Functional Characterization and Synthetic Application of Is2-SDR, a Novel Thermostable and Promiscuous Ketoreductase from a Hot Spring Metagenome
Abstract
:1. Introduction
2. Results
2.1. Functional Characterization of Is2-SDR
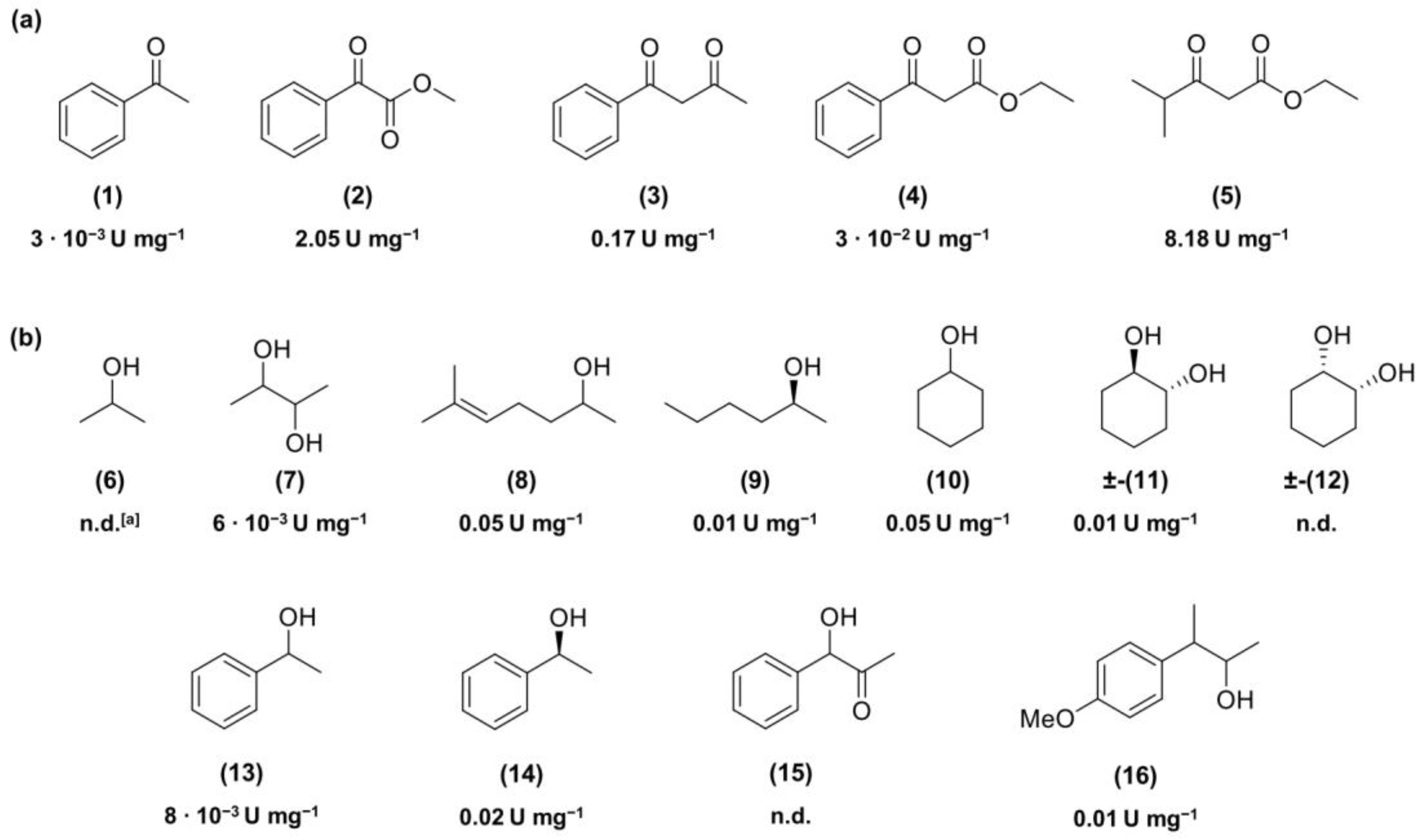
2.1.1. Spectrophotometric Screening of Is2-SDR Reductive/Oxidative Activity in the Presence of Different Substrates
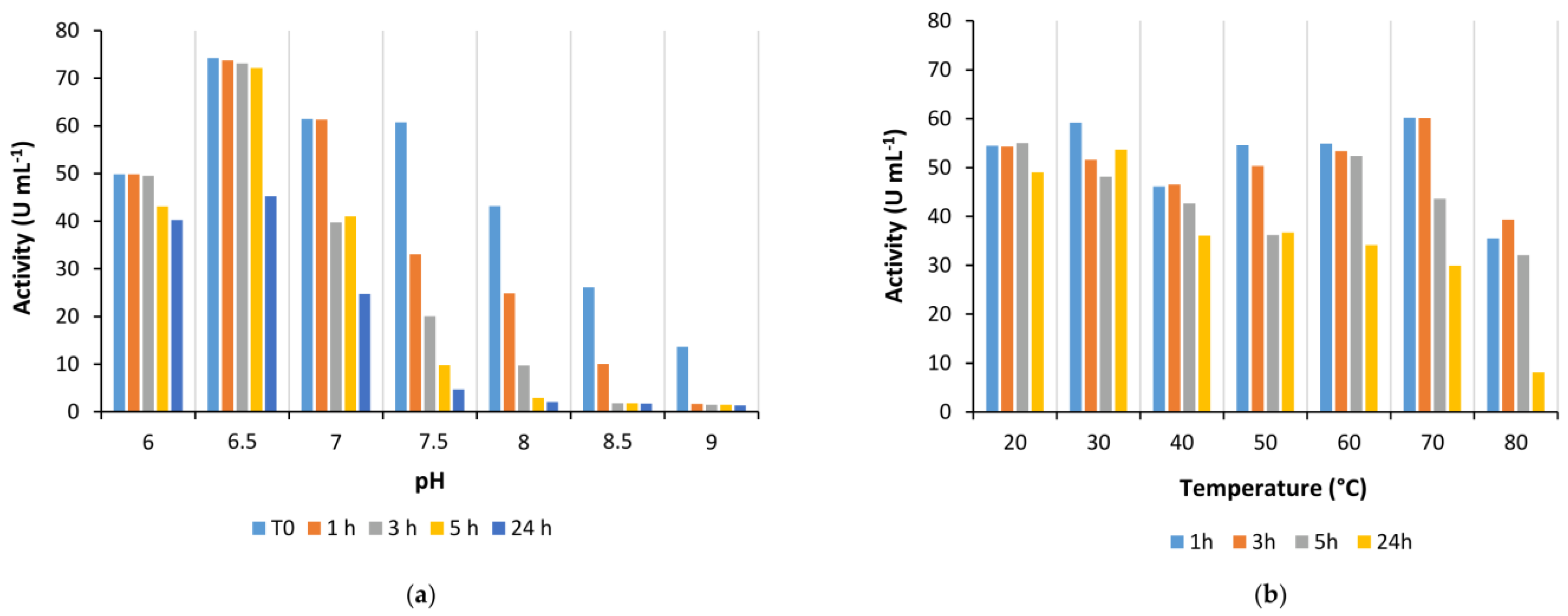
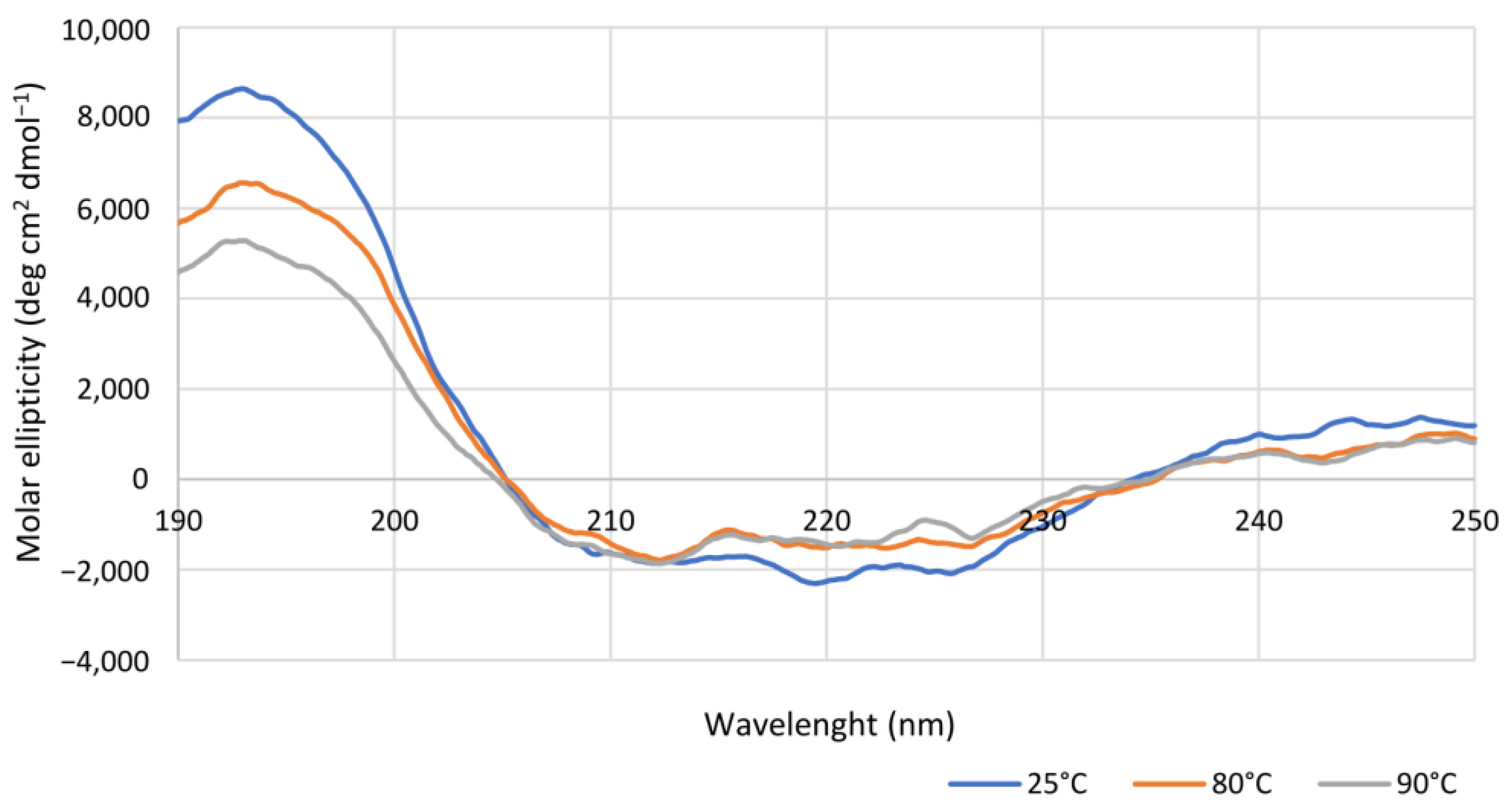
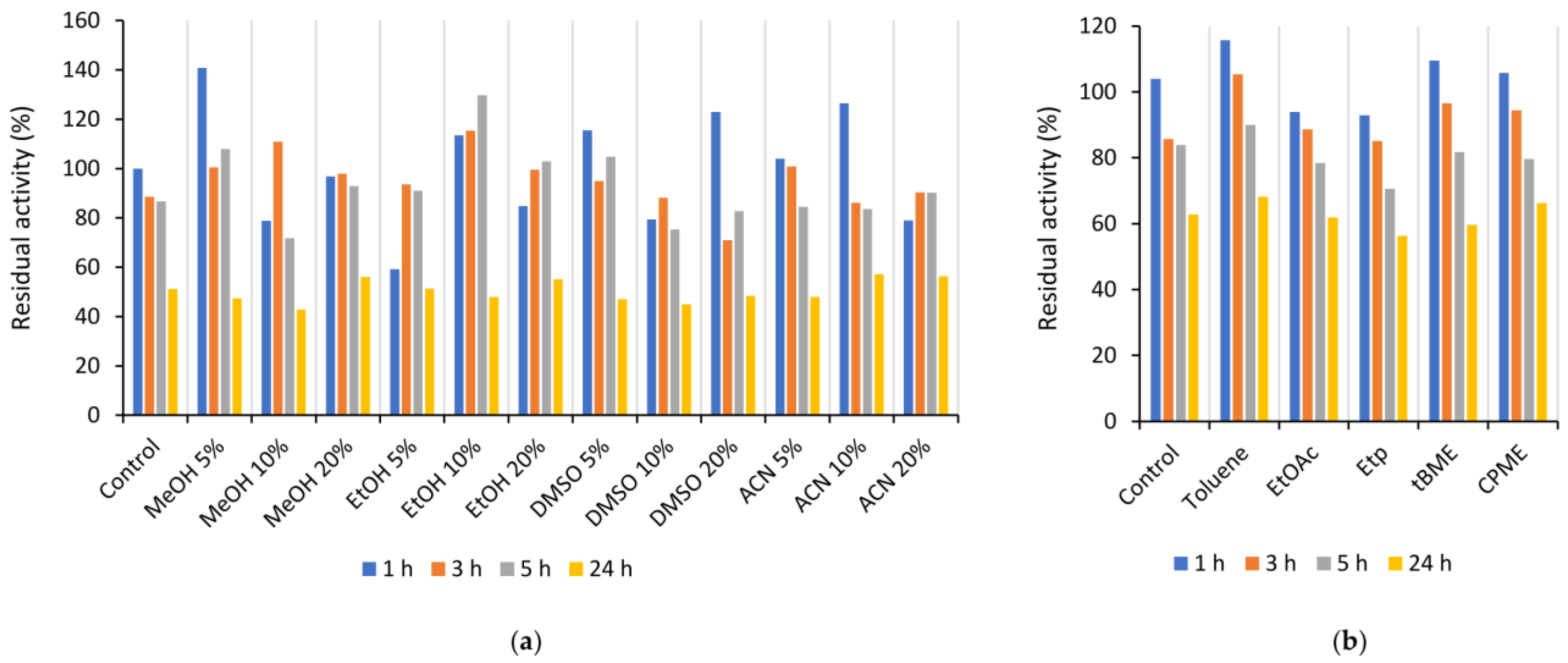
2.1.2. Influence of pH, Temperature, and Organic Solvents on Is2-SDR Activity and Stability
2.2. Screening of Bulky Ketones, α- and β-Ketoesters, and α-Diketones: Activity and Selectivity
2.3. Promiscuous Catalytic Activity of Is2-SDR in the Regio- and Stereoselective Reduction of Oxidized Bile Acids/Steroids
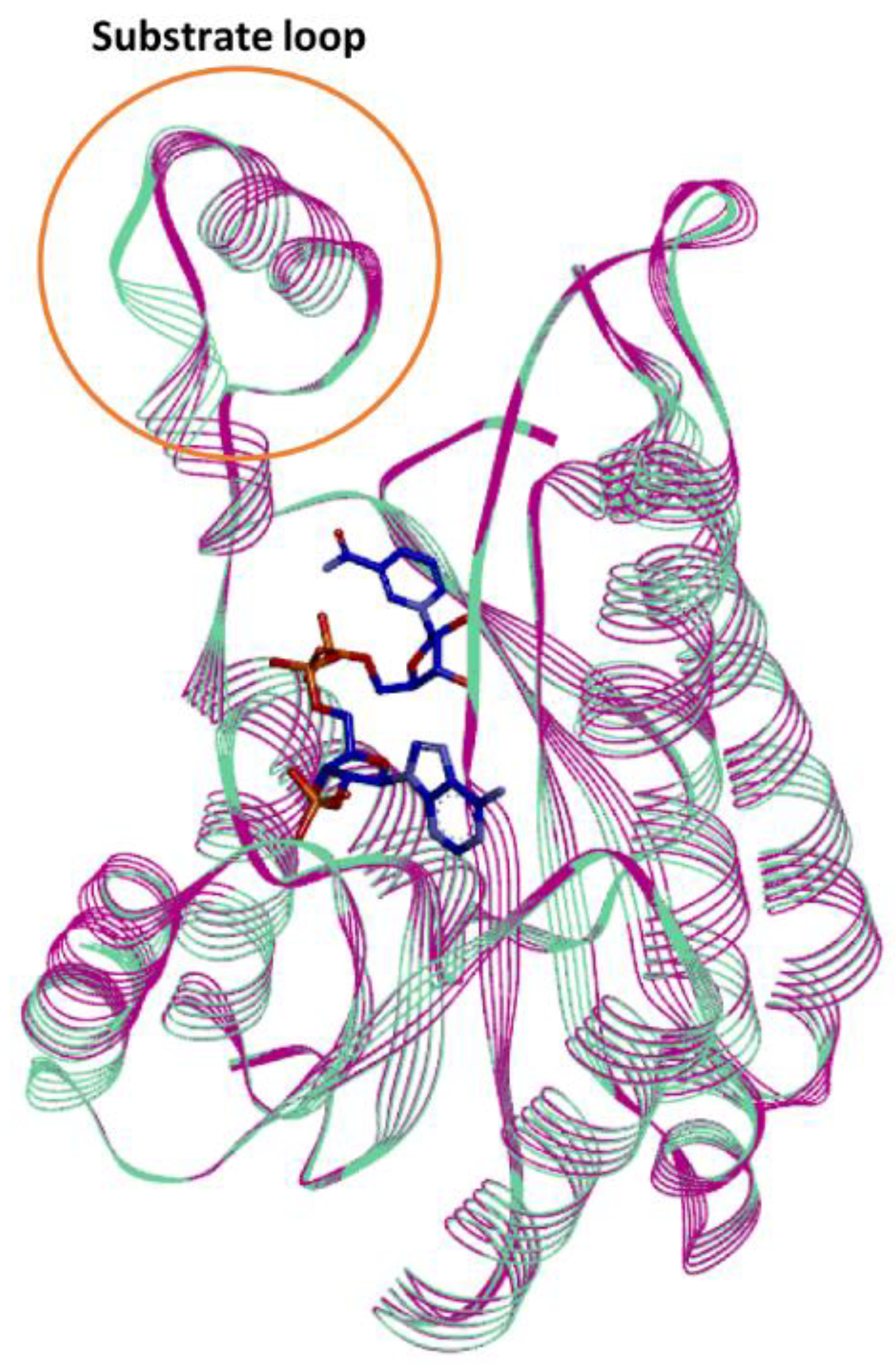
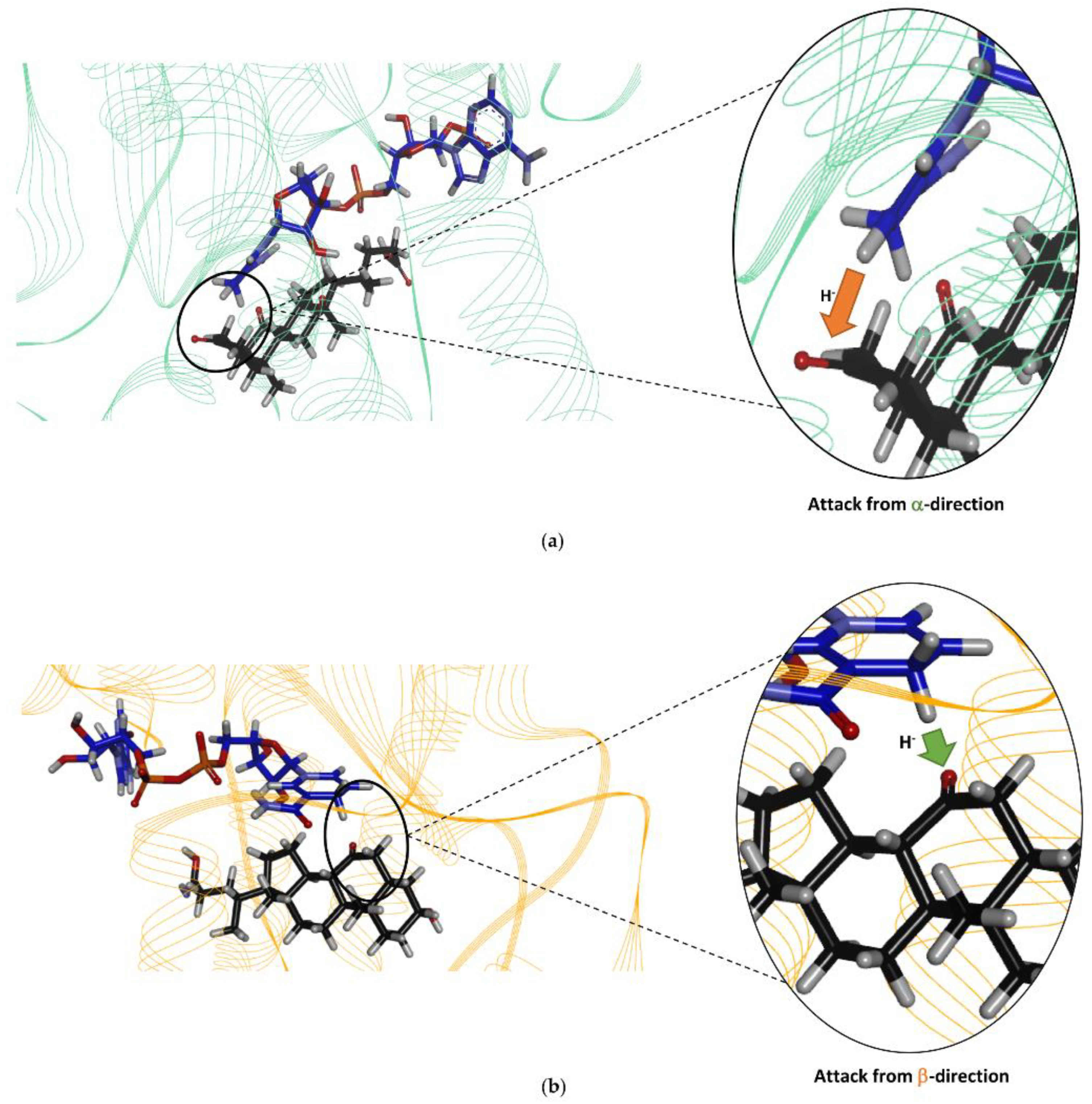
2.4. Homology Modelling of Is2-SDR Protein Structure and Active Site Docking Analysis
3. Discussion
4. Materials and Methods
4.1. General
4.2. Is2-SDR Expression and Purification
4.3. Enzyme Assays and Is2-SDR Characterization
4.4. Bioinformatic and Protein Structure Analysis
4.5. Analytical Methods
4.6. Preparation of Standard Racemates
4.7. Analytical Scale, Biocatalyzed Reduction of Ketone Substrates with Is2-SDR
4.7.1. Enzymatic Reduction of 17
4.7.2. Enzymatic Reduction of 18
4.7.3. Enzymatic Reduction of 19
4.7.4. Enzymatic Reduction of 22
4.7.5. Enzymatic Reduction of 23
4.7.6. Enzymatic Reduction of 24
4.7.7. Enzymatic Reduction of 29
4.8. Is2-SDR Biocatalyzed Reduction of Bile Acids and Steroids
4.8.1. Enzymatic Reduction of Dehydrocholic Acid (30)
4.8.2. Enzymatic Reduction of 5α-Dihydrotestosterone (31)
4.8.3. Enzymatic Reduction of Androsterone (32)
