TMAO Upregulates Members of the miR-17/92 Cluster and Impacts Targets Associated with Atherosclerosis
Abstract
1. Introduction
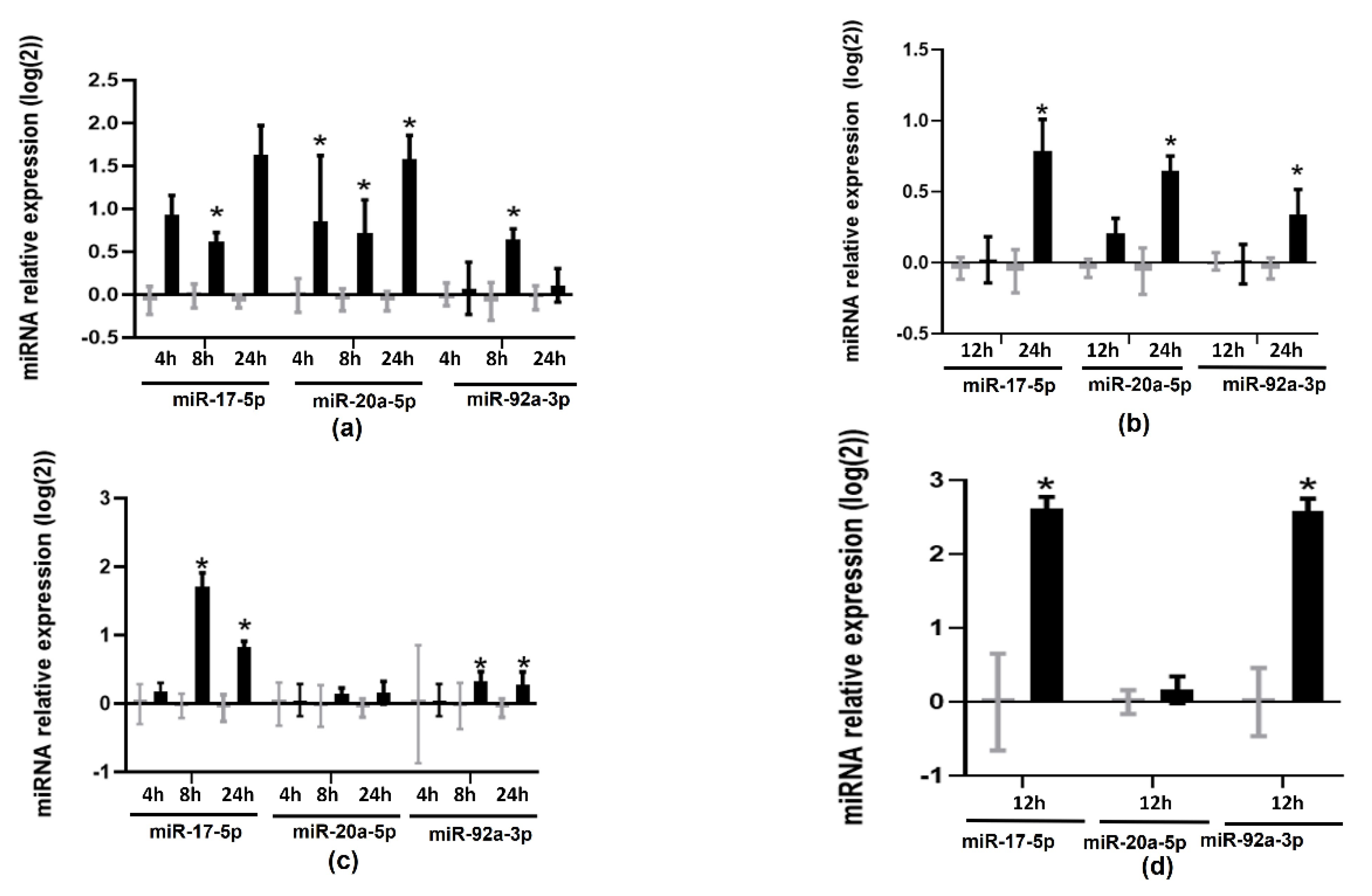
2. Results
3. Discussion
4. Materials and Methods
4.1. Cellular Models
4.2. Treatments
4.3. microRNA-Enriched RNA Isolation and Amplification
4.4. Gene Expression Measurement
4.5. Measurement of Protein Levels
4.6. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khandelwal, S.; Kurpad, A.; Narayan, K.M.V. Global Non-Communicable Diseases—The Nutrition Conundrum. Front. Public Health 2018, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Pan, A.; Sun, Q.; Bernstein, A.M.; Schulze, M.B.; Manson, J.E.; Stampfer, M.J.; Willett, W.C.; Hu, F.B. Red Meat Consumption and Mortality: Results from 2 Prospective Cohort Studies. Arch. Intern. Med. 2012, 172, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Vernooij, R.W.M.; Zeraatkar, D.; Han, M.A.; El Dib, R.; Zworth, M.; Milio, K.; Sit, D.; Lee, Y.; Gomaa, H.; Valli, C.; et al. Patterns of Red and Processed Meat Consumption and Risk for Cardiometabolic and Cancer Outcomes: A Systematic Review and Meta-Analysis of Cohort Studies. Ann. Intern. Med. 2019, 171, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal Microbiota Metabolism of L-Carnitine, a Nutrient in Red Meat, Promotes Atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef]
- Stock, J. Gut Microbiota: An Environmental Risk Factor for Cardiovascular Disease. Atherosclerosis 2013, 229, 440–442. [Google Scholar] [CrossRef]
- Geng, J.; Yang, C.; Wang, B.; Zhang, X.; Hu, T.; Gu, Y.; Li, J. Trimethylamine N-Oxide Promotes Atherosclerosis via CD36-Dependent MAPK/JNK Pathway. Biomed. Pharmacother. 2018, 97, 941–947. [Google Scholar] [CrossRef]
- Sheng, Z.; Tan, Y.; Liu, C.; Zhou, P.; Li, J.; Zhou, J.; Chen, R.; Chen, Y.; Song, L.; Zhao, H.; et al. Relation of Circulating Trimethylamine N-Oxide With Coronary Atherosclerotic Burden in Patients With ST-Segment Elevation Myocardial Infarction. Am. J. Cardiol. 2019, 123, 894–898. [Google Scholar] [CrossRef]
- Costabile, G.; Vetrani, C.; Bozzetto, L.; Giacco, R.; Bresciani, L.; Del Rio, D.; Vitale, M.; Della Pepa, G.; Brighenti, F.; Riccardi, G.; et al. Plasma TMAO Increase after Healthy Diets: Results from 2 Randomized Controlled Trials with Dietary Fish, Polyphenols, and Whole-Grain Cereals. Am. J. Clin. Nutr. 2021, 114, 1342–1350. [Google Scholar] [CrossRef]
