL21(DE3) Cell Strain. PLoS ONE 2013, 8, e57035. [Google Scholar] [CrossRef] [Green Version]
- Fukunaga, J.; Yokogawa, T.; Ohno, S.; Nishikawa, K. Misacylation of Yeast Amber Suppressor tRNATyr by E. Coli Lysyl-tRNA Synthetase and Its Effective Repression by Genetic Engineering of the tRNA Sequence. J. Biochem. 2006, 139, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.; Wang, P.; Tirrell, D.A. Design of a Bacterial Host for Site-Specific Incorporation of p-Bromophenylalanine into Recombinant Proteins. J. Am. Chem. Soc. 2006, 128, 11778–11783. [Google Scholar] [CrossRef]
- Taira, H.; Fukushima, M.; Hohsaka, T.; Sisido, M. Four-Base Codon-Mediated Incorporation of Nonnatural Amino Acids into Proteins in a Eukaryotic Cell-Free Translation System. J. Biosci. Bioeng. 2005, 99, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Hohsaka, T.; Ashizuka, Y.; Taira, H.; Murakami, H.; Sisido, M. Incorporation of Nonnatural Amino Acids into Proteins by Using Various Four-Base Codons in an Escherichia coli In Vitro Translation System. Biochemistry 2001, 40, 11060–11064. [Google Scholar] [CrossRef] [PubMed]
- Kiga, D.; Sakamoto, K.; Kodama, K.; Kigawa, T.; Matsuda, T.; Yabuki, T.; Shirouzu, M.; Harada, Y.; Nakayama, H.; Takio, K.; et al. An Engineered Escherichia coli Tyrosyl-tRNA Synthetase for Site-Specific Incorporation of an Unnatural Amino Acid into Proteins in Eukaryotic Translation and Its Application in a Wheat Germ Cell-Free System. Proc. Natl. Acad. Sci. USA 2002, 99, 9715–9720. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, K.; Hayashi, A.; Sakamoto, A.; Kiga, D.; Nakayama, H.; Soma, A.; Kobayashi, T.; Kitabatake, M.; Takio, K.; Saito, K.; et al. Site-Specific Incorporation of an Unnatural Amino Acid into Proteins in Mammalian Cells. Nucleic Acids Res. 2002, 30, 4692–4699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukai, T.; Wakiyama, M.; Sakamoto, K.; Yokoyama, S. Genetic Encoding of Non-Natural Amino Acids in Drosophila melanogaster Schneider 2 Cells. Protein Sci. 2010, 19, 440–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, J.W.; Cropp, T.A.; Anderson, J.C.; Mukherji, M.; Zhang, Z.; Schultz, P.G. An Expanded Eukaryotic Genetic Code. Science 2003, 301, 964–967. [Google Scholar] [CrossRef]
- Shao, N.; Singh, N.S.; Slade, S.E.; Jones, A.M.E.; Balasubramanian, M.K. Site Specific Genetic Incorporation of Azidophenylalanine in Schizosaccharomyces pombe. Sci. Rep. 2015, 5, 17196. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Deiters, A.; Cropp, T.A.; King, D.; Schultz, P.G. A Genetically Encoded Photocaged Amino Acid. J. Am. Chem. Soc. 2004, 126, 14306–14307. [Google Scholar] [CrossRef]
- Zhang, Z.; Alfonta, L.; Tian, F.; Bursulaya, B.; Uryu, S.; King, D.S.; Schultz, P.G. Selective Incorporation of 5-Hydroxytryptophan into Proteins in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2004, 101, 8882–8887. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Brock, A.; Chen, S.; Chen, S.; Schultz, P.G. Genetic Incorporation of Unnatural Amino Acids into Proteins in Mammalian Cells. Nat. Methods 2007, 4, 239–244. [Google Scholar] [CrossRef]
- Warner, J.B. Multifunctionalization of Proteins: Strategies for Combining Semi-Synthetic and Bioorthogonal Techniques. Ph.D. Thesis, University of Pennsylvania, Philadelphia, PA, USA, 2014. [Google Scholar]
- Arranz-Gibert, P.; Vanderschuren, K.; Isaacs, F.J. Next-Generation Genetic Code Expansion. Curr. Opin. Chem. Biol. 2018, 46, 203–211. [Google Scholar] [CrossRef]
- Benner, S.A. Expanding the Genetic Lexicon: Incorporating Non-Standard Amino Acids into Proteins by Ribosome-Based Synthesis. Trends Biotechnol. 1994, 12, 158–163. [Google Scholar] [CrossRef]
- D’Aquino, A.E.; Kim, D.S.; Jewett, M.C. Engineered Ribosomes for Basic Science and Synthetic Biology. Annu. Rev. Chem. Biomol. Eng. 2018, 9, 311–340. [Google Scholar] [CrossRef]
- Anderson, J.C.; Schultz, P.G. Adaptation of an Orthogonal Archaeal Leucyl-tRNA and Synthetase Pair for Four-Base, Amber, and Opal Suppression. Biochemistry 2003, 42, 9598–9608. [Google Scholar] [CrossRef] [PubMed]
- Köhrer, C.; Xie, L.; Kellerer, S.; Varshney, U.; RajBhandary, U.L. Import of Amber and Ochre Suppressor tRNAs into Mammalian Cells: A General Approach to Site-Specific Insertion of Amino Acid Analogues into Proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 14310–14315. [Google Scholar] [CrossRef] [Green Version]
- Köhrer, C.; Yoo, J.-H.; Bennett, M.; Schaack, J.; RajBhandary, U.L. A Possible Approach to Site-Specific Insertion of Two Different Unnatural Amino Acids into Proteins in Mammalian Cells via Nonsense Suppression. Chem. Biol. 2003, 10, 1095–1102. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Brock, A.; Schultz, P.G. Adding L-3-(2-Naphthyl)Alanine to the Genetic Code of E. coli. J. Am. Chem. Soc. 2002, 124, 1836–1837. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.W.; Martin, A.B.; King, D.S.; Wang, L.; Schultz, P.G. Addition of a Photocrosslinking Amino Acid to the Genetic Code of Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 11020–11024. [Google Scholar] [CrossRef] [Green Version]
- Scolnick, E.; Tompkins, R.; Caskey, T.; Nirenberg, M. Release Factors Differing in Specificity for Terminator Codons. Proc. Natl. Acad. Sci. USA 1968, 61, 768–774. [Google Scholar] [CrossRef] [Green Version]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.-R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically Recoded Organisms Expand Biological Functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukai, T.; Yamaguchi, A.; Ohtake, K.; Takahashi, M.; Hayashi, A.; Iraha, F.; Kira, S.; Yanagisawa, T.; Yokoyama, S.; Hoshi, H.; et al. Reassignment of a Rare Sense Codon to a Non-Canonical Amino Acid in Escherichia coli. Nucleic Acids Res. 2015, 43, 8111–8122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.; Wang, W.; Liu, W.R. Towards Reassigning the Rare AGG Codon in Escherichia coli. Chembiochem 2014, 15, 1750–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohlke, N.; Budisa, N. Sense Codon Emancipation for Proteome-Wide Incorporation of Noncanonical Amino Acids: Rare Isoleucine Codon AUA as a Target for Genetic Code Expansion. FEMS Microbiol. Lett. 2014, 351, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirao, I.; Ohtsuki, T.; Fujiwara, T.; Mitsui, T.; Yokogawa, T.; Okuni, T.; Nakayama, H.; Takio, K.; Yabuki, T.; Kigawa, T.; et al. An Unnatural Base Pair for Incorporating Amino Acid Analogs into Proteins. Nat. Biotechnol. 2002, 20, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.C.; Wu, N.; Santoro, S.W.; Lakshman, V.; King, D.S.; Schultz, P.G. An Expanded Genetic Code with a Functional Quadruplet Codon. Proc. Natl. Acad. Sci. USA 2004, 101, 7566–7571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, E.A.; Lester, H.A.; Dougherty, D.A. In Vivo Incorporation of Multiple Unnatural Amino Acids through Nonsense and Frameshift Suppression. Proc. Natl. Acad. Sci. USA 2006, 103, 8650–8655. [Google Scholar] [CrossRef] [Green Version]
