De Novo Assembly of a Sarcocarp Transcriptome Set Identifies AaMYB1 as a Regulator of Anthocyanin Biosynthesis in Actinidia arguta var. purpurea
Abstract
:1. Introduction
2. Results
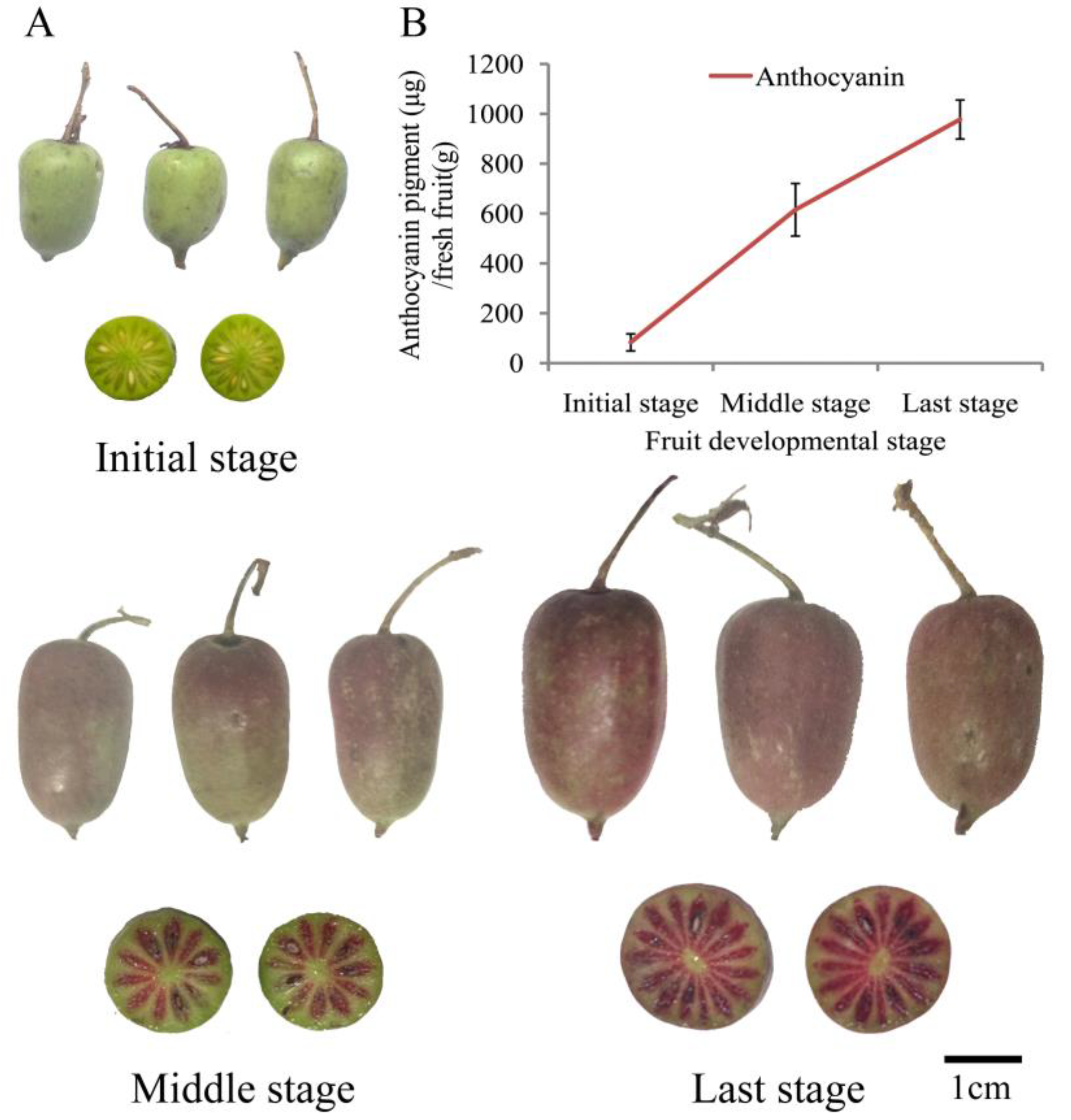
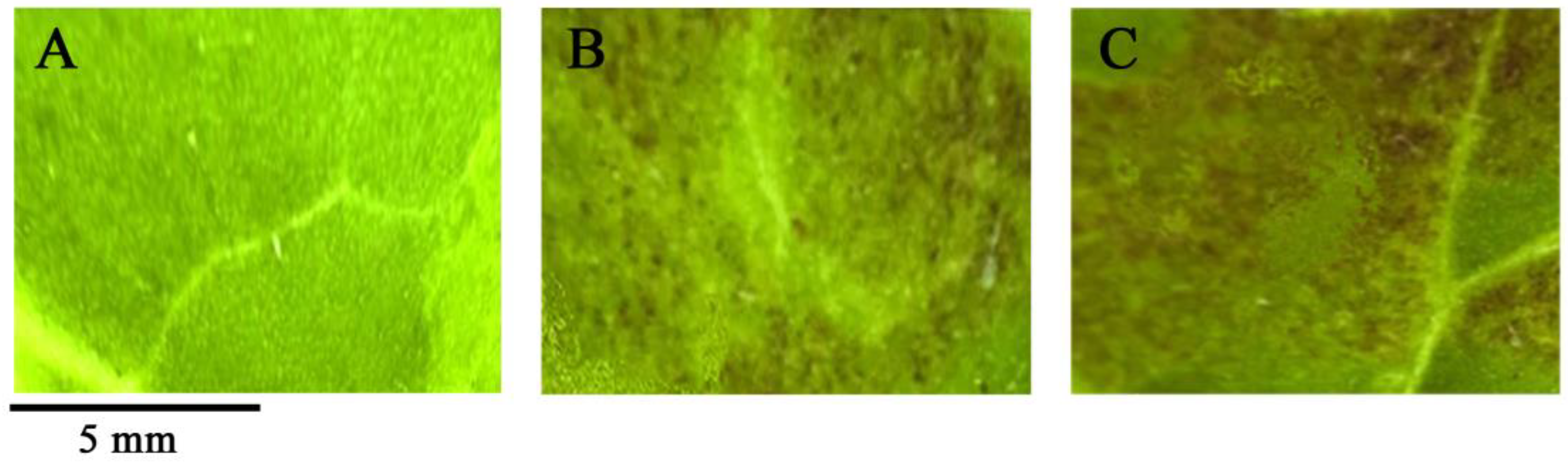
2.1. Anthocyanin Pigment Measurement during Fruit Development
2.2. RNA-Seq and De Novo Transcriptome Assembly
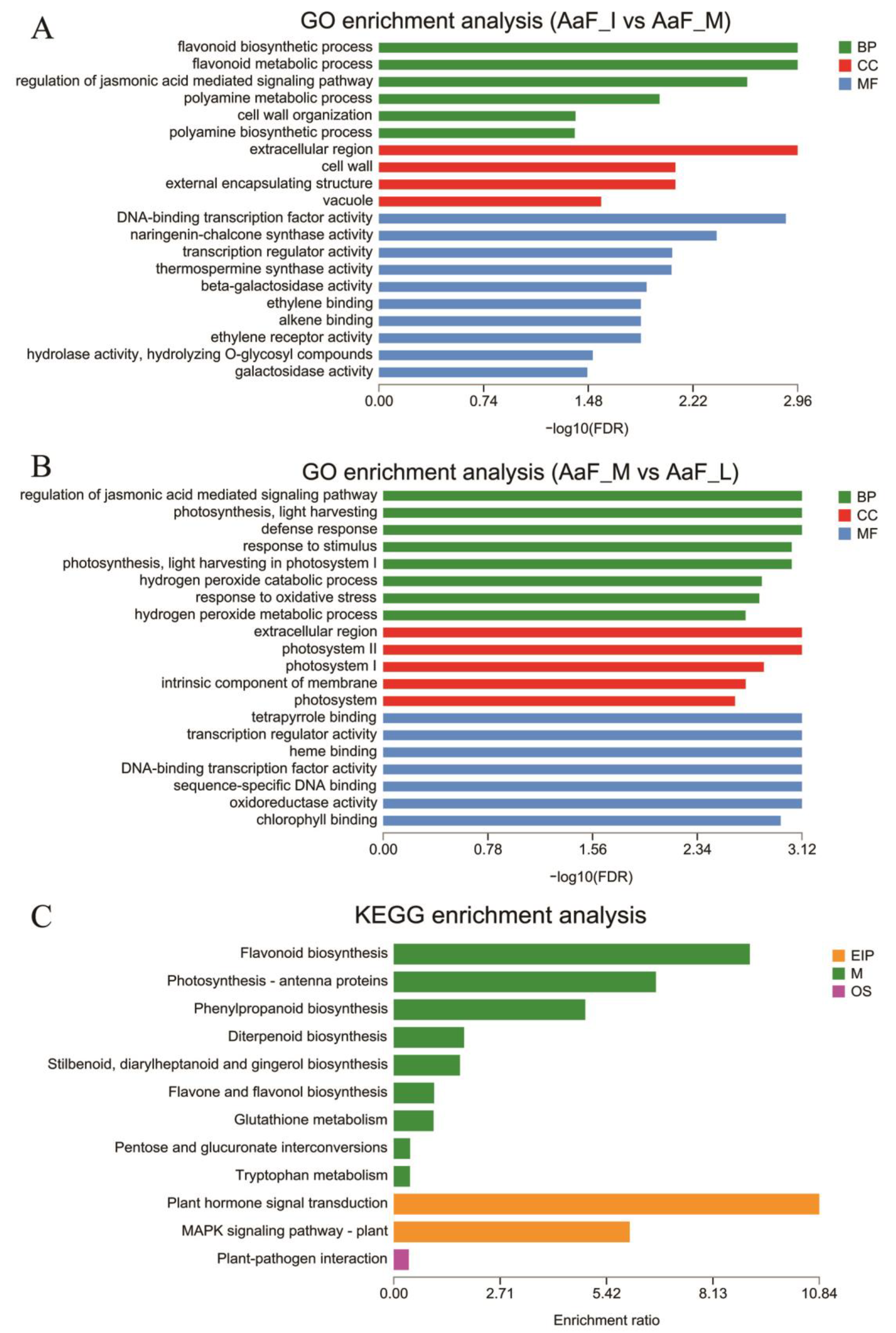
2.3. Functional Annotation and Enrichment Analysis
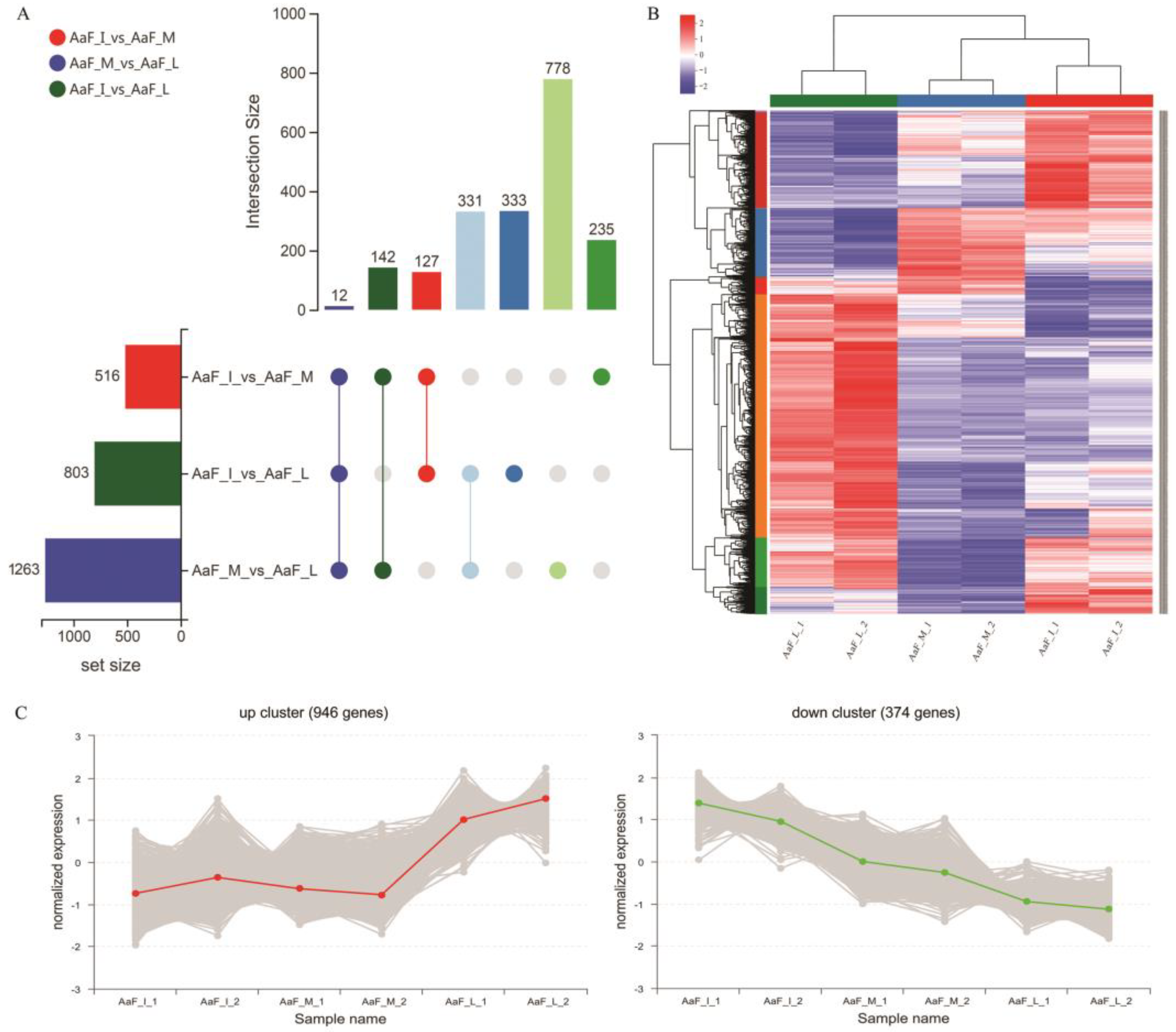
2.4. Identification of Differently Expressed Genes (DEGs) Functional Categorization
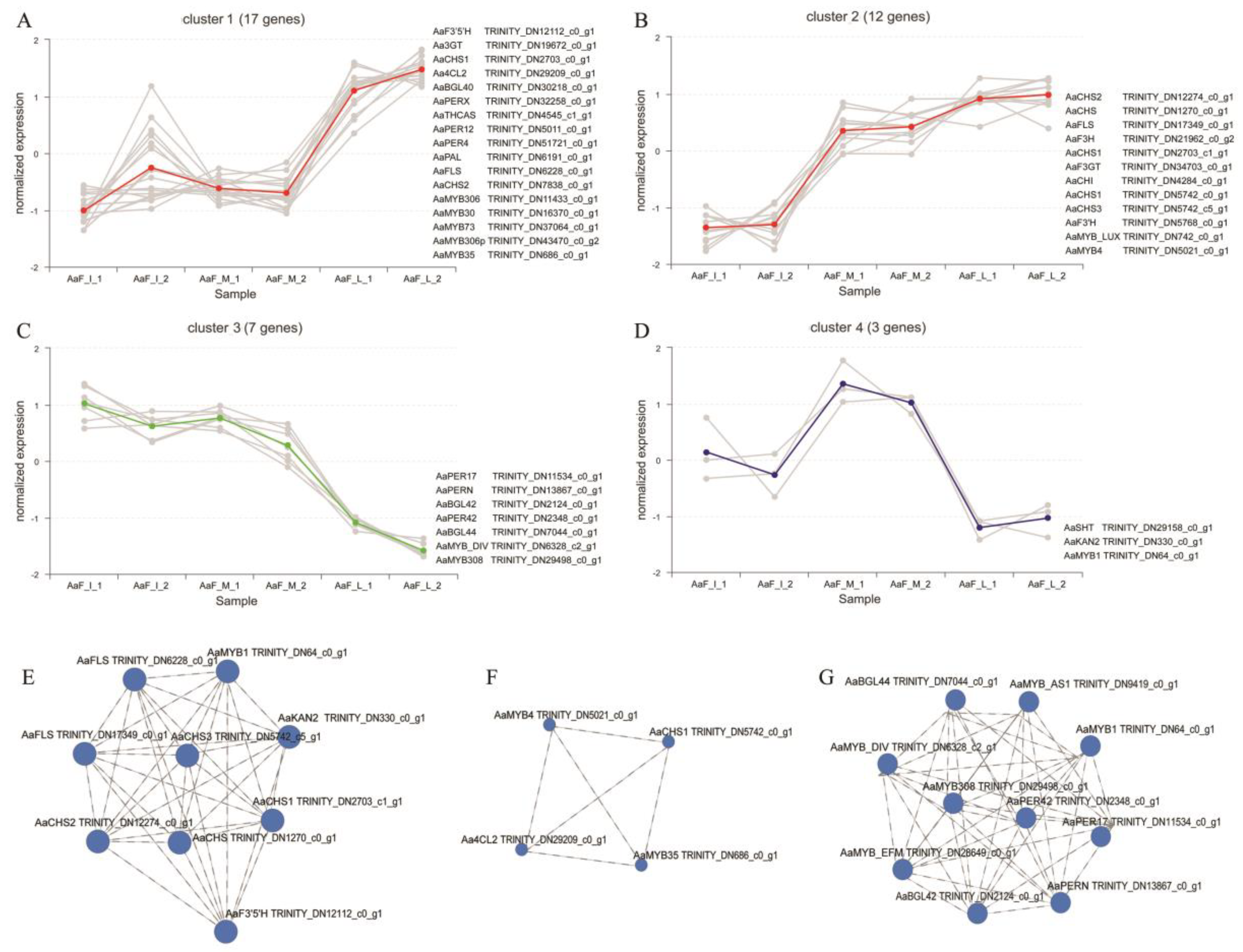
2.5. DEGs Involved in Anthocyanin Biosynthesis
2.6. Correlation Analysis of Genes Involved in Anthocyanin Biosynthesis
2.7. Quantitative RT-PCR Verification and Correlation Analysis of Genes Involved in Anthocyanin Biosynthesis
2.8. Identification of AaMYB1 as a Regulator of Anthocyanin Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Anthocyanin Content
4.3. RNA Extraction, Library Preparation, and Sequencing
4.4. De Novo Assembly and Annotation
4.5. Differential Expression Analysis and Functional Enrichment
4.6. Quantitative RT-PCR Analysis
4.7. Transient Transformation of Nicotiana Benthamiana Leaves
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Qin, X.; Zhang, M.; Li, Q.; Chen, D.; Sun, L.; Qi, X.; Cao, K.; Fang, J. Transcriptional Analysis on Resistant and Susceptible Kiwifruit Genotypes Activating Different Plant-Immunity Processes against Pseudomonas syringae Pv. Actinidiae. Int. J. Mol. Sci. 2022, 23, 7643. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.R.; Seal, A.G. Kiwifruit. In Temperate Fruit Crop Breeding: Germplasm to Genomics; James, F.H., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 235–264. ISBN 978-1-4020-6906-2. [Google Scholar]