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hollmann, F.; Opperman, D.J.; Paul, C.E. Biocatalytic Reduction Reactions from a Chemist’s Perspective. Angew. Chem. Int. Ed. 2021, 60, 5644–5665. [Google Scholar] [CrossRef] [PubMed]
- Simić, S.; Zukić, E.; Schmermund, L.; Faber, K.; Winkler, C.K.; Kroutil, W. Shortening Synthetic Routes to Small Molecule Active Pharmaceutical Ingredients Employing Biocatalytic Methods. Chem. Rev. 2022, 122, 1052–1126. [Google Scholar] [CrossRef]
- Monti, D.; Ottolina, G.; Carrea, G.; Riva, S. Redox Reactions Catalyzed by Isolated Enzymes. Chem. Rev. 2011, 111, 4111–4140. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, A.S.; Milagre, C.D.F.; Hollmann, F. Alcohol Dehydrogenases as Catalysts in Organic Synthesis. Front. Catal. 2022, 2, 900554. [Google Scholar] [CrossRef]
- Kavanagh, K.L.; Jörnvall, H.; Persson, B.; Oppermann, U. Medium- and Short-Chain Dehydrogenase/Reductase Gene and Protein Families. Cell. Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef] [Green Version]
- Penning, T.M. The Aldo-Keto Reductases (AKRs): Overview. Chem. Biol. Interact. 2015, 234, 236–246. [Google Scholar] [CrossRef] [Green Version]
- Radianingtyas, H.; Wright, P.C. Alcohol Dehydrogenases from Thermophilic and Hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 2003, 27, 593–616. [Google Scholar] [CrossRef]
- Littlechild, J.A. Improving the ‘Tool Box’ for Robust Industrial Enzymes. J. Ind. Microbiol. Biotechnol. 2017, 44, 711–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berini, F.; Casciello, C.; Marcone, G.L.; Marinelli, F. Metagenomics: Novel Enzymes from Non-Culturable Microbes. FEMS Microbiol. Lett. 2017, 364, fnx211. [Google Scholar] [CrossRef]
- Sysoev, M.; Grötzinger, S.W.; Renn, D.; Eppinger, J.; Rueping, M.; Karan, R. Bioprospecting of Novel Extremozymes From Prokaryotes—The Advent of Culture-Independent Methods. Front. Microbiol. 2021, 12, 630013. [Google Scholar] [CrossRef]
