Microbial Precipitation of Pb(II) with Wild Strains of Paraclostridium bifermentans and Klebsiella pneumoniae Isolated from an Industrially Obtained Microbial Consortium
Abstract
1. Introduction
2. Results and Discussions
2.1. Isolation of Bacterial Strain P. bifermentans
2.2. Precipitation Study—Long Duration Study
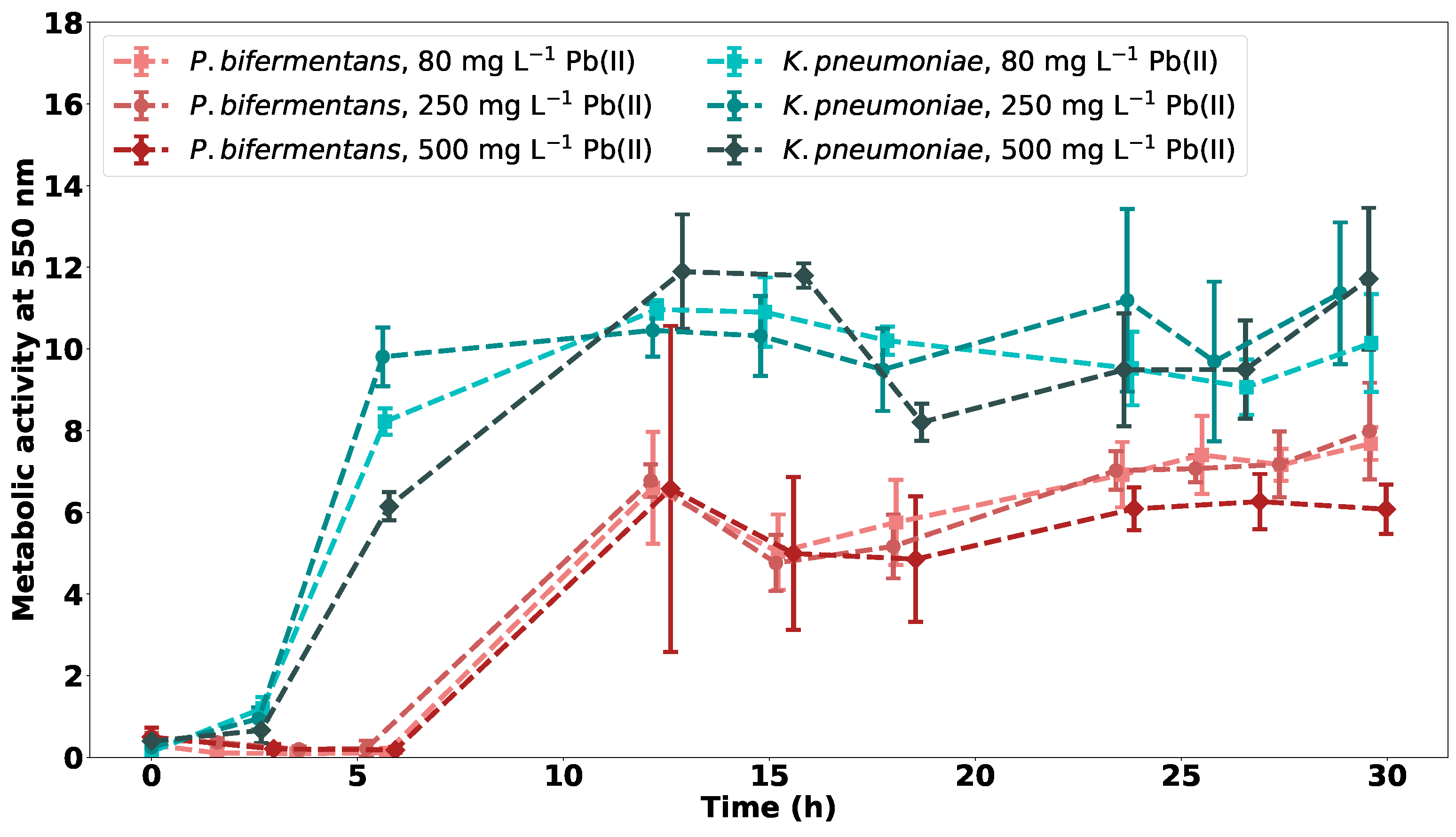
2.2.1. Metabolic Activity
2.2.2. Residual Pb(II) Concentration
2.2.3. Nitrate Concentration
2.3. Precipitation Study—Short-Term Study
2.3.1. Visual Changes
2.3.2. Metabolic Activity for Varying Initial Concentrations of Pb(II)
2.3.3. Residual Pb(II) Concentration
2.3.4. Nitrate Concentration
2.3.5. Comparison between Metabolic Activity Measurements, Nitrate Concentration and Residual Pb(II) Concentration
2.3.6. Determination of Extracellular and Intracellular Pb(II) Concentration
2.3.7. Specific Growth Rate
2.4. Precipitate Identity
3. Methods and Materials
3.1. Materials
3.2. Bacterial Strain Isolation
3.3. Batch Reactor Preparation
3.4. Metabolic Activity
3.5. Nitrate Concentration
3.6. Lead Concentration
3.7. Determination of Extracellular and Intracellular Pb(II)
3.8. Precipitate Identity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rigoletto, M.; Calxa, P.; Gaggero, E.; Malandrino, M.; Fabbri, D. Bioremediation Methods for the Recovery of Lead-Contaminated Soils: A Review. Appl. Sci. 2020, 2, 3528. [Google Scholar] [CrossRef]
- Lombó, C.G.; Posadas, Y.; Quintanar, L.Y.; Eugenia, M. Neurotoxicity linked to dysfunctional metal ion homeostasis and xenobiotic metal exposure: Redox signaling and oxidative stress. Antioxid. Redox Signal. 2018, 28, 1669–1703. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhang, C.; Lü, T. Removal of Pb(II) Ions from Wastewater by Using Magnetic Nanoparticles. Appl. Sci. 2020, 10, 948. [Google Scholar] [CrossRef]
- Dongre, R.S. Lead: Toxicological Profile, Pollution Aspects and Remedial Solutions. In Lead Chemistry; Chooto, P., Ed.; Intech Press: London, UK, 2020; pp. 1–18. [Google Scholar] [CrossRef]
- Naik, M.M.; Dubey, S.K. Ecotoxicology and Environmental Safety Lead resistant bacteria: Lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol. Environ. Saf. 2013, 98, 1–7. [Google Scholar] [CrossRef]
- Shamuyarira, K.; Gumbo, J. Assessment of Heavy Metals in Municipal Sewage Sludge: A Case Study of Limpopo Province, South Africa. Int. J. Environ. Res. Public Health 2014, 11, 2569–2579. [Google Scholar] [CrossRef]