- He, J.; Giusti, M.M. Anthocyanins: Natural Colorants with Health-Promoting Properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Babu, P.V.A.; Liu, D.; Gilbert, E.R. Recent Advances in Understanding the Anti-Diabetic Actions of Dietary Flavonoids. J. Nutr. Biochem. 2013, 24, 1777–1789. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and Anthocyanins: Colored Pigments as Food, Pharmaceutical Ingredients, and the Potential Health Benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [Green Version]
- Qin, X. Research Advances of Anthocyanin Accumulation in Plants Tissues. J. Bot. Res. 2022, 4, 26–28. [Google Scholar] [CrossRef]
- Crifò, T.; Puglisi, I.; Petrone, G.; Recupero, G.R.; Lo Piero, A.R. Expression Analysis in Response to Low Temperature Stress in Blood Oranges: Implication of the Flavonoid Biosynthetic Pathway. Gene 2011, 476, 1–9. [Google Scholar] [CrossRef]
- Feng, C.; Chen, M.; Xu, C.; Bai, L.; Yin, X.; Li, X.; Allan, A.C.; Ferguson, I.B.; Chen, K. Transcriptomic Analysis of Chinese Bayberry (Myrica rubra) Fruit Development and Ripening Using RNA-Seq. BMC Genom. 2012, 13, 19. [Google Scholar] [CrossRef] [Green Version]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome Analysis of an Apple (Malus × Domestica) Yellow Fruit Somatic Mutation Identifies a Gene Network Module Highly Associated with Anthocyanin and Epigenetic Regulation. EXBOTJ 2015, 66, 7359–7376. [Google Scholar] [CrossRef] [Green Version]
- Jaakola, L. New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Jørgensen, K.; Rasmussen, A.V.; Morant, M.; Nielsen, A.H.; Bjarnholt, N.; Zagrobelny, M.; Bak, S.; Møller, B.L. Metabolon Formation and Metabolic Channeling in the Biosynthesis of Plant Natural Products. Curr. Opin. Plant Biol. 2005, 8, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Shuai, Q.; Chen, H.; Fan, L.; Zeng, Q.; He, N. Cloning and Expression Analyses of the Anthocyanin Biosynthetic Genes in Mulberry Plants. Mol. Genet. Genom. 