- Sousa, J.; Silvério, S.C.; Costa, A.M.A.; Rodrigues, L.R. Metagenomic Approaches as a Tool to Unravel Promising Biocatalysts from Natural Resources: Soil and Water. Catalysts 2022, 12, 385. [Google Scholar] [CrossRef]
- Ferrandi, E.E.; Sayer, C.; Isupov, M.N.; Annovazzi, C.; Marchesi, C.; Iacobone, G.; Peng, X.; Bonch-Osmolovskaya, E.; Wohlgemuth, R.; Littlechild, J.A.; et al. Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries. FEBS J. 2015, 282, 2879–2894. [Google Scholar] [CrossRef] [PubMed]
- Wohlgemuth, R.; Littlechild, J.; Monti, D.; Schnorr, K.; van Rossum, T.; Siebers, B.; Menzel, P.; Kublanov, I.V.; Rike, A.G.; Skretas, G.; et al. Discovering Novel Hydrolases from Hot Environments. Biotechnol. Adv. 2018, 36, 2077–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrandi, E.E.; Sayer, C.; De Rose, S.A.; Guazzelli, E.; Marchesi, C.; Saneei, V.; Isupov, M.N.; Littlechild, J.A.; Monti, D. New Thermophilic α/β Class Epoxide Hydrolases Found in Metagenomes from Hot Environments. Front. Bioeng. Biotechnol. 2018, 6, 144. [Google Scholar] [CrossRef] [Green Version]
- Ye, M.; Li, G.; Liang, W.Q.; Liu, Y.H. Molecular Cloning and Characterization of a Novel Metagenome-Derived Multicopper Oxidase with Alkaline Laccase Activity and Highly Soluble Expression. Appl. Microbiol. Biotechnol. 2010, 87, 1023–1031. [Google Scholar] [CrossRef]
- Fang, Z.; Li, T.; Wang, Q.; Zhang, X.; Peng, H.; Fang, W.; Hong, Y.; Ge, H.; Xiao, Y. A Bacterial Laccase from Marine Microbial Metagenome Exhibiting Chloride Tolerance and Dye Decolorization Ability. Appl. Microbiol. Biotechnol. 2011, 89, 1103–1110. [Google Scholar] [CrossRef]
- Ferrandi, E.E.; Previdi, A.; Bassanini, I.; Riva, S.; Peng, X.; Monti, D. Novel Thermostable Amine Transferases from Hot Spring Metagenomes. Appl. Microbiol. Biotechnol. 2017, 101, 4963–4979. [Google Scholar] [CrossRef]