- Magliery, T.J.; Anderson, J.C.; Schultz, P.G. Expanding the Genetic Code: Selection of Efficient Suppressors of Four-Base Codons and Identification of “Shifty” Four-Base Codons with a Library Approach in Escherichia coli. J. Mol. Biol. 2001, 307, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.C.; Magliery, T.J.; Schultz, P.G. Exploring the Limits of Codon and Anticodon Size. Chem. Biol. 2002, 9, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Taki, M.; Matsushita, J.; Sisido, M. Expanding the Genetic Code in a Mammalian Cell Line by the Introduction of Four-Base Codon/Anticodon Pairs. ChemBioChem 2006, 7, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Xiao, H.; Schultz, P.G. Evolution of Multiple, Mutually Orthogonal Prolyl-tRNA Synthetase/tRNA Pairs for Unnatural Amino Acid Mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2012, 109, 14841–14846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Yu, A.C.S.; Chan, T.F. Efforts and Challenges in Engineering the Genetic Code. Life 2017, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtsuki, T.; Manabe, T.; Sisido, M. Multiple Incorporation of Non-Natural Amino Acids into a Single Protein Using tRNAs with Non-Standard Structures. FEBS Lett. 2005, 579, 6769–6774. [Google Scholar] [CrossRef] [Green Version]
- Xiao, H.; Chatterjee, A.; Choi, S.; Bajjuri, K.M.; Sinha, S.C.; Schultz, P.G. Genetic Incorporation of Multiple Unnatural Amino Acids into Proteins in Mammalian Cells. Angew. Chem. 2013, 125, 14330–14333. [Google Scholar] [CrossRef]
- Oh, S.-J.; Lee, K.-H.; Kim, H.-C.; Catherine, C.; Yun, H.; Kim, D.-M. Translational Incorporation of Multiple Unnatural Amino Acids in a Cell-Free Protein Synthesis System. Biotechnol. Bioprocess Eng. 2014, 19, 426–432. [Google Scholar] [CrossRef]
- Wang, L.; Xie, J.; Schultz, P.G. Expanding the Genetic Code. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 225–249. [Google Scholar] [CrossRef]
- Niu, W.; Guo, J. Expanding the Chemistry of Fluorescent Protein Biosensors through Genetic Incorporation of Unnatural Amino Acids. Mol. Biosyst. 2013, 9, 2961–2970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Sachdeva, A.; Cox, D.J.; Wilf, N.M.; Lang, K.; Wallace, S.; Mehl, R.A.; Chin, J.W. Optimized Orthogonal Translation of Unnatural Amino Acids Enables Spontaneous Protein Double-Labelling and FRET. Nat. Chem. 2014, 6, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Hamann, C.S.; Hou, Y.-M. Probing a tRNA Core That Contributes to Aminoacylation. J. Mol. Biol. 2000, 295, 777–789. [Google Scholar] [CrossRef]
- Agris, P.F.; Vendeix, F.A.P.; Graham, W.D. tRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Decoding the Genome: A Modified View. Nucleic Acids Res. 2004, 32, 223–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weixlbaumer, A.; Murphy, F.V.; Dziergowska, A.; Malkiewicz, A.; Vendeix, F.A.P.; Agris, P.F.; Ramakrishnan, V. Mechanism for Expanding the Decoding Capacity of Transfer RNAs by Modification of Uridines. Nat. Struct. Mol. Biol. 2007, 14, 498–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, M.; Francisco, S.; Varanda, A.S.; Santos, M.; Santos, M.A.S.; Soares, A.R. Impact of tRNA Modifications and tRNA-Modifying Enzymes on Proteostasis and Human Disease. Int. J. Mol. Sci. 2018, 19, 3738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biddle, W.; Schmitt, M.A.; Fisk, J.D. Modification of Orthogonal tRNAs: Unexpected Consequences for Sense Codon Reassignment. Nucleic Acids Res. 2016, 44, 10042–10050. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA Modifications Controls the Oxidative Stress Response by Codon-Biased Translation of Proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [Green Version]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Babu, M.M. Cells Alter Their tRNA Abundance to Selectively Regulate Protein Synthesis during Stress Conditions. Sci. Signal. 2018, 11, eaat6409. [Google Scholar] [CrossRef] [Green Version]
- Alings, F.; Sarin, L.P.; Fufezan, C.; Drexler, H.C.A.; Leidel, S.A. An Evolutionary Approach Uncovers a Diverse Response of tRNA 2-Thiolation to Elevated Temperatures in Yeast. RNA 2015, 21, 202–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuorto, F.; Lyko, F. Genome Recoding by tRNA Modifications. Open Biol. 2016, 6, 160287. [Google Scholar] [CrossRef] [Green Version]
- Giegé, R.; Sissler, M.; Florentz, C. Universal Rules and Idiosyncratic Features in tRNA Identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [Green Version]
- Kisselev, L.L. The Role of the Anticodon in Recognition of tRNA by Aminoacyl-tRNA Synthetases. In Progress in Nucleic Acid Research and Molecular Biology; Cohn, W.E., Moldave, K., Eds.; Academic Press: Cambridge, MA, USA, 1985; Volume 32, pp. 237–266. [Google Scholar]
- Kwon, I.; Kirshenbaum, K.; Tirrell, D.A. Breaking the Degeneracy of the Genetic Code. J. Am. Chem. Soc. 2003, 125, 7512–7513. [Google Scholar] [CrossRef] [Green Version]
- Noma, A.; Kirino, Y.; Ikeuchi, Y.; Suzuki, T. Biosynthesis of Wybutosine, a Hyper-Modified Nucleoside in Eukaryotic Phenylalanine tRNA. EMBO J. 2006, 25, 2142–2154. [Google Scholar] [CrossRef]
- Lee, B.S.; Shin, S.; Jeon, J.Y.; Jang, K.-S.; Lee, B.Y.; Choi, S.; Yoo, T.H. Incorporation of Unnatural Amino Acids in Response to the AGG Codon. ACS Chem. Biol. 2015, 10, 1648–1653. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.M.; Reynolds, N.M.; Rivera, K.; Connolly, M.; Guo, L.-T.; Ling, J.; Pappin, D.J.; Church, G.M.; Söll, D. Efficient Reassignment of a Frequent Serine Codon in Wild-Type Escherichia coli. ACS Synth. Biol. 2016, 5, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, R.; Prat, L.; Aerni, H.-R.; Ling, J.; Merryman, C.; Glass, J.I.; Rinehart, J.; Söll, D. Transfer RNA Misidentification Scrambles Sense Codon Recoding. ChemBioChem 2013, 14, 1967–1972. [Google Scholar] [CrossRef] [Green Version]
- Biddle, W.; Schmitt, M.A.; Fisk, J.D. Evaluating Sense Codon Reassignment with a Simple Fluorescence Screen. Biochemistry 2015, 54, 7355–7364. [Google Scholar] [CrossRef]
- Yokogawa, T.; Hassan, H.M.B.C.; Yokota, Y.; Ohno, S.; Nishikawa, K. Preparation of an Ochre Suppressor tRNA Recognizing Exclusively UAA Codon by Using the Molecular Surgery Technique. Nucleic Acids Symp. Ser. 2009, 53, 295–296. [Google Scholar] [CrossRef]
- Bouadloun, F.; Srichaiyo, T.; Isaksson, L.A.; Björk, G.R. Influence of Modification next to the Anticodon in tRNA on Codon Context Sensitivity of Translational Suppression and Accuracy. J. Bacteriol. 1986, 166, 1022–1027. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Walvekar, A.S.; Liang, S.; Rashida, Z.; Shah, P.; Laxman, S. A tRNA Modification Balances Carbon and Nitrogen Metabolism by Regulating Phosphate Homeostasis. eLife 2019, 8, e44795. [Google Scholar] [CrossRef] [PubMed]
- Marchand, V.; Pichot, F.; Thüring, K.; Ayadi, L.; Freund, I.; Dalpke, A.; Helm, M.; Motorin, Y. Next-Generation Sequencing-Based RiboMethSeq Protocol for Analysis of tRNA 2′-O-Methylation. Biomolecules 2017, 7, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barraud, P.; Tisné, C. To Be or Not to Be Modified: Miscellaneous Aspects Influencing Nucleotide Modifications in tRNAs. IUBMB Life 2019, 71, 1126–1140. [Google Scholar] [CrossRef]