- Andraos, S.; Jones, B.; Lange, K.; Clifford, S.A.; Thorstensen, E.B.; Kerr, J.A.; Wake, M.; Saffery, R.; Burgner, D.P.; O’Sullivan, J.M. Trimethylamine N-Oxide (TMAO) Is Not Associated with Cardiometabolic Phenotypes and Inflammatory Markers in Children and Adults. Curr. Dev. Nutr. 2021, 5, nzaa179. [Google Scholar] [CrossRef]
- Meyer, K.A.; Benton, T.Z.; Bennett, B.J.; Jacobs, D.R.J.; Lloyd-Jones, D.M.; Gross, M.D.; Carr, J.J.; Gordon-Larsen, P.; Zeisel, S.H. Microbiota-Dependent Metabolite Trimethylamine N-Oxide and Coronary Artery Calcium in the Coronary Artery Risk Development in Young Adults Study (CARDIA). J. Am. Heart Assoc. 2016, 5, e003970. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, K.; Hering, D.; Mosieniak, G.; Bielak-Zmijewska, A.; Pilz, M.; Konwerski, M.; Gasecka, A.; Kapłon-Cieślicka, A.; Filipiak, K.; Sikora, E.; et al. TMA, A Forgotten Uremic Toxin, but Not TMAO, Is Involved in Cardiovascular Pathology. Toxins 2019, 11, 490. [Google Scholar] [CrossRef]
- Díez-Ricote, L.; San-Cristobal, R.; Concejo, M.J.; Martínez-González, M.Á.; Corella, D.; Salas-Salvadó, J.; Goday, A.; Martínez, J.A.; Alonso-Gómez, Á.M.; Wärnberg, J.; et al. One-Year Longitudinal Association between Changes in Dietary Choline or Betaine Intake Association with Cardiometabolic Variables in the PREDIMED-Plus Trial. Am. J. Clin. Nutr. 2022, nqac255. [Google Scholar] [CrossRef]
- Soh, J.; Iqbal, J.; Queiroz, J.; Fernandez-Hernando, C.; Hussain, M.M. MicroRNA-30c Reduces Hyperlipidemia and Atherosclerosis in Mice by Decreasing Lipid Synthesis and Lipoprotein Secretion. Nat. Med. 2013, 19, 892–900. [Google Scholar] [CrossRef]
- Díez-Ricote, L.; Ruiz-Valderrey, P.; Micó, V.; Blanco-Rojo, R.; Tomé-Carneiro, J.; Dávalos, A.; Ordovás, J.M.; Daimiel, L. Trimethylamine N-Oxide (TMAO) Modulates the Expression of Cardiovascular Disease-Related MicroRNAs and Their Targets. Int. J. Mol. Sci. 2021, 22, 11145. [Google Scholar] [CrossRef]
- Canfrán-Duque, A.; Rotllan, N.; Zhang, X.; Fernández-Fuertes, M.; Ramírez-Hidalgo, C.; Araldi, E.; Daimiel, L.; Busto, R.; Fernández-Hernando, C.; Suárez, Y. Macrophage Deficiency of MiR-21 Promotes Apoptosis, Plaque Necrosis, and Vascular Inflammation during Atherogenesis. EMBO Mol. Med. 2017, 9, 1244–1262. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Liu, Z.; Zhou, L. Roles of MiR-17-92 Cluster in Cardiovascular Development and Common Diseases. Biomed. Res. Int. 2017, 2017, 9102909. [Google Scholar] [CrossRef] [PubMed]
- Danielson, L.S.; Park, D.S.; Rotllan, N.; Chamorro-Jorganes, A.; Guijarro, M.V.; Fernandez-Hernando, C.; Fishman, G.I.; Phoon, C.K.L.; Hernando, E. Cardiovascular Dysregulation of MiR-17-92 Causes a Lethal Hypertrophic Cardiomyopathy and Arrhythmogenesis. FASEB J. 2013, 27, 1460–1467. [Google Scholar] [CrossRef]