- Ferrandi, E.E.; Bassanini, I.; Sechi, B.; Vanoni, M.; Tessaro, D.; Guðbergsdóttir, S.R.; Riva, S.; Peng, X.; Monti, D. Discovery and Characterization of a Novel Thermostable β-Amino Acid Transaminase from a Meiothermus Strain Isolated in an Icelandic Hot Spring. Biotechnol. J. 2020, 15, 2000125. [Google Scholar] [CrossRef]
- Ferrandi, E.E.; Bertuletti, S.; Monti, D.; Riva, S. Hydroxysteroid Dehydrogenases: An Ongoing Story. Eur. J. Org. Chem. 2020, 2020, 4463–4473. [Google Scholar] [CrossRef]
- Bertuletti, S.; Ferrandi, E.E.; Marzorati, S.; Vanoni, M.; Riva, S.; Monti, D. Insights into the Substrate Promiscuity of Novel Hydroxysteroid Dehydrogenases. Adv. Synth. Catal. 2020, 362, 2474–2485. [Google Scholar] [CrossRef]
- Menzel, P.; Gudbergsdóttir, S.R.; Rike, A.G.; Lin, L.; Zhang, Q.; Contursi, P.; Moracci, M.; Kristjansson, J.K.; Bolduc, B.; Gavrilov, S.; et al. Comparative Metagenomics of Eight Geographically Remote Terrestrial Hot Springs. Microb. Ecol. 2015, 70, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Bertuletti, S.; Bayout, I.; Bassanini, I.; Ferrandi, E.E.; Bouzemi, N.; Monti, D.; Riva, S. Biocatalytic Approaches to the Enantiomers of Wieland–Miescher Ketone and Its Derivatives. Eur. J. Org. Chem. 2021, 2021, 3992–3998. [Google Scholar] [CrossRef]
- Nasti, R.; Bassanini, I.; Ferrandi, E.E.; Linguardo, F.; Bertuletti, S.; Vanoni, M.; Riva, S.; Verotta, L.; Monti, D. Stereoselective Biocatalyzed Reductions of Ginger Active Components Recovered from Industrial Wastes. ChemBioChem 2022, 23, e202200105. [Google Scholar] [CrossRef]
- Bertuletti, S.; Ferrandi, E.E.; Monti, D.; Fronza, G.; Bassanini, I.; Riva, S. Synthesis of ω-Muricholic Acid by One-Pot Enzymatic Mitsunobu Inversion Using Hydroxysteroid Dehydrogenases. ChemCatChem 2021, 13, 4948–4953. [Google Scholar] [CrossRef]