2014, 289, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB–BHLH–WD40 Protein Complex and the Evolution of Cellular Diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation Tagging Identifies a Conserved MYB Regulator of Phenylpropanoid Biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef] [Green Version]
- Quattrocchio, F.; Wing, J.; van der Woude, K.; Souer, E.; de Vetten, N.; Mol, J.; Koes, R. Molecular Analysis of the Anthocyanin2 Gene of Petunia and Its Role in the Evolution of Flower Color. Plant Cell 1999, 11, 1433–1444. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-Induced Mutations in Grape Skin Color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White Grapes Arose through the Mutation of Two Similar and Adjacent Regulatory Genes: White Grape Genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Mano, H.; Ogasawara, F.; Sato, K.; Higo, H.; Minobe, Y. Isolation of a Regulatory Gene of Anthocyanin Biosynthesis in Tuberous Roots of Purple-Fleshed Sweet Potato. Plant Physiol. 2007, 143, 1252–1268. [Google Scholar] [CrossRef] [Green Version]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-Induced Expression of a MYB Gene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [Green Version]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and Functional Analysis of a MYB Transcription Factor Gene That Is a Key Regulator for the Development of Red Coloration in Apple Skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red Colouration in Apple Fruit Is Due to the Activity of the MYB Transcription Factor, MdMYB10: A MYB Transcription Factor Controlling Apple Fruit Colour. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peel, G.J.; Pang, Y.; Modolo, L.V.; Dixon, R.A. The LAP1 MYB Transcription Factor Orchestrates Anthocyanidin Biosynthesis and Glycosylation in Medicago. Plant J. 2009, 59, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, C.S.; Giovannoni, J.J. Ripening in the Tomato Green-Ripe Mutant Is Inhibited by Ectopic Expression of a Protein That Disrupts Ethylene Signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 7923–7928. [Google Scholar] [CrossRef] [Green Version]
- Oms-Oliu, G.; Hertog, M.L.A.T.M.; Van de Poel, B.; Ampofo-Asiama, J.; Geeraerd, A.H.; Nicolaï, B.M. Metabolic Characterization of Tomato Fruit during Preharvest Development, Ripening, and Postharvest Shelf-Life. Postharvest Biol. Technol. 2011, 62, 7–16. [Google Scholar] [CrossRef]
- Carrari, F.; Baxter, C.; Usadel, B.; Urbanczyk-Wochniak, E.; Zanor, M.-I.; Nunes-Nesi, A. Integrated Analysis of Metabolite and Transcript Levels Reveals the Metabolic Shifts That Underlie Tomato Fruit Development and Highlight Regulatory Aspects of Metabolic Network Behavior. Plant Physiol. 2006, 142, 1380–1396. [Google Scholar] [CrossRef] [Green Version]
- Feng, F.; Li, M.; Ma, F.; Cheng, L. Phenylpropanoid Metabolites and Expression of Key Genes Involved in Anthocyanin Biosynthesis in the Shaded Peel of Apple Fruit in Response to Sun Exposure. Plant Physiol. Biochem. 2013, 69, 54–61. [Google Scholar] [CrossRef]
- Liu, Y.; Che, F.; Wang, L.; Meng, R.; Zhang, X.; Zhao, Z. Fruit Coloration and Anthocyanin Biosynthesis after Bag Removal in Non-Red and Red Apples (Malus × Domestica Borkh.). Molecules 2013, 18, 1549–1563. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin Biosynthesis in Pears Is Regulated by a R2R3-MYB Transcription Factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Soubeyrand, E.; Basteau, C.; Hilbert, G.; van Leeuwen, C.; Delrot, S.; Gomès, E. Nitrogen Supply Affects Anthocyanin Biosynthetic and Regulatory Genes in Grapevine Cv. Cabernet-Sauvignon Berries. Phytochemistry 2014, 103, 38–49. [Google Scholar] [CrossRef]