- Ai, F.; Zhang, Y.; Peng, B. MiR-20a Regulates Proliferation, Differentiation and Apoptosis in P19 Cell Model of Cardiac Differentiation by Targeting Smoothened. Biol. Open 2016, 5, 1260–1265. [Google Scholar] [CrossRef]
- Bonauer, A.; Carmona, G.; Iwasaki, M.; Mione, M.; Koyanagi, M.; Fischer, A.; Burchfield, J.; Fox, H.; Doebele, C.; Ohtani, K.; et al. MicroRNA-92a Controls Angiogenesis and Functional Recovery of Ischemic Tissues in Mice. Science 2009, 324, 1710–1713. [Google Scholar] [CrossRef]
- Du, W.W.; Li, X.; Li, T.; Li, H.; Khorshidi, A.; Liu, F.; Yang, B.B. The MicroRNA MiR-17-3p Inhibits Mouse Cardiac Fibroblast Senescence by Targeting Par4. J. Cell Sci. 2015, 128, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, R.; Zhang, Y.; Qiu, J.; Ling, W. Association of Plasma MiR-17-92 with Dyslipidemia in Patients with Coronary Artery Disease. Medicine 2014, 93, e98. [Google Scholar] [CrossRef]
- Gong, R.; Lv, X.; Liu, F. MiRNA-17 Encoded by the MiR-17-92 Cluster Increases the Potential for Steatosis in Hepatoma Cells by Targeting CYP7A1. Cell. Mol. Biol. Lett. 2018, 23, 16. [Google Scholar] [CrossRef] [PubMed]
- Daimiel, L.; Micó, V.; Valls, R.M.; Pedret, A.; Motilva, M.J.; Rubió, L.; Fitó, M.; Farrás, M.; Covas, M.I.; Solá, R.; et al. Impact of Phenol-Enriched Virgin Olive Oils on the Postprandial Levels of Circulating MicroRNAs Related to Cardiovascular Disease. Mol. Nutr. Food Res. 2020, 64, e2000049. [Google Scholar] [CrossRef] [PubMed]
- Daimiel, L.; Micó, V.; Díez-Ricote, L.; Ruiz-Valderrey, P.; Istas, G.; Rodríguez-Mateos, A.; Ordovás, J.M. Alcoholic and Non-Alcoholic Beer Modulate Plasma and Macrophage MicroRNAs Differently in a Pilot Intervention in Humans with Cardiovascular Risk. Nutrients 2020, 13, 69. [Google Scholar] [CrossRef]
- Schneiderman, J.; Sawdey, M.S.; Keeton, M.R.; Bordin, G.M.; Bernstein, E.F.; Dilley, R.B.; Loskutoff, D.J. Increased Type 1 Plasminogen Activator Inhibitor Gene Expression in Atherosclerotic Human Arteries. Proc. Natl. Acad. Sci. USA 1992, 89, 6998–7002. [Google Scholar] [CrossRef]
- Raghunath, P.N.; Tomaszewski, J.E.; Brady, S.T.; Caron, R.J.; Okada, S.S.; Barnathan, E.S. Plasminogen Activator System in Human Coronary Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 1995, 15, 1432–1443. [Google Scholar] [CrossRef]