- Benach, J.; Filling, C.; Oppermann, U.C.T.; Roversi, P.; Bricogne, G.; Berndt, K.D.; Jörnvall, H.; Ladenstein, R. Structure of Bacterial 3β/17β-Hydroxysteroid Dehydrogenase at 1.2 Å Resolution: A Model for Multiple Steroid Recognition. Biochemistry 2002, 41, 14659–14668. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, A.P. FabG: From a Core to Circumstantial Catalyst. Biotechnol. Lett. 2019, 41, 675–688. [Google Scholar] [CrossRef]
- Savino, S.; Ferrandi, E.E.; Forneris, F.; Rovida, S.; Riva, S.; Monti, D.; Mattevi, A. Structural and Biochemical Insights into 7β-Hydroxysteroid Dehydrogenase Stereoselectivity. Proteins Struct. Funct. Bioinf. 2016, 84, 859–865. [Google Scholar] [CrossRef]
- Gao, M.; Nie, K.; Qin, M.; Xu, H.; Wang, F.; Liu, L. Molecular Mechanism Study on Stereo-Selectivity of α or β Hydroxysteroid Dehydrogenases. Crystals 2021, 11, 224. [Google Scholar] [CrossRef]
- Kallberg, Y.; Oppermann, U.; Persson, B. Classification of the Short-Chain Dehydrogenase ⁄ Reductase Superfamily Using Hidden Markov Models. FEBS J. 2010, 277, 2375–2386. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Chen, J.; Liu, Z.-L.; Shou, L.-B.; Lin, D.-D.; Zhou, L.; Yang, S.-Z.; Liu, J.-F.; Li, W.; Gu, J.-D.; et al. Anaerobic Degradation of Paraffins by Thermophilic Actinobacteria under Methanogenic Conditions. Environ. Sci. Technol. 2020, 54, 10610–10620. [Google Scholar] [CrossRef]
- Littlechild, J.A. Archaeal Enzymes and Applications in Industrial Biocatalysts. Archaea 2015, 2015, 147671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Zhang, C.; Orita, I.; Imanaka, T.; Fukui, T.; Xing, X.-H. Thermostable Alcohol Dehydrogenase from Thermococcus kodakarensis KOD1 for Enantioselective Bioconversion of Aromatic Secondary Alcohols. Appl. Environ. Microbiol. 2013, 79, 2209–2217. [Google Scholar] [CrossRef] [Green Version]