- Asakura, T.; Sasaki, T.; Nagano, F.; Satoh, A.; Obaishi, H.; Nishioka, H.; Imamura, H.; Hotta, K.; Tanaka, K.; Nakanishi, H.; et al. Isolation and Characterization of a Novel Actin Filament-Binding Protein from Saccharomyces cerevisiae. Oncogene 1998, 16, 121–130. [Google Scholar] [CrossRef] [Green Version]
- De Crécy-Lagard, V.; Jaroch, M. Functions of Bacterial tRNA Modifications: From Ubiquity to Diversity. Trends Microbiol. 2021, 29, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Gaston, K.W.; Rubio, M.A.T.; Spears, J.L.; Pastar, I.; Papavasiliou, F.N.; Alfonzo, J.D. C to U Editing at Position 32 of the Anticodon Loop Precedes tRNA 5’ Leader Removal in Trypanosomatids. Nucleic Acids Res. 2007, 35, 6740–6749. [Google Scholar] [CrossRef] [Green Version]
- Phizicky, E.M.; Hopper, A.K. tRNA Biology Charges to the Front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Brockenbrough, J.S.; Paddy, M.R.; Aris, J.P. NCL1, a Novel Gene for a Non-Essential Nuclear Protein in Saccharomyces cerevisiae. Gene 1998, 220, 109–117. [Google Scholar] [CrossRef]
- Simos, G.; Tekotte, H.; Grosjean, H.; Segref, A.; Sharma, K.; Tollervey, D.; Hurt, E.C. Nuclear Pore Proteins Are Involved in the Biogenesis of Functional tRNA. EMBO J. 1996, 15, 2270–2284. [Google Scholar] [CrossRef]
- Joshi, K.; Bhatt, M.J.; Farabaugh, P.J. Codon-Specific Effects of tRNA Anticodon Loop Modifications on Translational Misreading Errors in the Yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, 10331–10339. [Google Scholar] [CrossRef] [Green Version]
- Björk, G.R.; Huang, B.; Persson, O.P.; Byström, A.S. A Conserved Modified Wobble Nucleoside (Mcm5s2U) in Lysyl-tRNA Is Required for Viability in Yeast. RNA 2007, 13, 1245–1255. [Google Scholar] [CrossRef] [Green Version]
- De Crécy-Lagard, V.; Ross, R.L.; Jaroch, M.; Marchand, V.; Eisenhart, C.; Brégeon, D.; Motorin, Y.; Limbach, P.A. Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp. Subtilis Strain 168. Biomolecules 2020, 10, 977. [Google Scholar] [CrossRef]
- Wolf, J.; Gerber, A.P.; Keller, W. TadA, an Essential tRNA-Specific Adenosine Deaminase from Escherichia coli. EMBO J. 2002, 21, 3841–3851. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.J.O.; Byström, A.S. Transfer RNA Modifications and Modifying Enzymes in Saccharomyces cerevisiae. In Fine-Tuning of RNA Functions by Modification and Editing; Grosjean, H., Ed.; Topics in Current Genetics; Springer: Berlin/Heidelberg, Germany, 2005; pp. 87–120. ISBN 978-3-540-31454-7. [Google Scholar]
- Lee, C.; Kramer, G.; Graham, D.E.; Appling, D.R. Yeast Mitochondrial Initiator tRNA Is Methylated at Guanosine 37 by the Trm5-Encoded tRNA (Guanine-N1-)-Methyltransferase. J. Biol. Chem. 2007, 282, 27744–27753. [Google Scholar] [CrossRef] [Green Version]
- Grosjean, H.; Edqvist, J.; Stråby, K.B.; Giegé, R. Enzymatic Formation of Modified Nucleosides in tRNA: Dependence on tRNA Architecture. J. Mol. Biol. 1996, 255, 67–85. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, U.; Bohleber, S.; Fradejas-Villar, N. The Modified Base Isopentenyladenosine and Its Derivatives in tRNA. RNA Biol. 2017, 14, 1197–1208. [Google Scholar] [CrossRef] [Green Version]
- Gunnelius, L.; Hakkila, K.; Kurkela, J.; Wada, H.; Tyystjärvi, E.; Tyystjärvi, T. The Omega Subunit of the RNA Polymerase Core Directs Transcription Efficiency in Cyanobacteria. Nucleic Acids Res. 2014, 42, 4606–4614. [Google Scholar] [CrossRef]
- Robb, N.C.; Cordes, T.; Hwang, L.C.; Gryte, K.; Duchi, D.; Craggs, T.D.; Santoso, Y.; Weiss, S.; Ebright, R.H.; Kapanidis, A.N. The Transcription Bubble of the RNA Polymerase–Promoter Open Complex Exhibits Conformational Heterogeneity and Millisecond-Scale Dynamics: Implications for Transcription Start-Site Selection. J. Mol. Biol. 2013, 425, 875–885. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A Novel Class of Small RNAs: tRNA-Derived RNA Fragments (TRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.F.; Abelson, J. The Yeast tRNATyr Gene Intron Is Essential for Correct Modification of Its tRNA Product. Nature 1983, 302, 681–687. [Google Scholar] [CrossRef]
- Nishikura, K.; De Robertis, E.M. RNA Processing in Microinjected Xenopus Oocytes. Sequential Addition of Base Modifications in the Spliced Transfer RNA. J. Mol. Biol. 1981, 145, 405–420. [Google Scholar] [CrossRef]
- Kessler, A.C.; Silveira d’Almeida, G.; Alfonzo, J.D. The Role of Intracellular Compartmentalization on tRNA Processing and Modification. RNA Biol. 2017, 15, 554–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svenningsen, S.L.; Kongstad, M.; Stenum, T.S.; Muñoz-Gómez, A.J.; Sørensen, M.A. Transfer RNA Is Highly Unstable during Early Amino Acid Starvation in Escherichia coli. Nucleic Acids Res. 2017, 45, 793–804. [Google Scholar] [CrossRef] [Green Version]
- Arts, G.J.; Kuersten, S.; Romby, P.; Ehresmann, B.; Mattaj, I.W. The Role of Exportin-t in Selective Nuclear Export of Mature tRNAs. EMBO J. 1998, 17, 7430–7441. [Google Scholar] [CrossRef] [Green Version]
- Yoshihisa, T.; Yunoki-Esaki, K.; Ohshima, C.; Tanaka, N.; Endo, T. Possibility of Cytoplasmic Pre-tRNA Splicing: The Yeast tRNA Splicing Endonuclease Mainly Localizes on the Mitochondria. Mol. Biol. Cell 2003, 14, 3266–3279. [Google Scholar] [CrossRef] [Green Version]
- Lopes, R.R.S.; Silveira, G.d.O.; Eitler, R.; Vidal, R.S.; Kessler, A.; Hinger, S.; Paris, Z.; Alfonzo, J.D.; Polycarpo, C. The Essential Function of the Trypanosoma Brucei Trl1 Homolog in Procyclic Cells Is Maturation of the Intron-Containing tRNATyr. RNA 2016, 22, 1190–1199. [Google Scholar] [CrossRef] [Green Version]
- Murthi, A.; Shaheen, H.H.; Huang, H.-Y.; Preston, M.A.; Lai, T.-P.; Phizicky, E.M.; Hopper, A.K. Regulation of tRNA Bidirectional Nuclear-Cytoplasmic Trafficking in Saccharomyces Cerevisiae. Mol. Biol. Cell 2010, 21, 639–649. [Google Scholar] [CrossRef] [Green Version]
- Arts, G.J.; Fornerod, M.; Mattaj, I.W. Identification of a Nuclear Export Receptor for tRNA. Curr. Biol. 1998, 8, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Stanford, D.R.; Whitney, M.L.; Hurto, R.L.; Eisaman, D.M.; Shen, W.-C.; Hopper, A.K. Division of Labor Among the Yeast Sol Proteins Implicated in tRNA Nuclear Export and Carbohydrate Metabolism. Genetics 2004, 168, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Green, M.R. Sequence-Specific Binding of Transfer RNA by Glyceraldehyde-3-Phosphate Dehydrogenase. Science 1993, 259, 365–368. [Google Scholar] [CrossRef]
- Nakai, Y.; Nakai, M.; Hayashi, H. Thio-Modification of Yeast Cytosolic tRNA Requires a Ubiquitin-Related System That Resembles Bacterial Sulfur Transfer Systems. J. Biol. Chem. 2008, 283, 27469–27476. [Google Scholar] [CrossRef] [Green Version]
- Umeda, N.; Suzuki, T.; Yukawa, M.; Ohya, Y.; Shindo, H.; Watanabe, K.; Suzuki, T. Mitochondria-Specific RNA-Modifying Enzymes Responsible for the Biosynthesis of the Wobble Base in Mitochondrial tRNAs. Implications for the Molecular Pathogenesis of Human Mitochondrial Diseases. J. Biol. Chem. 2005, 280, 1613–1624. [Google Scholar] [CrossRef] [Green Version]