- Bastos, K.R.; Barboza, R.; Sardinha, L.; Russo, M.; Alvarez, J.M.; Lima, M.R. Role of Endogenous IFN-Gamma in Macrophage Programming Induced by IL-12 and IL-18. J. Interferon Cytokine Res. 2007, 27, 399–410. [Google Scholar] [CrossRef]
- Chen, T.; Huang, J.B.; Dai, J.; Zhou, Q.; Raj, J.U.; Zhou, G. PAI-1 Is a Novel Component of the MiR-17~92 Signaling That Regulates Pulmonary Artery Smooth Muscle Cell Phenotypes. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 315, L149–L161. [Google Scholar] [CrossRef]
- Mestdagh, P.; Boström, A.-K.; Impens, F.; Fredlund, E.; Van Peer, G.; De Antonellis, P.; von Stedingk, K.; Ghesquière, B.; Schulte, S.; Dews, M.; et al. The MiR-17-92 MicroRNA Cluster Regulates Multiple Components of the TGF-β Pathway in Neuroblastoma. Mol. Cell 2010, 40, 762–773. [Google Scholar] [CrossRef]
- Izreig, S.; Samborska, B.; Johnson, R.M.; Sergushichev, A.; Ma, E.H.; Lussier, C.; Loginicheva, E.; Donayo, A.O.; Poffenberger, M.C.; Sagan, S.M.; et al. The MiR-17∼92 MicroRNA Cluster Is a Global Regulator of Tumor Metabolism. Cell Rep. 2016, 16, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tang, S.; Ji-Yan, C.; Huang, C.; Li, J.; Cai, A.P.; Feng, Y.Q. Circulating MiR-92a Expression Level in Patients with Essential Hypertension: A Potential Marker of Atherosclerosis. J. Hum. Hypertens. 2017, 31, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Zhang, S.; Yan, S.; Wang, C.; Zhang, X. MiR-30e and MiR-92a Are Related to Atherosclerosis by Targeting ABCA1. Mol. Med. Rep. 2019, 19, 3298–3304. [Google Scholar] [CrossRef]
- Telkoparan-Akillilar, P.; Cevik, D. Identification of MiR-17, MiR-21, MiR-27a, MiR-106b and MiR-222 as Endoplasmic Reticulum Stress-Related Potential Biomarkers in Circulation of Patients with Atherosclerosis. Mol. Biol. Rep. 2021, 48, 3503–3513. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Liu, D.; Zhu, W.; Su, Z.; Zhang, L.; Zhou, C.; Li, P. Circulating MiR-17-5p, MiR-126-5p and MiR-145-3p Are Novel Biomarkers for Diagnosis of Acute Myocardial Infarction. Front. Physiol. 2019, 10, 123. [Google Scholar] [CrossRef]
- Zhao, L.; Jiang, S.; Wu, N.; Shi, E.; Yang, L.; Li, Q. MiR-17-5p-Mediated Endoplasmic Reticulum Stress Promotes Acute Myocardial Ischemia Injury through Targeting Tsg101. Cell Stress Chaperones 2021, 26, 77–90. [Google Scholar] [CrossRef]
- Gao, G.; Chen, W.; Liu, M.; Yan, X.; Yang, P. Circulating MicroRNAs as Novel Potential Biomarkers for Left Ventricular Remodeling in Postinfarction Heart Failure. Dis. Markers 2019, 2019, 5093803. [Google Scholar] [CrossRef]