- Herselman, J.E.; Snyman, H.G. Guidelines for the Utilisation and Disposal of Wastewater Sludge, Volume 3: Requirements of the On-Site and Off-Site Disposal of Sludge; Water Research Commission: Pretoria, South Africa, 2009. [Google Scholar]
- Arbabi, M.; Hemati, S.; Amiri, M. Removal of lead ions from industrial wastewater: A review of removal methods. Int. J. Epidemiol. Res. 2015, 2, 105–109. [Google Scholar]
- Sylwan, I.; Thorin, E. Removal of heavy metals during primary treatment of municipal wastewater and possibilities of enhanced removal: A review. Water 2021, 13, 1121. [Google Scholar] [CrossRef]
- Boryczko, B.; Hołda, A.; Kolenda, Z. Depletion of the non-renewable natural resource reserved in copper, zinc, lead and aluminium production. J. Clean. Prod. 2014, 84, 313–321. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, H. Microbial Consortia Are Needed to Degrade Soil Pollutants. Microorganisms 2022, 10, 261. [Google Scholar] [CrossRef]
- Patel, S.K.S.; Gupta, R.K.; Has, D.; Lee, J.; Kalia, V.C. Continuous biohydrogen production from poplar biomass hydrolysate by a defined bacterial mixture immobilized on lignocellulosic materials under non-sterile conditions. J. Clean. Prod. 2021, 287, 125037. [Google Scholar] [CrossRef]
- Patel, S.K.S.; Kumar, P.; Mehariya, S.; Purohit, H.J.; Lee, J.; Kalia, V.C. Enhancement in hydrogen production by co-cultures of Bacillus and Enterobacter. Int. J. Hydrogen Energy 2014, 39, 14663–14668. [Google Scholar] [CrossRef]
- Padhi, S.K.; Tripathy, S.; Sen, R.; Mahapatra, A.S.; Mohanty, S.; Maiti, N.K. Characterisation of heterotrophic nitrifying and aerobic denitrifying Klebsiella pneumoniae CF-S9 strain for bioremediation of wastewater. Int. Biodeterior. Biodegrad. 2013, 78, 67–73. [Google Scholar] [CrossRef]
- Binish, M.B.; Binu, V.G.; Gopikrishna, V.G.; Mohan, M. Potential of anaerobic bacteria in bioremediation of metal-contaminated marine and estuarine environment. In Microbial Biodegradation and Bioremediation; Das, S., Dash, H.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 305–341. [Google Scholar]
- Hörstmann, C.; Brink, H.G.; Chirwa, E.M.N. Pb(II) Bio-Removal, Viability, and Population Distribution of an Industrial Microbial Consortium: The Effect of Pb(II) and Nutrient Concentrations. Sustainability 2020, 6, 2511. [Google Scholar] [CrossRef]