- Sun, W.; Meng, X.; Liang, L.; Jiang, W.; Huang, Y.; He, J. Molecular and Biochemical Analysis of Chalcone Synthase from Freesia Hybrid in Flavonoid Biosynthetic Pathway. PLoS ONE 2015, 10, e0119054. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.; Zhang, X.; Han, X.; Zhang, Y.; Gao, S.; Liu, C.; Lou, H. Identification of Chalcone Isomerase in the Basal Land Plants Reveals an Ancient Evolution of Enzymatic Cyclization Activity for Synthesis of Flavonoids. New Phytol. 2018, 217, 909–924. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic Expression of Apple F3′H Genes Contributes to Anthocyanin Accumulation in the Arabidopsis Tt7 Mutant Grown Under Nitrogen Stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Fan, W.; Li, H.; Yang, J.; Huang, J.; Zhang, P. Functional Characterization of Dihydroflavonol-4-Reductase in Anthocyanin Biosynthesis of Purple Sweet Potato Underlies the Direct Evidence of Anthocyanins Function against Abiotic Stresses. PLoS ONE 2013, 8, e78484. [Google Scholar] [CrossRef] [PubMed]
- Wilmouth, R.C.; Turnbull, J.J.; Welford, R.W.D.; Clifton, I.J.; Prescott, A.G.; Schofield, C.J. Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis thaliana. Structure 2002, 10, 93–103. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The Methylation of the PcMYB10 Promoter Is Associated with Green-Skinned Sport in Max Red Bartlett Pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The Involvement of PybZIPa in Light-Induced Anthocyanin Accumulation via the Activation of PyUFGT through Binding to Tandem G-Boxes in Its Promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.C.; Hu, G.B.; Hu, F.C.; Wang, H.C.; Yang, Z.Y.; Lai, B. The UDP Glucose: Flavonoid-3-O-Glucosyltransferase (UFGT) Gene Regulates Anthocyanin Biosynthesis in Litchi (Litchi chinesis Sonn.) during Fruit Coloration. Mol. Biol. Rep. 2012, 39, 6409–6415. [Google Scholar] [CrossRef]