- Li, J.M.; Hopper, A.K.; Martin, N.C. N2,N2-Dimethylguanosine-Specific tRNA Methyltransferase Contains Both Nuclear and Mitochondrial Targeting Signals in Saccharomyces cerevisiae. J. Cell Biol. 1989, 109, 1411–1419. [Google Scholar] [CrossRef]
- Lecointe, F.; Simos, G.; Sauer, A.; Hurt, E.C.; Motorin, Y.; Grosjean, H. Characterization of Yeast Protein Deg1 as Pseudouridine Synthase (Pus3) Catalyzing the Formation of Ψ38 and Ψ39 in tRNA Anticodon Loop. J. Biol. Chem. 1998, 273, 1316–1323. [Google Scholar] [CrossRef] [Green Version]
- Behm-Ansmant, I.; Grosjean, H.; Massenet, S.; Motorin, Y.; Branlant, C. Pseudouridylation at Position 32 of Mitochondrial and Cytoplasmic tRNAs Requires Two Distinct Enzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 52998–53006. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, S.K.; Bujnicki, J.M.; Grosjean, H.; Lapeyre, B. Trm11p and Trm112p Are Both Required for the Formation of 2-Methylguanosine at Position 10 in Yeast tRNA. Mol. Cell. Biol. 2005, 25, 4359–4370. [Google Scholar] [CrossRef] [Green Version]
- Pintard, L.; Lecointe, F.; Bujnicki, J.M.; Bonnerot, C.; Grosjean, H.; Lapeyre, B. Trm7p Catalyses the Formation of Two 2’-O-Methylriboses in Yeast tRNA Anticodon Loop. EMBO J. 2002, 21, 1811–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Lyons, S.M.; Fay, M.M.; Ivanov, P. The Role of RNA Modifications in the Regulation of tRNA Cleavage. FEBS Lett. 2018, 592, 2828–2844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, C.; Lünse, C.E.; Mörl, M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björk, G.R.; Ericson, J.U.; Gustafsson, C.E.; Hagervall, T.G.; Jönsson, Y.H.; Wikström, P.M. Transfer RNA Modification. Annu. Rev. Biochem. 1987, 56, 263–287. [Google Scholar] [CrossRef]
- Han, L.; Guy, M.P.; Kon, Y.; Phizicky, E.M. Lack of 2’-O-Methylation in the tRNA Anticodon Loop of Two Phylogenetically Distant Yeast Species Activates the General Amino Acid Control Pathway. PLoS Genet. 2018, 14, e1007288. [Google Scholar] [CrossRef] [PubMed]
- Zinshteyn, B.; Gilbert, W.V. Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling. PLoS Genet. 2013, 9, e1003675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laxman, S.; Sutter, B.M.; Wu, X.; Kumar, S.; Guo, X.; Trudgian, D.C.; Mirzaei, H.; Tu, B.P. Sulfur Amino Acids Regulate Translational Capacity and Metabolic Homeostasis through Modulation of tRNA Thiolation. Cell 2013, 154, 416–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarian, C.; Townsend, H.; Czestkowski, W.; Sochacka, E.; Malkiewicz, A.J.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Accurate Translation of the Genetic Code Depends on tRNA Modified Nucleosides. J. Biol. Chem. 2002, 277, 16391–16395. [Google Scholar] [CrossRef] [Green Version]
- Rezgui, V.A.N.; Tyagi, K.; Ranjan, N.; Konevega, A.L.; Mittelstaet, J.; Rodnina, M.V.; Peter, M.; Pedrioli, P.G.A. tRNA TKUUU, TQUUG, and TEUUC Wobble Position Modifications Fine-Tune Protein Translation by Promoting Ribosome A-Site Binding. Proc. Natl. Acad. Sci. USA 2013, 110, 12289–12294. [Google Scholar] [CrossRef] [Green Version]
- Szatkowska, R.; Garcia-Albornoz, M.; Roszkowska, K.; Holman, S.W.; Furmanek, E.; Hubbard, S.J.; Beynon, R.J.; Adamczyk, M. Glycolytic Flux in Saccharomyces Cerevisiae Is Dependent on RNA Polymerase III and Its Negative Regulator Maf1. Biochem. J. 2019, 476, 1053–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cieśla, M.; Boguta, M. Regulation of RNA Polymerase III Transcription by Maf1 Protein. Acta Biochim. Pol. 2008, 55, 215–225. [Google Scholar] [CrossRef] [Green Version]
- Arimbasseri, A.G.; Blewett, N.H.; Iben, J.R.; Lamichhane, T.N.; Cherkasova, V.; Hafner, M.; Maraia, R.J. RNA Polymerase III Output Is Functionally Linked to tRNA Dimethyl-G26 Modification. PLoS Genet. 2015, 11, e1005671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierrel, F.; Björk, G.R.; Fontecave, M.; Atta, M. Enzymatic Modification of tRNAs: MiaB Is an Iron-Sulfur Protein. J. Biol. Chem. 2002, 277, 13367–13370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wettstein, F.O.; Stent, G.S. Physiologically Induced Changes in the Property of Phenylalanine tRNA in Escherichia coli. J. Mol. Biol. 1968, 38, 25–40. [Google Scholar] [CrossRef]
- Gall, A.R.; Datsenko, K.A.; Figueroa-Bossi, N.; Bossi, L.; Masuda, I.; Hou, Y.-M.; Csonka, L.N. Mg2+ Regulates Transcription of MgtA in Salmonella typhimurium via Translation of Proline Codons during Synthesis of the MgtL Peptide. Proc. Natl. Acad. Sci. USA 2016, 113, 15096–15101. [Google Scholar] [CrossRef] [Green Version]
- Serebrov, V.; Vassilenko, K.; Kholod, N.; Gross, H.J.; Kisselev, L. Mg2+ Binding and Structural Stability of Mature and In Vitro Synthesized Unmodified Escherichia coli tRNAPhe. Nucleic Acids Res. 1998, 26, 2723–2728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarin, L.P.; Leidel, S.A. Modify or Die?—RNA Modification Defects in Metazoans. RNA Biol. 2014, 11, 1555–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvanin, A.; Vogt, L.-M.; Grober, A.; Freund, I.; Ayadi, L.; Bourguignon-Igel, V.; Bessler, L.; Jacob, D.; Eigenbrod, T.; Marchand, V.; et al. Bacterial tRNA 2′-O-Methylation Is Dynamically Regulated under Stress Conditions and Modulates Innate Immune Response. Nucleic Acids Res. 2020, 48, 12833–12844. [Google Scholar] [CrossRef] [PubMed]
- Rana, A.K.; Ankri, S. Reviving the RNA World: An Insight into the Appearance of RNA Methyltransferases. Front. Genet. 2016, 7, 99. [Google Scholar] [CrossRef] [Green Version]
- Cavaillé, J.; Chetouani, F.; Bachellerie, J.P. The Yeast Saccharomyces Cerevisiae YDL112w ORF Encodes the Putative 2’-O-Ribose Methyltransferase Catalyzing the Formation of Gm18 in tRNAs. RNA 1999, 5, 66–81. [Google Scholar] [CrossRef] [Green Version]
- Bjork, G.R.; Wikstrom, P.M.; Bystrom, A.S. Prevention of Translational Frameshifting by the Modified Nucleoside 1-Methylguanosine. Science 1989, 244, 986–989. [Google Scholar] [CrossRef] [PubMed]
- Rimbach, K.; Kaiser, S.; Helm, M.; Dalpke, A.H.; Eigenbrod, T. 2′-O-Methylation within Bacterial RNA Acts as Suppressor of TLR7/TLR8 Activation in Human Innate Immune Cells. J. Innate Immun. 2015, 7, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Kawai, G.; Yamamoto, Y.; Kamimura, T.; Masegi, T.; Sekine, M.; Hata, T.; Iimori, T.; Watanabe, T.; Miyazawa, T.; Yokoyama, S. Conformational Rigidity of Specific Pyrimidine Residues in tRNA Arises from Posttranscriptional Modifications That Enhance Steric Interaction between the Base and the 2’-Hydroxyl Group. Biochemistry 1992, 31, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Kawai, G.; Ue, H.; Yasuda, M.; Sakamoto, K.; Hashizume, T.; McCloskey, J.A.; Miyazawa, T.; Yokoyama, S. Relation between Functions and Conformational Characteristics of Modified Nucleosides Found in tRNAs. Nucleic Acids Symp. Ser. 1991, 25, 49–50. [Google Scholar]