- van Veenhuyzen, B.; Chirwa, E.M.N.; Brink, H.G. Microbial Pb(II) Precipitation: The Role of Biosorption as a Pb(II) Removal Mechanism. Chem. Eng. Trans. 2021, 86, 181–186. [Google Scholar] [CrossRef]
- Chimhundi, J.; Hörstmann, C.; Chirwa, E.M.N.; Brink, H.G. Microbial Removal of Pb(II) Using an Upflow Anaerobic Sludge Blanket (UASB) Reactor. Catalysts 2021, 11, 512. [Google Scholar] [CrossRef]
- Neveling, O.; van Veenhuyzen, B.; Cilliers, C.; Chirwa, E.M.N.; Brink, H.G. Microbial Pb(II) Removal by Precipitation and Adsorption Mechanisms with Klebsiella Pneumoniae Isolated from an Industrially Obtained Consortium. Chem. Eng. Trans. 2022, 92, 103–108. [Google Scholar] [CrossRef]
- Cilliers, C.; Neveling, O.; Tichapondwa, S.M.; Chirwa, E.M.N.; Brink, H.G. Microbial Pb(II)-bioprecipitation: Characterising Responsible Biotransformation Mechanisms. J. Clean. Prod. 2022, 374, 133973. [Google Scholar] [CrossRef]
- Kirchman, D.L. Calculating microbial growth rates from data on production and standing stocks. Mar. Ecol. Prog. Ser. 2002, 233, 303–306. [Google Scholar] [CrossRef]
- NIST X-ray Photoelectron Spectroscopy Database, NIST Standard Reference Database Number 20; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2000; p. 20899. [CrossRef]
- Brink, H.G.; Hörstmann, C.; Peens, J. Microbial Pb(II)-precipitation: The influence of oxygen on Pb(II)-removal from aqueous environment and the resulting precipitate identity. Int. J. Environ. Sci. Technol. 2019, 17, 409–420. [Google Scholar] [CrossRef]
- Lal, A.; Cheeptham, N. Eosin-Methylene Blue Agar Plates Protocol. Am. Soc. Microbiol. Protocol 2007. [Google Scholar]
- Ogbu, K.; Anyika, K.C.; Ochai, S.O.; Gyendeng, J.; Olaolu, O. Determination of bacterial load in dog milk with its associated risk factors in Jos, Plateau State. World J. Pharm. Med. Res. 2017, 3, 130–134. [Google Scholar]
- Sanders, E.R. Aseptic laboratory techniques: Plating methods. J. Vis. Exp. 2012, 63, e3064. [Google Scholar] [CrossRef] [PubMed]
- Kupesik, L. Estimation of cell number based on metabolic activity: The MTT reduction assay. Methods Mol. Biol. 2011, 740, 13–19. [Google Scholar] [CrossRef]
- Zhang, K.T.; Zhang, D.; Li, X.; Xue, Y. Biomineralization of lead in wastewater: Bacterial reutilization and metal recovery. J. Hazard. Mater. 2021, 421, 126765. [Google Scholar] [CrossRef]