- van der Oost, J.; Voorhorst, W.G.B.; Kengen, S.W.M.; Geerling, A.C.M.; Wittenhorst, V.; Gueguen, Y.; de Vos, W.M. Genetic and Biochemical Characterization of a Short-Chain Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Pyrococcus furiosus. Eur. J. Biochem. 2001, 268, 3062–3068. [Google Scholar] [CrossRef]
- Lavandera, I.; Kern, A.; Ferreira-Silva, B.; Glieder, A.; de Wildeman, S.; Kroutil, W. Stereoselective Bioreduction of Bulky-Bulky Ketones by a Novel ADH from Ralstonia sp. J. Org. Chem. 2008, 73, 6003–6005. [Google Scholar] [CrossRef]
- Carrea, G.; Riva, S.; Bovara, R.; Pasta, P. Enzymatic Oxidoreduction of Steroids in Two-Phase Systems: Effects of Organic Solvents on Enzyme Kinetics and Evaluation of the Performance of Different Reactors. Enzyme Microb. Technol. 1988, 10, 333–340. [Google Scholar] [CrossRef]
- Carrea, G. Biocatalysis in Water-Organic Solvent Two-Phase Systems. Trends Biotechnol. 1984, 2, 102–106. [Google Scholar] [CrossRef]
- Crotti, M.; Parmeggiani, F.; Ferrandi, E.E.; Gatti, F.G.; Sacchetti, A.; Riva, S.; Brenna, E.; Monti, D. Stereoselectivity Switch in the Reduction of α-Alkyl-β-Arylenones by Structure-Guided Designed Variants of the Ene Reductase OYE1. Front. Bioeng. Biotechnol. 2019, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life v2: Online Annotation and Display of Phylogenetic Trees Made Easy. Nucleic Acids Res. 2011, 39, W475–W478. [Google Scholar] [CrossRef] [PubMed]
- Łaskowski, A.; MacArthur, M.W.; Moss, D.S.; Thorton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A Method to Identify Protein Sequences That Fold into a Known Three-Dimensional Structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [Green Version]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- de Gonzalo, G. Biocatalysed Reductions of α-Ketoesters Employing Cyrene TM as Cosolvent. Biocatal. Biotransf. 2022, 40, 252–257. [Google Scholar] [CrossRef]