- Sprinzl, M.; Vassilenko, K.S. Compilation of tRNA Sequences and Sequences of tRNA Genes. Nucleic Acids Res. 2005, 33, D139–D140. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, R.; Mark, P.; Prevost, C.T.; AlKindi, A.; Alhag, A.; Estwani, F.; Al-Sheddi, T.; Alobeid, E.; Alenazi, M.M.; Ewida, N.; et al. Biallelic Variants in CTU2 Cause DREAM-PL Syndrome and Impair Thiolation of tRNA Wobble U34. Hum. Mutat. 2019, 40, 2108–2120. [Google Scholar] [CrossRef] [PubMed]
- Leimkühler, S.; Bühning, M.; Beilschmidt, L. Shared Sulfur Mobilization Routes for tRNA Thiolation and Molybdenum Cofactor Biosynthesis in Prokaryotes and Eukaryotes. Biomolecules 2017, 7, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tükenmez, H.; Xu, H.; Esberg, A.; Byström, A.S. The Role of Wobble Uridine Modifications in +1 Translational Frameshifting in Eukaryotes. Nucleic Acids Res. 2015, 43, 9489–9499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klassen, R.; Bruch, A.; Schaffrath, R. Independent Suppression of Ribosomal +1 Frameshifts by Different tRNA Anticodon Loop Modifications. RNA Biol. 2017, 14, 1252–1259. [Google Scholar] [CrossRef] [Green Version]
- Tigano, M.; Ruotolo, R.; Dallabona, C.; Fontanesi, F.; Barrientos, A.; Donnini, C.; Ottonello, S. Elongator-Dependent Modification of Cytoplasmic tRNALysUUU Is Required for Mitochondrial Function under Stress Conditions. Nucleic Acids Res. 2015, 43, 8368–8380. [Google Scholar] [CrossRef] [Green Version]
- Pandey, A.; Pain, J.; Dziuba, N.; Pandey, A.K.; Dancis, A.; Lindahl, P.A.; Pain, D. Mitochondria Export Sulfur Species Required for Cytosolic tRNA Thiolation. Cell Chem. Biol. 2018, 25, 738–748.e3. [Google Scholar] [CrossRef]
- Pandey, A.K.; Pain, J.; Dancis, A.; Pain, D. Mitochondria Export Iron–Sulfur and Sulfur Intermediates to the Cytoplasm for Iron–Sulfur Cluster Assembly and tRNA Thiolation in Yeast. J. Biol. Chem. 2019, 294, 9489–9502. [Google Scholar] [CrossRef]
- Arragain, S.; Bimai, O.; Legrand, P.; Caillat, S.; Ravanat, J.-L.; Touati, N.; Binet, L.; Atta, M.; Fontecave, M.; Golinelli-Pimpaneau, B. Nonredox Thiolation in tRNA Occurring via Sulfur Activation by a [4Fe-4S] Cluster. Proc. Natl. Acad. Sci. USA 2017, 114, 7355–7360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Vinyard, D.J.; Reesbeck, M.E.; Suzuki, T.; Manakongtreecheep, K.; Holland, P.L.; Brudvig, G.W.; Söll, D. A [3Fe-4S] Cluster Is Required for tRNA Thiolation in Archaea and Eukaryotes. Proc. Natl. Acad. Sci. USA 2016, 113, 12703–12708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agris, P.F.; Narendran, A.; Sarachan, K.; Väre, V.Y.P.; Eruysal, E. The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity. Enzymes 2017, 41, 1–50. [Google Scholar] [CrossRef]
- Bruch, A.; Laguna, T.; Butter, F.; Schaffrath, R.; Klassen, R. Misactivation of Multiple Starvation Responses in Yeast by Loss of tRNA Modifications. Nucleic Acids Res. 2020, 48, 7307–7320. [Google Scholar] [CrossRef]
- Damon, J.R.; Pincus, D.; Ploegh, H.L. tRNA Thiolation Links Translation to Stress Responses in Saccharomyces Cerevisiae. Mol. Biol. Cell 2015, 26, 270–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurėnas, D.; Chatterjee, S.; Konijnenberg, A.; Sobott, F.; Droogmans, L.; Garcia-Pino, A.; Van Melderen, L. AtaT Blocks Translation Initiation by N-Acetylation of the Initiator tRNAfMet. Nat. Chem. Biol. 2017, 13, 640–646. [Google Scholar] [CrossRef]
- Taniguchi, T.; Miyauchi, K.; Sakaguchi, Y.; Yamashita, S.; Soma, A.; Tomita, K.; Suzuki, T. Acetate-Dependent tRNA Acetylation Required for Decoding Fidelity in Protein Synthesis. Nat. Chem. Biol. 2018, 14, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Cheverton, A.M.; Gollan, B.; Przydacz, M.; Wong, C.T.; Mylona, A.; Hare, S.A.; Helaine, S. A Salmonella Toxin Promotes Persister Formation through Acetylation of tRNA. Mol. Cell 2016, 63, 86–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, F.V.; Ramakrishnan, V.; Malkiewicz, A.; Agris, P.F. The Role of Modifications in Codon Discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004, 11, 1186–1191. [Google Scholar] [CrossRef]
- Persson, B.C.; Esberg, B.; Olafsson, O.; Björk, G.R. Synthesis and Function of Isopentenyl Adenosine Derivatives in tRNA. Biochimie 1994, 76, 1152–1160. [Google Scholar] [CrossRef]
- Gefter, M.L.; Russell, R.L. Role of Modifications in Tyrosine Transfer RNA: A Modified Base Affecting Ribosome Binding. J. Mol. Biol. 1969, 39, 145–157. [Google Scholar] [CrossRef]
- Eisenberg, S.P.; Yarus, M.; Soll, L. The Effect of an Escherichia coli Regulatory Mutation on Transfer RNA Structure. J. Mol. Biol. 1979, 135, 111–126. [Google Scholar] [CrossRef]
- Diaz, I.; Pedersen, S.; Kurland, C.G. Effects of MiaA on Translation and Growth Rates. Mol. Gen. Genet. MGG 1987, 208, 373–376. [Google Scholar] [CrossRef]
- Ericson, J.U.; Björk, G.R. Pleiotropic Effects Induced by Modification Deficiency next to the Anticodon of tRNA from Salmonella typhimurium LT2. J. Bacteriol. 1986, 166, 1013–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, M.A.T.; Paris, Z.; Gaston, K.W.; Fleming, I.M.C.; Sample, P.; Trotta, C.R.; Alfonzo, J.D. Unusual Noncanonical Intron Editing Is Important for tRNA Splicing in Trypanosoma Brucei. Mol. Cell 2013, 52, 184–192. [Google Scholar] [CrossRef] [Green Version]
- Tsutsumi, S.; Sugiura, R.; Ma, Y.; Tokuoka, H.; Ohta, K.; Ohte, R.; Noma, A.; Suzuki, T.; Kuno, T. Wobble Inosine tRNA Modification Is Essential to Cell Cycle Progression in G1/S and G2/M Transitions in Fission Yeast. J. Biol. Chem. 2007, 282, 33459–33465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerber, A.P.; Keller, W. An Adenosine Deaminase That Generates Inosine at the Wobble Position of tRNAs. Science 1999, 286, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Piñeyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I Editing on tRNAs: Biochemical, Biological and Evolutionary Implications. FEBS Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef]
- Rafels-Ybern, À.; Torres, A.G.; Grau-Bove, X.; Ruiz-Trillo, I.; Ribas de Pouplana, L. Codon Adaptation to tRNAs with Inosine Modification at Position 34 Is Widespread among Eukaryotes and Present in Two Bacterial Phyla. RNA Biol. 2017, 15, 500–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endres, L.; Dedon, P.C.; Begley, T.J. Codon-Biased Translation Can Be Regulated by Wobble-Base tRNA Modification Systems during Cellular Stress Responses. RNA Biol. 2015, 12, 603–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novoselov, S.V.; Calvisi, D.F.; Labunskyy, V.M.; Factor, V.M.; Carlson, B.A.; Fomenko, D.E.; Moustafa, M.E.; Hatfield, D.L.; Gladyshev, V.N. Selenoprotein Deficiency and High Levels of Selenium Compounds Can Effectively Inhibit Hepatocarcinogenesis in Transgenic Mice. Oncogene 2005, 24, 8003–8011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA Modifications Regulate Translation during Cellular Stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.S.; Brunner, S.F.; Huguenin-Dezot, N.; Liang, A.D.; Schmied, W.H.; Rogerson, D.T.; Chin, J.W. Biosynthesis and Genetic Encoding of Phosphothreonine through Parallel Selection and Deep Sequencing. Nat. Methods 2017, 14, 729–736. [Google Scholar] [CrossRef]