- Mirimanoff, N.; Wilkinson, K.J. Regulation of Zn Accumulation by a Freshwater Gram-Positive Bacterium (Rhodococcus opacus). Environ. Sci. Technol. 2000, 34, 616–622. [Google Scholar] [CrossRef]
- Bates, S.S.; Tessier, A.; Campbell, P.G.C.; Buffle, J. Zinc adsorption and transport by Chlamydomonas variabilis and Scenedesmus subspicatus (Chlorophyceae) grown in semicontinuous culture. J. Phycol. 1982, 18, 521–529. [Google Scholar] [CrossRef]
- D’Agostino, R.B.; Sullivan, L.M.; Beiser, A.S. Introductory Applied Biostatistics, 1st ed.; Cengage Learning Location: Belmont, CA, USA, 2006; p. 239. [Google Scholar]









| Strain | Initial Pb(II) Concentration (mg L) | (d) | (d) |
|---|---|---|---|
| P. bifermentans | 80 | 7.94 | 0.0873 |
| 250 | 15.2 | 0.0441 | |
| 500 | 280 | 0.00247 | |
| K. pneumoniae | 80 | 36.1 | 0.0192 |
| 250 | 45.6 | 0.0152 | |
| 500 | 87.2 | 0.00795 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neveling, O.; Ncube, T.M.C.; Ngxongo, Z.P.; Chirwa, E.M.N.; Brink, H.G. Microbial Precipitation of Pb(II) with Wild Strains of Paraclostridium bifermentans and Klebsiella pneumoniae Isolated from an Industrially Obtained Microbial Consortium. Int. J. Mol. Sci. 2022, 23, 12255. https://doi.org/10.3390/ijms232012255
Neveling O, Ncube TMC, Ngxongo ZP, Chirwa EMN, Brink HG. Microbial Precipitation of Pb(II) with Wild Strains of Paraclostridium bifermentans and Klebsiella pneumoniae Isolated from an Industrially Obtained Microbial Consortium. International Journal of Molecular Sciences. 2022; 23(20):12255. https://doi.org/10.3390/ijms232012255
Chicago/Turabian StyleNeveling, Olga, Thato M. C. Ncube, Ziyanda P. Ngxongo, Evans M. N. Chirwa, and Hendrik G. Brink. 2022. "Microbial Precipitation of Pb(II) with Wild Strains of Paraclostridium bifermentans and Klebsiella pneumoniae Isolated from an Industrially Obtained Microbial Consortium" International Journal of Molecular Sciences 23, no. 20: 12255. https://doi.org/10.3390/ijms232012255
APA StyleNeveling, O., Ncube, T. M. C., Ngxongo, Z. P., Chirwa, E. M. N., & Brink, H. G. (2022). Microbial Precipitation of Pb(II) with Wild Strains of Paraclostridium bifermentans and Klebsiella pneumoniae Isolated from an Industrially Obtained Microbial Consortium. International Journal of Molecular Sciences, 23(20), 12255. https://doi.org/10.3390/ijms232012255