- Zhao, J.; Dixon, R.A. The ‘Ins’ and ‘Outs’ of Flavonoid Transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [Green Version]
- Broun, P. Transcription Factors as Tools for Metabolic Engineering in Plants. Curr. Opin. Plant Biol. 2004, 7, 202–209. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A Conserved Network of Transcriptional Activators and Repressors Regulates Anthocyanin Pigmentation in Eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyagawa, Y.; Tobimatsu, Y.; Lam, P.Y.; Mizukami, T.; Sakurai, S.; Kamitakahara, H.; Takano, T. Possible Mechanisms for the Generation of Phenyl Glycoside-type Lignin–Carbohydrate Linkages in Lignification with Monolignol Glucosides. Plant J. 2020, 104, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.-J.; Xu, J.-K.; Wu, S.-T.; Gao, S.-Q.; Wen, G.-B.; Tan, X.; Lin, Y.-W. Design and Engineering of an Efficient Peroxidase Using Myoglobin for Dye Decolorization and Lignin Bioconversion. Int. J. Mol. Sci. 2021, 23, 413. [Google Scholar] [CrossRef]
- Gu, J.-N.; Zhu, J.; Yu, Y.; Teng, X.-D.; Lou, Y.; Xu, X.-F.; Liu, J.-L.; Yang, Z.-N. DYT1 Directly Regulates the Expression of TDF1 for Tapetum Development and Pollen Wall Formation in Arabidopsis. Plant J. 2014, 80, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Hemm, M.R.; Herrmann, K.M.; Chapple, C. AtMYB4: A Transcription Factor General in the Battle against UV. Trends Plant Sci. 2001, 6, 135–136. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive Identification of Arabidopsis Thaliana MYB Transcription Factors Interacting with R/B-like BHLH Proteins: Systematic Analysis of MYB/BHLH-Interactions. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, X.; Zhang, Y.; Peng, W.; Wang, Z.; Xie, D. Molecular Mechanism for Jasmonate-Induction of Anthocyanin Accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, M.; Zhang, G.; Li, P.; Ma, F. Differential Regulation of Anthocyanin Synthesis in Apple Peel under Different Sunlight Intensities. Int. J. Mol. Sci. 2019, 20, 6060. [Google Scholar] [CrossRef]
- Xie, S.; Lei, Y.; Chen, H.; Li, J.; Chen, H.; Zhang, Z. R2R3-MYB Transcription Factors Regulate Anthocyanin Biosynthesis in Grapevine Vegetative Tissues. Front. Plant Sci. 2020, 11, 527. [Google Scholar] [CrossRef]
- de Wit, M.; Galvão, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Schneider, S.; Ziegler, C.; Melzer, A. Growth towards Light as an Adaptation to High Light Conditions in Chara Branches. New Phytol. 2006, 172, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Tian, C.; Yin, X.; Li, K.; Yang, Y.; Guo, J.; Miao, Y.; et al. Negative Roles of Strigolactone-Related SMXL6, 7 and 8 Proteins in Drought Resistance in Arabidopsis. Biomolecules 2020, 10, 607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.-C.; Zheng, X.-T.; Lin, W.; He, W.; Shao, L.; Peng, C.-L. Photoprotection of Arabidopsis Leaves under Short-Term High Light Treatment: The Antioxidant Capacity Is More Important than the Anthocyanin Shielding Effect. Plant Physiol. Biochem. 2021, 166, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-T.; Yu, Z.-C.; Tang, J.-W.; Cai, M.-L.; Chen, Y.-L.; Yang, C.-W.; Chow, W.S.; Peng, C.-L. The Major Photoprotective Role of Anthocyanins in Leaves of Arabidopsis Thaliana under Long-Term High Light Treatment: Antioxidant or Light Attenuator? Photosynth. Res. 2021, 149, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]

| Sample | Raw Reads | Clean Reads | Mapped Reads | Q20 (%) | Q30 (%) | GC Content (%) | Mapped Ratio to Assembling |