- Luo, X.; Fu, G.; Wang, R.E.; Zhu, X.; Zambaldo, C.; Liu, R.; Liu, T.; Lyu, X.; Du, J.; Xuan, W.; et al. Genetically Encoding Phosphotyrosine and Its Nonhydrolyzable Analog in Bacteria. Nat. Chem. Biol. 2017, 13, 845–849. [Google Scholar] [CrossRef] [Green Version]
- Ambrogelly, A.; Palioura, S.; Söll, D. Natural Expansion of the Genetic Code. Nat. Chem. Biol. 2007, 3, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Xuan, W.; Shao, S.; Liu, T.; Schultz, P.G. Genetic Incorporation of ε-N-2-Hydroxyisobutyryl-Lysine into Recombinant Histones. ACS Chem. Biol. 2015, 10, 1599–1603. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.H.; Kang, M.; Kim, H.J.; Chatterjee, A.; Schultz, P.G. Site-Specific Incorporation of ε-N-Crotonyllysine into Histones. Angew. Chem. Int. Ed. 2012, 51, 7246–7249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Song, H.; He, D.; Zhang, S.; Dai, S.; Lin, S.; Meng, R.; Wang, C.; Chen, P.R. Genetically Encoded Protein Photocrosslinker with a Transferable Mass Spectrometry-Identifiable Label. Nat. Commun. 2016, 7, 12299. [Google Scholar] [CrossRef]
- Roy, S.; Ghosh, P.; Ahmed, I.; Chakraborty, M.; Naiya, G.; Ghosh, B. Constrained α-Helical Peptides as Inhibitors of Protein-Protein and Protein-DNA Interactions. Biomedicines 2018, 6, 118. [Google Scholar] [CrossRef] [Green Version]
- Hoppmann, C.; Wong, A.; Yang, B.; Li, S.; Hunter, T.; Shokat, K.M.; Wang, L. Site-Specific Incorporation of Phosphotyrosine Using an Expanded Genetic Code. Nat. Chem. Biol. 2017, 13, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Ip, K.; Söll, D. Expanding the Genetic Code of Escherichia coli with Phosphotyrosine. FEBS Lett. 2016, 590, 3040–3047. [Google Scholar] [CrossRef] [Green Version]
- Rogerson, D.T.; Sachdeva, A.; Wang, K.; Haq, T.; Kazlauskaite, A.; Hancock, S.M.; Huguenin-Dezot, N.; Muqit, M.M.K.; Fry, A.M.; Bayliss, R.; et al. Efficient Genetic Encoding of Phosphoserine and Its Non-Hydrolyzable Analog. Nat. Chem. Biol. 2015, 11, 496–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Mejías, G.; Velázquez-Cruz, A.; Guerra-Castellano, A.; Baños-Jaime, B.; Díaz-Quintana, A.; González-Arzola, K.; Ángel De la Rosa, M.; Díaz-Moreno, I. Exploring Protein Phosphorylation by Combining Computational Approaches and Biochemical Methods. Comput. Struct. Biotechnol. J. 2020, 18, 1852–1863. [Google Scholar] [CrossRef] [PubMed]
- Elsässer, S.J.; Ernst, R.J.; Walker, O.S.; Chin, J.W. Genetic Code Expansion in Stable Cell Lines Enables Encoded Chromatin Modification. Nat. Methods 2016, 13, 158–164. [Google Scholar] [CrossRef]
- Neumann, H.; Hancock, S.M.; Buning, R.; Routh, A.; Chapman, L.; Somers, J.; Owen-Hughes, T.; van Noort, J.; Rhodes, D.; Chin, J.W. A Method for Genetically Installing Site-Specific Acetylation in Recombinant Histones Defines the Effects of H3 K56 Acetylation. Mol. Cell 2009, 36, 153–163. [Google Scholar] [CrossRef]
- Liu, T.; Wang, Y.; Luo, X.; Li, J.; Reed, S.A.; Xiao, H.; Young, T.S.; Schultz, P.G. Enhancing Protein Stability with Extended Disulfide Bonds. Proc. Natl. Acad. Sci. USA 2016, 113, 5910–5915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, W.; Li, J.; Luo, X.; Schultz, P.G. Genetic Incorporation of a Reactive Isothiocyanate Group into Proteins. Angew. Chem. 2016, 128, 10219–10222. [Google Scholar] [CrossRef]
- Brustad, E.; Bushey, M.L.; Lee, J.W.; Groff, D.; Liu, W.; Schultz, P.G. A Genetically Encoded Boronate-Containing Amino Acid. Angew. Chem. Int. Ed. 2008, 47, 8220–8223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deiters, A.; Schultz, P.G. In Vivo Incorporation of an Alkyne into Proteins in Escherichia coli. Bioorg. Med. Chem. Lett. 2005, 15, 1521–1524. [Google Scholar] [CrossRef]
- Xiang, Z.; Lacey, V.K.; Ren, H.; Xu, J.; Burban, D.J.; Jennings, P.A.; Wang, L. Proximity-Enabled Protein Crosslinking through Genetically Encoding Haloalkane Unnatural Amino Acids. Angew. Chem. Int. Ed. 2014, 53, 2190–2193. [Google Scholar] [CrossRef]
- Chin, J.W.; Santoro, S.W.; Martin, A.B.; King, D.S.; Wang, L.; Schultz, P.G. Addition of P-Azido-l-Phenylalanine to the Genetic Code of Escherichia coli. J. Am. Chem. Soc. 2002, 124, 9026–9027. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Wu, B.; Raymond, J.E.; Zeng, Y.; Fang, X.; Wooley, K.L.; Liu, W.R. A Genetically Encoded Acrylamide Functionality. ACS Chem. Biol. 2013, 8, 1664–1670. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.-H.; Han, J.; Jin, Z.; Lee, D.-W.; Park, H.-S.; Kim, H.-S. Controlled and Oriented Immobilization of Protein by Site-Specific Incorporation of Unnatural Amino Acid. Anal. Chem. 2011, 83, 2841–2845. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Jaworski, J. A Portable and Chromogenic Enzyme-Based Sensor for Detection of Abrin Poisoning. Biosens. Bioelectron. 2014, 54, 667–673. [Google Scholar] [CrossRef]
- Guan, D.; Kurra, Y.; Liu, W.; Chen, Z. A Click Chemistry Approach to Site-Specific Immobilization of a Small Laccase Enables Efficient Direct Electron Transfer in a Biocathode. Chem. Commun. 2015, 51, 2522–2525. [Google Scholar] [CrossRef] [PubMed]
- Amir, L.; Carnally, S.A.; Rayo, J.; Rosenne, S.; Melamed Yerushalmi, S.; Schlesinger, O.; Meijler, M.M.; Alfonta, L. Surface Display of a Redox Enzyme and Its Site-Specific Wiring to Gold Electrodes. J. Am. Chem. Soc. 2013, 135, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Guo, J. Chapter Seven—Novel Fluorescence-Based Biosensors Incorporating Unnatural Amino Acids. In Methods in Enzymology. Enzymes as Sensors; Thompson, R.B., Fierke, C.A., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 589, pp. 191–219. [Google Scholar]
- Youssef, S.; Zhang, S.; Ai, H. A Genetically Encoded, Ratiometric Fluorescent Biosensor for Hydrogen Sulfide. ACS Sens. 2019, 4, 1626–1632. [Google Scholar] [CrossRef]
- Drienovská, I.; Mayer, C.; Dulson, C.; Roelfes, G. A Designer Enzyme for Hydrazone and Oxime Formation Featuring an Unnatural Catalytic Aniline Residue. Nat. Chem. 2018, 10, 946–952. [Google Scholar] [CrossRef]
- Ma, H.; Yang, X.; Lu, Z.; Liu, N.; Chen, Y. The “Gate Keeper” Role of Trp222 Determines the Enantiopreference of Diketoreductase toward 2-Chloro-1-Phenylethanone. PLoS ONE 2014, 9, e103792. [Google Scholar] [CrossRef]
- Drienovská, I.; Roelfes, G. Expanding the Enzyme Universe with Genetically Encoded Unnatural Amino Acids. Nat. Catal. 2020, 3, 193–202. [Google Scholar] [CrossRef]
- Xiao, H.; Nasertorabi, F.; Choi, S.; Han, G.W.; Reed, S.A.; Stevens, R.C.; Schultz, P.G. Exploring the Potential Impact of an Expanded Genetic Code on Protein Function. Proc. Natl. Acad. Sci. USA 2015, 112, 6961–6966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tack, D.S.; Ellefson, J.W.; Thyer, R.; Wang, B.; Gollihar, J.; Forster, M.T.; Ellington, A.D. Addicting Diverse Bacteria to a Noncanonical Amino Acid. Nat. Chem. Biol. 2016, 12, 138–140. [Google Scholar] [CrossRef]