- Jolley, K.E.; Zanotti-Gerosa, A.; Hancock, F.; Dyke, A.; Grainger, D.M.; Medlock, J.A.; Nedden, H.G.; Le Paih, J.J.M.; Roseblade, S.J.; Seger, A.; et al. Application of Tethered Ruthenium Catalysts to Asymmetric Hydrogenation of Ketones, and the Selective Hydrogenation of Aldehydes. Adv. Synth. Catal. 2012, 354, 2545–2555. [Google Scholar] [CrossRef]
- Gajewski, P.; Renom-Carrasco, M.; Facchini, S.V.; Pignataro, L.; Lefort, L.; de Vries, J.G.; Ferraccioli, R.; Forni, A.; Piarulli, U.; Gennari, C. Chiral (Cyclopentadienone)Iron Complexes for the Catalytic Asymmetric Hydrogenation of Ketones. Eur. J. Org. Chem. 2015, 2015, 1887–1893. [Google Scholar] [CrossRef]
- Renom-Carrasco, M.; Gajewski, P.; Pignataro, L.; de Vries, J.G.; Piarulli, U.; Gennari, C.; Lefort, L. Asymmetric Transfer Hydrogenation of Ketones with Modified Grubbs Metathesis Catalysts: On the Way to a Tandem Process. Adv. Synth. Catal. 2016, 358, 515–519. [Google Scholar] [CrossRef]
- Hunter, A.C.; Collins, C.; Dodd, H.T.; Dedi, C.; Koussoroplis, S.-J. Transformation of a Series of Saturated Isomeric Steroidal Diols by Aspergillus tamarii KITA Reveals a Precise Stereochemical Requirement for Entrance into the Lactonization Pathway. J. Steroid Biochem. Mol. Biol. 2010, 122, 352–358. [Google Scholar] [CrossRef]


| Substrate | Conversion (%) | Enantiomeric Excess (e.e., %) | Reference | |
|---|---|---|---|---|
| 17 |  | >99 | -- | This work |
| 18 |  | 72 | 91.0 (S) | [20] |
| 19 |  | 25 | -- | This work |
| 20 |  | >99 | 82.0 (R) | [20] |
| 21 |  | >99 | 98.4 (R) | [20] |
| 22 |  | 52 1-OH, 2-keto (PAC): 67 1-keto, 2-OH: 25 Diols: 8 | n.d. 1 | This work |
| 23 | 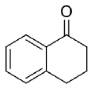 | 93 | 98 (R) | This work |
| 24 |  | 26 | 22 (R) | This work |