|---|---|---|---|---|---|---|---|
| AaF_I_1 | 55,090,506 | 55,082,814 | 46,864,370 | 100 | 100 | 46.42 | 85.08% |
| AaF_I_2 | 54,921,752 | 54,912,276 | 46,617,616 | 100 | 100 | 46.34 | 84.89% |
| AaF_M_1 | 54,853,482 | 54,844,046 | 46,723,014 | 100 | 100 | 46.59 | 85.19% |
| AaF_M_2 | 55,063,856 | 55,056,096 | 46,740,038 | 100 | 100 | 46.46 | 84.90% |
| AaF_L_1 | 54,691,116 | 54,680,852 | 46,323,952 | 100 | 100 | 46.52 | 84.72% |
| AaF_L_2 | 54,776,194 | 54,767,628 | 45,765,756 | 100 | 100 | 46.21 | 83.56% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, B.; Li, Q.; Fan, L.; Shi, X.; Liu, Y.; Zhuang, Q.; Qin, X. De Novo Assembly of a Sarcocarp Transcriptome Set Identifies AaMYB1 as a Regulator of Anthocyanin Biosynthesis in Actinidia arguta var. purpurea. Int. J. Mol. Sci. 2022, 23, 12120. https://doi.org/10.3390/ijms232012120
Niu B, Li Q, Fan L, Shi X, Liu Y, Zhuang Q, Qin X. De Novo Assembly of a Sarcocarp Transcriptome Set Identifies AaMYB1 as a Regulator of Anthocyanin Biosynthesis in Actinidia arguta var. purpurea. International Journal of Molecular Sciences. 2022; 23(20):12120. https://doi.org/10.3390/ijms232012120
Chicago/Turabian StyleNiu, Bei, Qiaohong Li, Lijuan Fan, Xiaodong Shi, Yuan Liu, Qiguo Zhuang, and Xiaobo Qin. 2022. "De Novo Assembly of a Sarcocarp Transcriptome Set Identifies AaMYB1 as a Regulator of Anthocyanin Biosynthesis in Actinidia arguta var. purpurea" International Journal of Molecular Sciences 23, no. 20: 12120. https://doi.org/10.3390/ijms232012120
APA StyleNiu, B., Li, Q., Fan, L., Shi, X., Liu, Y., Zhuang, Q., & Qin, X. (2022). De Novo Assembly of a Sarcocarp Transcriptome Set Identifies AaMYB1 as a Regulator of Anthocyanin Biosynthesis in Actinidia arguta var. purpurea. International Journal of Molecular Sciences, 23(20), 12120. https://doi.org/10.3390/ijms232012120