- Liang, B.; Wang, X.; Song, X.; Bai, R.; Yang, H.; Yang, Z.; Xiao, C.; Bian, Y. MicroRNA-20a/b Regulates Cholesterol Efflux through Post-Transcriptional Repression of ATP-Binding Cassette Transporter A1. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 929–938. [Google Scholar] [CrossRef]
- Chen, M.; Li, W.; Zhang, Y.; Yang, J. MicroRNA-20a Protects Human Aortic Endothelial Cells from Ox-LDL-Induced Inflammation through Targeting TLR4 and TXNIP Signaling. Biomed. Pharmacother. 2018, 103, 191–197. [Google Scholar] [CrossRef]
- Gong, X.-Y.; Zhang, Y. Protective Effect of MiR-20a against Hypoxia/Reoxygenation Treatment on Cardiomyocytes Cell Viability and Cell Apoptosis by Targeting TLR4 and Inhibiting P38 MAPK/JNK Signaling. In Vitro Cell. Dev. Biol. Anim. 2019, 55, 793–800. [Google Scholar] [CrossRef]
- Lupu, F.; Bergonzelli, G.E.; Heim, D.A.; Cousin, E.; Genton, C.Y.; Bachmann, F.; Kruithof, E.K. Localization and Production of Plasminogen Activator Inhibitor-1 in Human Healthy and Atherosclerotic Arteries. Arterioscler. Thromb. 1993, 13, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, D.E. PAI-1 and Atherothrombosis. J. Thromb. Haemost. 2005, 3, 1879–1883. [Google Scholar] [CrossRef] [PubMed]
- Henkel, A.S.; Khan, S.S.; Olivares, S.; Miyata, T.; Vaughan, D.E. Inhibition of Plasminogen Activator Inhibitor 1 Attenuates Hepatic Steatosis but Does Not Prevent Progressive Nonalcoholic Steatohepatitis in Mice. Hepatol. Commun. 2018, 2, 1479–1492. [Google Scholar] [CrossRef] [PubMed]
- De Larrañaga, G.; Wingeyer, S.P.; Graffigna, M.; Belli, S.; Bendezú, K.; Alvarez, S.; Levalle, O.; Fainboim, H. Plasma Plasminogen Activator Inhibitor-1 Levels and Nonalcoholic Fatty Liver in Individuals with Features of Metabolic Syndrome. Clin. Appl. Thromb. Hemost. 2008, 14, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Nakada, T.A.; Takahashi, W.; Nakada, E.; Shimada, T.; Russell, J.A.; Walley, K.R. Genetic Polymorphisms in Sepsis and Cardiovascular Disease: Do Similar Risk Genes Suggest Similar Drug Targets? Chest 2019, 155, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Marchand, A.; Proust, C.; Morange, P.E.; Lompré, A.M.; Trégouët, D.A. MiR-421 and MiR-30c Inhibit SERPINE 1 Gene Expression in Human Endothelial Cells. PLoS ONE 2012, 7, e44532. [Google Scholar] [CrossRef] [PubMed]