| 25 |  | >99 | Cis-OH: >99 Trans-OH: 61.2 | [20] |
| 26 |  | 29 (4aS,5S-OH) 32 (4aR,5S-keto) Traces of: (4aR,5S-OH) and (4aR,5R-OH) | 91 (4aS,5S-OH) 96.5 (4aR,5S-keto) | [22] |
| 27 | 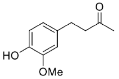 | 24 | 83 (S) | [23] |
| 28 | 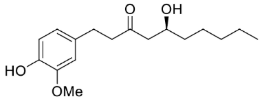 | 50 | >99 (R) | [23] |
| 29 | 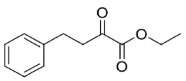 | >99 | >99 (R) | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrandi, E.E.; Bassanini, I.; Bertuletti, S.; Riva, S.; Tognoli, C.; Vanoni, M.; Monti, D. Functional Characterization and Synthetic Application of Is2-SDR, a Novel Thermostable and Promiscuous Ketoreductase from a Hot Spring Metagenome. Int. J. Mol. Sci. 2022, 23, 12153. https://doi.org/10.3390/ijms232012153
Ferrandi EE, Bassanini I, Bertuletti S, Riva S, Tognoli C, Vanoni M, Monti D. Functional Characterization and Synthetic Application of Is2-SDR, a Novel Thermostable and Promiscuous Ketoreductase from a Hot Spring Metagenome. International Journal of Molecular Sciences. 2022; 23(20):12153. https://doi.org/10.3390/ijms232012153
Chicago/Turabian StyleFerrandi, Erica Elisa, Ivan Bassanini, Susanna Bertuletti, Sergio Riva, Chiara Tognoli, Marta Vanoni, and Daniela Monti. 2022. "Functional Characterization and Synthetic Application of Is2-SDR, a Novel Thermostable and Promiscuous Ketoreductase from a Hot Spring Metagenome" International Journal of Molecular Sciences 23, no. 20: 12153. https://doi.org/10.3390/ijms232012153
APA StyleFerrandi, E. E., Bassanini, I., Bertuletti, S., Riva, S., Tognoli, C., Vanoni, M., & Monti, D. (2022). Functional Characterization and Synthetic Application of Is2-SDR, a Novel Thermostable and Promiscuous Ketoreductase from a Hot Spring Metagenome. International Journal of Molecular Sciences, 23(20), 12153. https://doi.org/10.3390/ijms232012153