- Wang, R.E.; Liu, T.; Wang, Y.; Cao, Y.; Du, J.; Luo, X.; Deshmukh, V.; Kim, C.H.; Lawson, B.R.; Tremblay, M.S.; et al. An Immunosuppressive Antibody–Drug Conjugate. J. Am. Chem. Soc. 2015, 137, 3229–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, T.S.; Young, D.D.; Ahmad, I.; Louis, J.M.; Benkovic, S.J.; Schultz, P.G. Evolution of Cyclic Peptide Protease Inhibitors. Proc. Natl. Acad. Sci. USA 2011, 108, 11052–11056. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Liu, T. Therapeutic Applications of Genetic Code Expansion. Synth. Syst. Biotechnol. 2018, 3, 150–158. [Google Scholar] [CrossRef]
- Bindman, N.A.; Bobeica, S.C.; Liu, W.R.; van der Donk, W.A. Facile Removal of Leader Peptides from Lanthipeptides by Incorporation of a Hydroxy Acid. J. Am. Chem. Soc. 2015, 137, 6975–6978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Toma, R.S.; Kuthning, A.; Exner, M.P.; Denisiuk, A.; Ziegler, J.; Budisa, N.; Süssmuth, R.D. Site-Directed and Global Incorporation of Orthogonal and Isostructural Noncanonical Amino Acids into the Ribosomal Lasso Peptide Capistruin. ChemBioChem 2015, 16, 503–509. [Google Scholar] [CrossRef]
- Piscotta, F.J.; Tharp, J.M.; Liu, W.R.; Link, A.J. Expanding the Chemical Diversity of Lasso Peptide MccJ25 with Genetically Encoded Noncanonical Amino Acids. Chem. Commun. 2014, 51, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Zambaldo, C.; Liu, T.; Zhang, Y.; Xuan, W.; Wang, C.; Reed, S.A.; Yang, P.-Y.; Wang, R.E.; Javahishvili, T.; et al. Recombinant Thiopeptides Containing Noncanonical Amino Acids. Proc. Natl. Acad. Sci. USA 2016, 113, 3615–3620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Yang, X.; Garg, N.; van der Donk, W.A. Production of Lantipeptides in Escherichia coli. J. Am. Chem. Soc. 2011, 133, 2338–2341. [Google Scholar] [CrossRef] [PubMed]
- Lopatniuk, M.; Myronovskyi, M.; Luzhetskyy, A. Streptomyces Albus: A New Cell Factory for Non-Canonical Amino Acids Incorporation into Ribosomally Synthesized Natural Products. ACS Chem. Biol. 2017, 12, 2362–2370. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Z.; Brock, A.; Schultz, P.G. Addition of the Keto Functional Group to the Genetic Code of Escherichia coli. Proc. Natl. Acad. Sci. USA 2003, 100, 56–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.H.; Axup, J.Y.; Schultz, P.G. Protein Conjugation with Genetically Encoded Unnatural Amino Acids. Curr. Opin. Chem. Biol. 2013, 17, 412–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Zhu, H.; Zhang, B.; Liu, F.; Chen, J.; Wang, Y.; Wang, Y.; Zhang, Z.; Wu, L.; Si, L.; et al. Synthesis of Site-Specific Radiolabeled Antibodies for Radioimmunotherapy via Genetic Code Expansion. Bioconjug. Chem. 2016, 27, 2460–2468. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, C.; Xu, H.; An, Y.; Xiao, S.; Zheng, M.; Liu, L.; Nie, L. Incorporation of Nonstandard Amino Acids into Proteins: Principles and Applications. World J. Microbiol. Biotechnol. 2020, 36, 60. [Google Scholar] [CrossRef]
- Axup, J.Y.; Bajjuri, K.M.; Ritland, M.; Hutchins, B.M.; Kim, C.H.; Kazane, S.A.; Halder, R.; Forsyth, J.S.; Santidrian, A.F.; Stafin, K.; et al. Synthesis of Site-Specific Antibody-Drug Conjugates Using Unnatural Amino Acids. Proc. Natl. Acad. Sci. USA 2012, 109, 16101–16106. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.J.; Kang, D.; Park, H.-S. Site-Specific Labeling of Proteins Using Unnatural Amino Acids. Mol. Cells 2019, 42, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Lieser, R.M.; Chen, W.; Sullivan, M.O. Controlled Epidermal Growth Factor Receptor Ligand Display on Cancer Suicide Enzymes via Unnatural Amino Acid Engineering for Enhanced Intracellular Delivery in Breast Cancer Cells. Bioconj. Chem. 2019, 30, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Kularatne, S.A.; Deshmukh, V.; Ma, J.; Tardif, V.; Lim, R.K.V.; Pugh, H.M.; Sun, Y.; Manibusan, A.; Sellers, A.J.; Barnett, R.S.; et al. A CXCR4-Targeted Site-Specific Antibody–Drug Conjugate. Angew. Chem. Int. Ed. 2014, 53, 11863–11867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, R.K.V.; Yu, S.; Cheng, B.; Li, S.; Kim, N.-J.; Cao, Y.; Chi, V.; Kim, J.Y.; Chatterjee, A.K.; Schultz, P.G.; et al. Targeted Delivery of LXR Agonist Using a Site-Specific Antibody–Drug Conjugate. Bioconj. Chem. 2015, 26, 2216–2222. [Google Scholar] [CrossRef]
- Yu, S.; Pearson, A.D.; Lim, R.K.; Rodgers, D.T.; Li, S.; Parker, H.B.; Weglarz, M.; Hampton, E.N.; Bollong, M.J.; Shen, J.; et al. Targeted Delivery of an Anti-Inflammatory PDE4 Inhibitor to Immune Cells via an Antibody–Drug Conjugate. Mol. Ther. 2016, 24, 2078–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho-Villegas, T.A.; Mata-González, M.T.; García-Ubbelohd, W.; Núñez-García, L.; Elosua, C.; Paniagua-Solis, J.F.; Licea-Navarro, A.F. Intraocular Penetration of a VNAR: In Vivo and In Vitro VEGF165 Neutralization. Mar. Drugs 2018, 16, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, M.; Bian, H.; Wu, X.; Fu, T.; Fu, Y.; Hong, J.; Fleming, B.D.; Flajnik, M.F.; Ho, M. Construction and Next-Generation Sequencing Analysis of a Large Phage-Displayed VNAR Single-Domain Antibody Library from Six Naïve Nurse Sharks. Antib. Ther. 2019, 2, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.H.; Axup, J.Y.; Dubrovska, A.; Kazane, S.A.; Hutchins, B.A.; Wold, E.D.; Smider, V.V.; Schultz, P.G. Synthesis of Bispecific Antibodies Using Genetically Encoded Unnatural Amino Acids. J. Am. Chem. Soc. 2012, 134, 9918–9921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Axup, J.Y.; Ma, J.S.Y.; Wang, R.E.; Choi, S.; Tardif, V.; Lim, R.K.V.; Pugh, H.M.; Lawson, B.R.; Welzel, G.; et al. Multiformat T-Cell-Engaging Bispecific Antibodies Targeting Human Breast Cancers. Angew. Chem. 2015, 127, 7128–7133. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Zhou, Q.; Deshmukh, V.; Phull, H.; Ma, J.; Tardif, V.; Naik, R.R.; Bouvard, C.; Zhang, Y.; Choi, S.; et al. Targeting Human C-Type Lectin-like Molecule-1 (CLL1) with a Bispecific Antibody for Immunotherapy of Acute Myeloid Leukemia. Angew. Chem. Int. Ed. 2014, 53, 9841–9845. [Google Scholar] [CrossRef] [Green Version]
- Ramadoss, N.S.; Schulman, A.D.; Choi, S.; Rodgers, D.T.; Kazane, S.A.; Kim, C.H.; Lawson, B.R.; Young, T.S. An Anti-B Cell Maturation Antigen Bispecific Antibody for Multiple Myeloma. J. Am. Chem. Soc. 2015, 137, 5288–5291. [Google Scholar] [CrossRef]
- Kim, C.H.; Axup, J.Y.; Lawson, B.R.; Yun, H.; Tardif, V.; Choi, S.H.; Zhou, Q.; Dubrovska, A.; Biroc, S.L.; Marsden, R.; et al. Bispecific Small Molecule–Antibody Conjugate Targeting Prostate Cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 17796–17801. [Google Scholar] [CrossRef] [Green Version]
- Kularatne, S.A.; Deshmukh, V.; Gymnopoulos, M.; Biroc, S.L.; Xia, J.; Srinagesh, S.; Sun, Y.; Zou, N.; Shimazu, M.; Pinkstaff, J.; et al. Recruiting Cytotoxic T Cells to Folate-Receptor-Positive Cancer Cells. Angew. Chem. 2013, 125, 12323–12326. [Google Scholar] [CrossRef]
- Lyu, Z.; Kang, L.; Buuh, Z.Y.; Jiang, D.; McGuth, J.C.; Du, J.; Wissler, H.L.; Cai, W.; Wang, R.E. A Switchable Site-Specific Antibody Conjugate. ACS Chem. Biol. 2018, 13, 958–964. [Google Scholar] [CrossRef]