- Posadas-Sánchez, R.; Pérez-Hernández, N.; Angeles-Martínez, J.; López-Bautista, F.; Villarreal-Molina, T.; Rodríguez-Pérez, J.M.; Fragoso, J.M.; Posadas-Romero, C.; Vargas-Alarcón, G. Interleukin 35 Polymorphisms Are Associated with Decreased Risk of Premature Coronary Artery Disease, Metabolic Parameters, and IL-35 Levels: The Genetics of Atherosclerotic Disease (GEA) Study. Mediators Inflamm. 2017, 2017, 6012795. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, X.; Fan, Y.; Hu, X.; Liu, X.; Xiang, M.; Wang, J. Dysregulated Expression of MicroRNAs and MRNAs in Myocardial Infarction. Am. J. Transl. Res. 2015, 7, 2291–2304. [Google Scholar]
- Zhang, X.; Smith, S.M.; Wang, X.; Zhao, B.; Wu, L.; Hu, X. Three Paralogous Clusters of the MiR-17~92 Family of MicroRNAs Restrain IL-12-Mediated Immune Defense. Cell. Mol. Immunol. 2021, 18, 1751–1760. [Google Scholar] [CrossRef]
- Zhang, L.; Xie, F.; Tang, H.; Zhang, X.; Hu, J.; Zhong, X.; Gong, N.; Lai, Y.; Zhou, M.; Tian, J.; et al. Gut Microbial Metabolite TMAO Increases Peritoneal Inflammation and Peritonitis Risk in Peritoneal Dialysis Patients. Transl. Res. 2022, 240, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, M.E.; Hov, J.R.; Ueland, T.; Dahl, T.B.; Kummen, M.; Otterdal, K.; Holm, K.; Berge, R.K.; Mollnes, T.E.; Trøseid, M.; et al. Gut Microbiota-Dependent Trimethylamine N-Oxide Associates With Inflammation in Common Variable Immunodeficiency. Front. Immunol. 2020, 11, 574500. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díez-Ricote, L.; Ruiz-Valderrey, P.; Micó, V.; Blanco, R.; Tomé-Carneiro, J.; Dávalos, A.; Ordovás, J.M.; Daimiel, L. TMAO Upregulates Members of the miR-17/92 Cluster and Impacts Targets Associated with Atherosclerosis. Int. J. Mol. Sci. 2022, 23, 12107. https://doi.org/10.3390/ijms232012107
Díez-Ricote L, Ruiz-Valderrey P, Micó V, Blanco R, Tomé-Carneiro J, Dávalos A, Ordovás JM, Daimiel L. TMAO Upregulates Members of the miR-17/92 Cluster and Impacts Targets Associated with Atherosclerosis. International Journal of Molecular Sciences. 2022; 23(20):12107. https://doi.org/10.3390/ijms232012107
Chicago/Turabian StyleDíez-Ricote, Laura, Paloma Ruiz-Valderrey, Víctor Micó, Ruth Blanco, Joao Tomé-Carneiro, Alberto Dávalos, José M. Ordovás, and Lidia Daimiel. 2022. "TMAO Upregulates Members of the miR-17/92 Cluster and Impacts Targets Associated with Atherosclerosis" International Journal of Molecular Sciences 23, no. 20: 12107. https://doi.org/10.3390/ijms232012107
APA StyleDíez-Ricote, L., Ruiz-Valderrey, P., Micó, V., Blanco, R., Tomé-Carneiro, J., Dávalos, A., Ordovás, J. M., & Daimiel, L. (2022). TMAO Upregulates Members of the miR-17/92 Cluster and Impacts Targets Associated with Atherosclerosis. International Journal of Molecular Sciences, 23(20), 12107. https://doi.org/10.3390/ijms232012107