- Weigle, W.O. The Induction of Autoimmunity in Rabbits Following Injection of Heterologous or Altered Homologous Thyroglobulin. J. Exp. Med. 1965, 121, 289–308. [Google Scholar] [CrossRef] [Green Version]
- Dalum, I.; Butler, D.M.; Jensen, M.R.; Hindersson, P.; Steinaa, L.; Waterston, A.M.; Grell, S.N.; Feldmann, M.; Elsner, H.I.; Mouritsen, S. Therapeutic Antibodies Elicited by Immunization against TNF-α. Nat. Biotechnol. 1999, 17, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Zuany-Amorim, C.; Manlius, C.; Dalum, I.; Jensen, M.R.; Gautam, A.; Pay, G.; Mouritsen, S.; Walker, C. Induction of TNF-α Autoantibody Production by AutoVac TNF106: A Novel Therapeutic Approach for the Treatment of Allergic Diseases. Int. Arch. Allergy Immunol. 2004, 133, 154–163. [Google Scholar] [CrossRef]
- Leitner, W.W.; Hwang, L.N.; deVeer, M.J.; Zhou, A.; Silverman, R.H.; Williams, B.R.G.; Dubensky, T.W.; Ying, H.; Restifo, N.P. Alphavirus-Based DNA Vaccine Breaks Immunological Tolerance by Activating Innate Antiviral Pathways. Nat. Med. 2003, 9, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, J.; Tsao, M.-L.; Perera, R.; Dong, L.; Niessen, F.; Wen, B.G.; Kubitz, D.M.; Smider, V.V.; Ruf, W.; Nasoff, M.; et al. Immunochemical Termination of Self-Tolerance. Proc. Natl. Acad. Sci. USA 2008, 105, 11276–11280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauba, V.; Grünewald, J.; Gorney, V.; Deaton, L.M.; Kang, M.; Bursulaya, B.; Ou, W.; Lerner, R.A.; Schmedt, C.; Geierstanger, B.H.; et al. Loss of CD4 T-Cell–Dependent Tolerance to Proteins with Modified Amino Acids. Proc. Natl. Acad. Sci. USA 2011, 108, 12821–12826. [Google Scholar] [CrossRef] [Green Version]
- Grünewald, J.; Hunt, G.S.; Dong, L.; Niessen, F.; Wen, B.G.; Tsao, M.-L.; Perera, R.; Kang, M.; Laffitte, B.A.; Azarian, S.; et al. Mechanistic Studies of the Immunochemical Termination of Self-Tolerance with Unnatural Amino Acids. Proc. Natl. Acad. Sci. USA 2009, 106, 4337–4342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
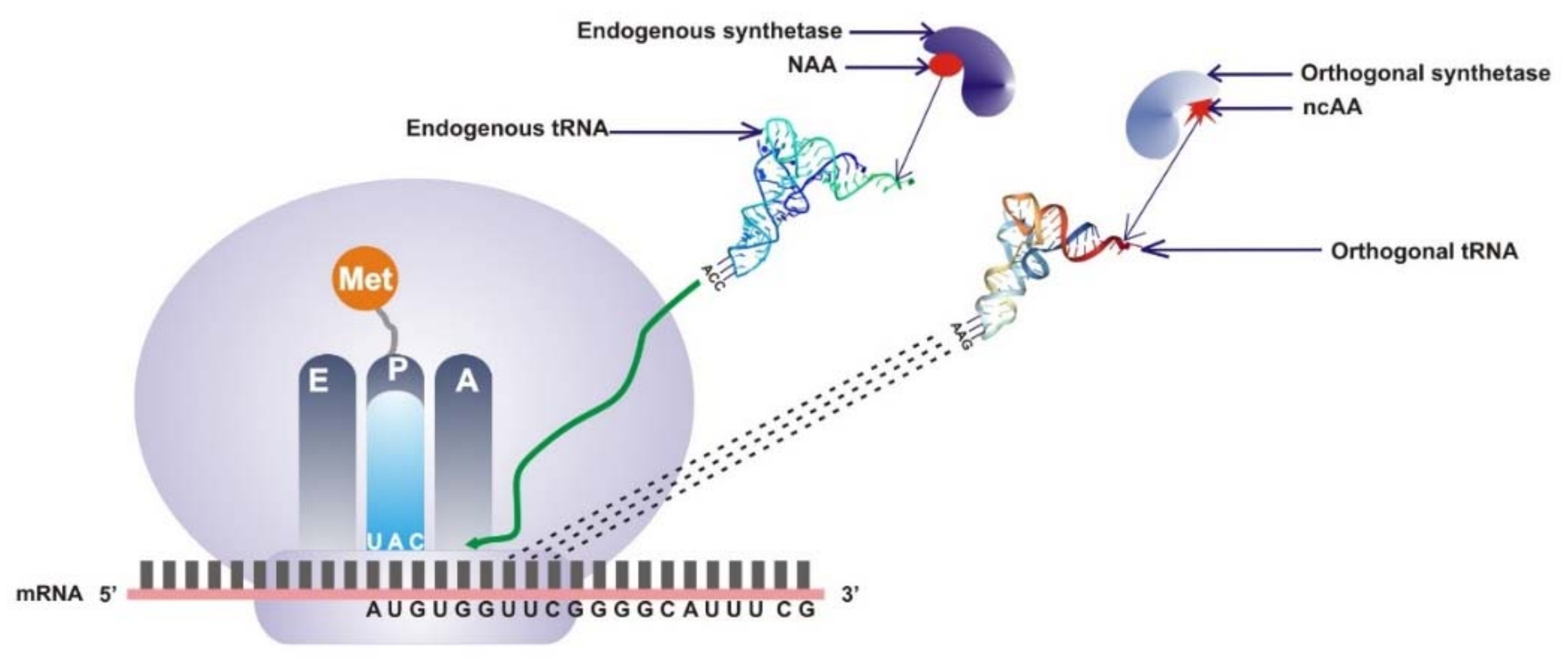

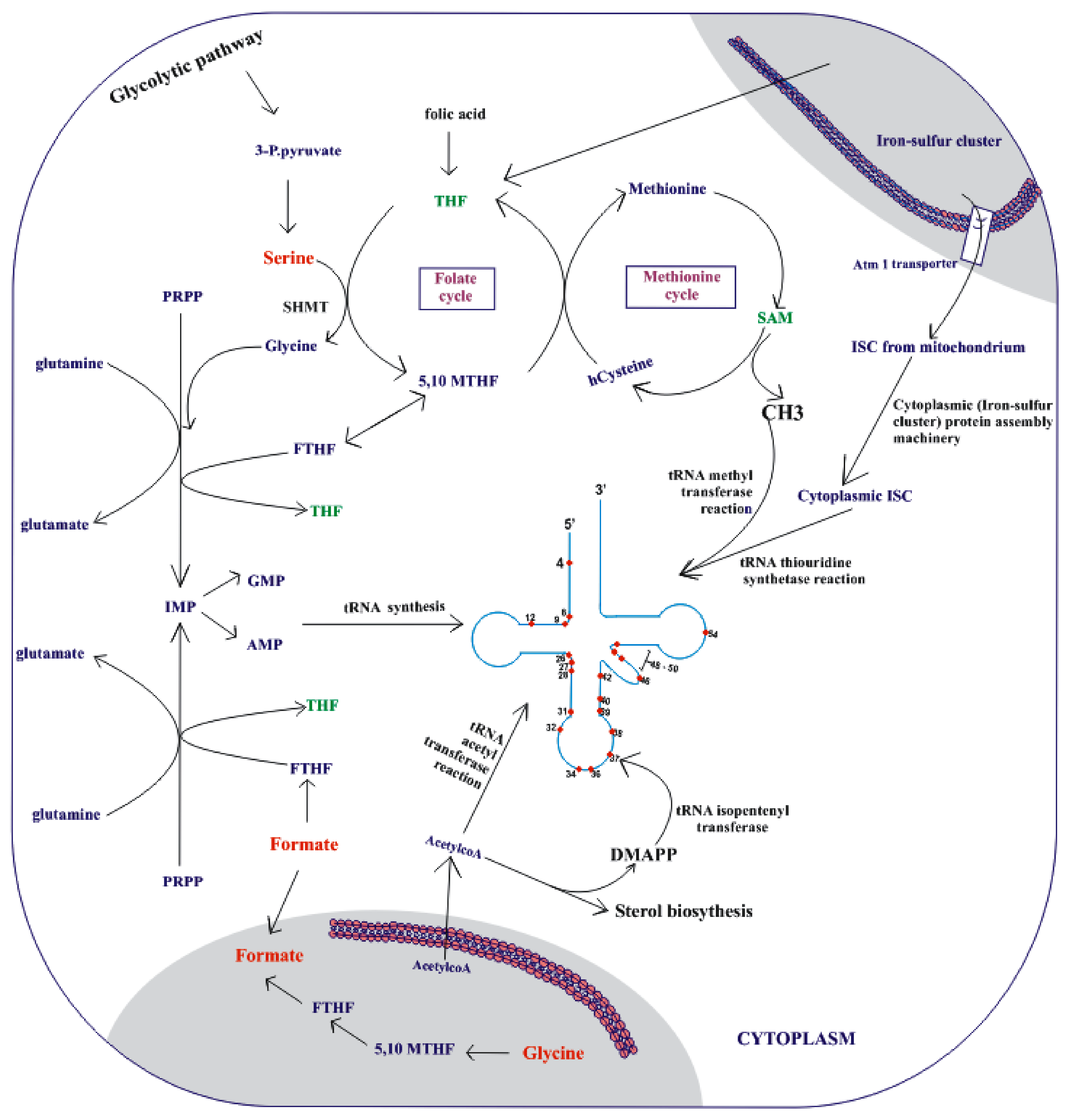

| Host of Orthogonal tRNA | tRNA | Anticodons | Noncanonical Amino Acids | Host Organism | Refs |
|---|---|---|---|---|---|
| Archaeal | |||||
| M. jannaschii | tRNATyr | CUA | O-methyl-L-tyrosine | E. coli | [35] |
| tRNACys | CUA | O-phosphoserine (Sep) | E. coli | [36] | |
| tRNAPyl | UCCU | N-3-(tert-butyloxycarbonyl)-L-lysine (Boc-Lys) | E. coli and mammalian cells | [37] | |
| tRNASer2 | UCCU | p-azido-L-phenylalanine | E. coli | [33] | |
| Methanosarcina. barkeri | tRNAPyl | CUA | Nε-acetyllysyl | E. coli | [38] |
| Nε-D-prolyl-L-lysine and N-Nε-cyclopentyloxycarbonyl-L-lysine | E. coli | [39] | |||
| Methanosarcina acetivorans | tRNAPyl | CGGA, CGGG | p-Nitrophenylalanine and 2-naphthylalanine | E. coli and rabbit reticulocyte cell-free systems | [40] |
| Methanosarcina mazei | tRNAPyl | CUA | O-methyl-L-tyrosine | E. coli | [41] |
| Nε-methyl-L-lysine | E. coli | [42] | |||
| Drosophila melanogaster | [20] | ||||
| Nε-tert-butyloxycarbonyl-L-lysine | Mammalian cells | [20] | |||
| Nε benzyloxycarbonyl-L-lysine | C. elegans | [43] | |||
| Nε-methyl L-lysine | [44] | ||||
| UCA | Nε-tert-butyloxycarbonyl-L-lysine | E. coli | [45] | ||
| Yeast | |||||
| S. cerevisiae | tRNATyr | CCCG | 2-naphthylalanine L-lysine | E. coli | [46] |
| tRNAPhe | CUA | p-bromophenylalanine | E. coli | [47] | |
| CUA | p-nitrophenylalanine | E. coli | [48] | ||
| ACCU, ACCG | [49] | ||||
| AGGG, AGAG | |||||
| AUAG, ACCC | |||||
| AGAG, CGGU | |||||
| CGCU, CCCU | |||||
| CUAU, GGGU | |||||
| Bacterial | |||||
| E. coli | tRNATyr | CUA | 3-iodo-L-tyrosine | E. coli | [50] |
| Mammalian cells | [51] | ||||
| Drosophila melanogaster | [52] | ||||
| 3-iodo-L-tyrosine | Mammalian cells | [53] | |||
| S. cerevisiae | |||||
| 4-azido-l-phenylalanine | |||||
| p-acetyl-L-phenylalanine | |||||
| p-benzoyl-L-phenylalanine | |||||
| p-azido-L-phenylalanine | |||||
| o-methyl-L-tyrosine | |||||
| p-iodo-L-phenylalanine | |||||
| tRNALeu | CUA | p-azido-L-phenylalanine | Schizosaccharomyces pombe | [54] | |
| o-methyltyrosine | S. cerevisiae | ||||
| α-amino-caprylic-acid | [55] | ||||
| o-nitrobenzyl cysteine | |||||
| B. subtilis | tRNATrp | UCA | 5-hydroxytryptophan | Mammalian cells | [56] |
| Geobacillus stearothermophilus | tRNATyr | CUA | p-methoxyphenylalanine (pMpa), | Mammalian cells | [57] |
| p-acetylphenylalanine (pApa), | [58] | ||||
| p-benzoylphenylalanine (pBpa), | |||||
| p-iodophenylalanine (pIpa), | |||||
| p-azidophenylalanine (pAzpa) | |||||
| p-propargyloxyphenylalanine |
| Anticodon Loop Position | S. cerevisiae | Enzymes | Refs | E. coli | Enzymes | Refs | Role during Incorporation |
|---|---|---|---|---|---|---|---|
| 26 | m22G | Trm1 | [106] | - | - | - | Improves translation fidelity efficiency |
| 32 | Cm | Trm7 | [107] | Cm, Um | TrmJ | [108] | |
| m3C, m3U | Trm140 | [109] | s2C | IscS, TtcA | [110] | ||
| Ψ | Pus8 | [111] | Ψ | RluA | [110] | ||
| 34 | Cm, Gm, cmnm5Um | Trm7/Trm7 | [107] | Cm, Um Cmnm5Um | TrmL TrmL | [108] [107] | |
| ncm5U mcm5U | Elp complex Trm9 | [112] | cmnm5Um | TrmL | [110] | ||
| m5C | Trm4 | [113] | ac4C | TmcA | [110] | ||
| ψ | Pus1 | [114] | mnm5se2U | TrmL | [110] | ||
| mcm5s2U | Uba4 Ncs2 Ncs6 | [115] [116] | s2U | IscS TusA | [117] [110] | ||
| A → I | Tad2,3 | [91] | A → I | TadA | [118] | ||
| 36 | Ψ | Pus1 | [119] | i6A | MiaA | [110] | |
| 37 | m1G | Trm5 | [90] | m1G | TrmD | [110] | |
| m2A | RlmN | [108] | |||||
| m1I | Trm5 | [120] | m6A | TrmN6 | [110] | ||
| i6A | MiaA | ||||||
| yW | Tyw1-4 | [121] | ms2i6A | MiaB | |||
| m6t6A | TrmO | ||||||
| I6A | Mod5 | [122] | |||||
| 38 | Ψ | Pus3 | [119] | Ψ | TruA | [110] | |
| 40 | m5C | Trm4 | [113] | Ψ | TruA | [110] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lateef, O.M.; Akintubosun, M.O.; Olaoba, O.T.; Samson, S.O.; Adamczyk, M. Making Sense of “Nonsense” and More: Challenges and Opportunities in the Genetic Code Expansion, in the World of tRNA Modifications. Int. J. Mol. Sci. 2022, 23, 938. https://doi.org/10.3390/ijms23020938
Lateef OM, Akintubosun MO, Olaoba OT, Samson SO, Adamczyk M. Making Sense of “Nonsense” and More: Challenges and Opportunities in the Genetic Code Expansion, in the World of tRNA Modifications. International Journal of Molecular Sciences. 2022; 23(2):938. https://doi.org/10.3390/ijms23020938
Chicago/Turabian StyleLateef, Olubodun Michael, Michael Olawale Akintubosun, Olamide Tosin Olaoba, Sunday Ocholi Samson, and Malgorzata Adamczyk. 2022. "Making Sense of “Nonsense” and More: Challenges and Opportunities in the Genetic Code Expansion, in the World of tRNA Modifications" International Journal of Molecular Sciences 23, no. 2: 938. https://doi.org/10.3390/ijms23020938
APA StyleLateef, O. M., Akintubosun, M. O., Olaoba, O. T., Samson, S. O., & Adamczyk, M. (2022). Making Sense of “Nonsense” and More: Challenges and Opportunities in the Genetic Code Expansion, in the World of tRNA Modifications. International Journal of Molecular Sciences, 23(2), 938. https://doi.org/10.3390/ijms23020938







