A Review: Highlighting the Links between Epigenetics, COVID-19 Infection, and Vitamin D
Abstract
:1. Introduction
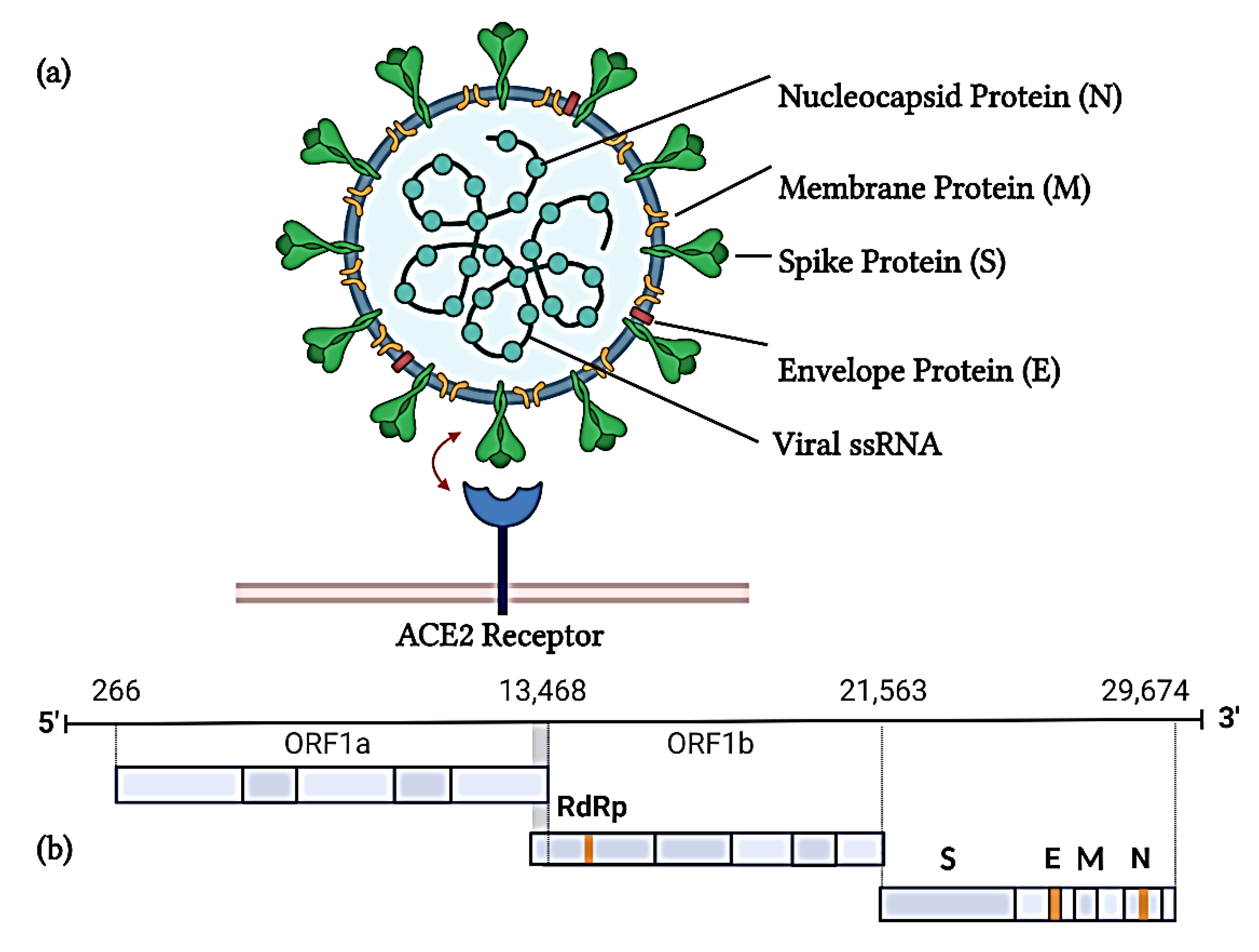
2. SARS-CoV-2 Structure
3. Epigenetics and COVID-19 Infection
3.1. DNA Methylation
3.2. Histone Modifications
3.3. miRNAs
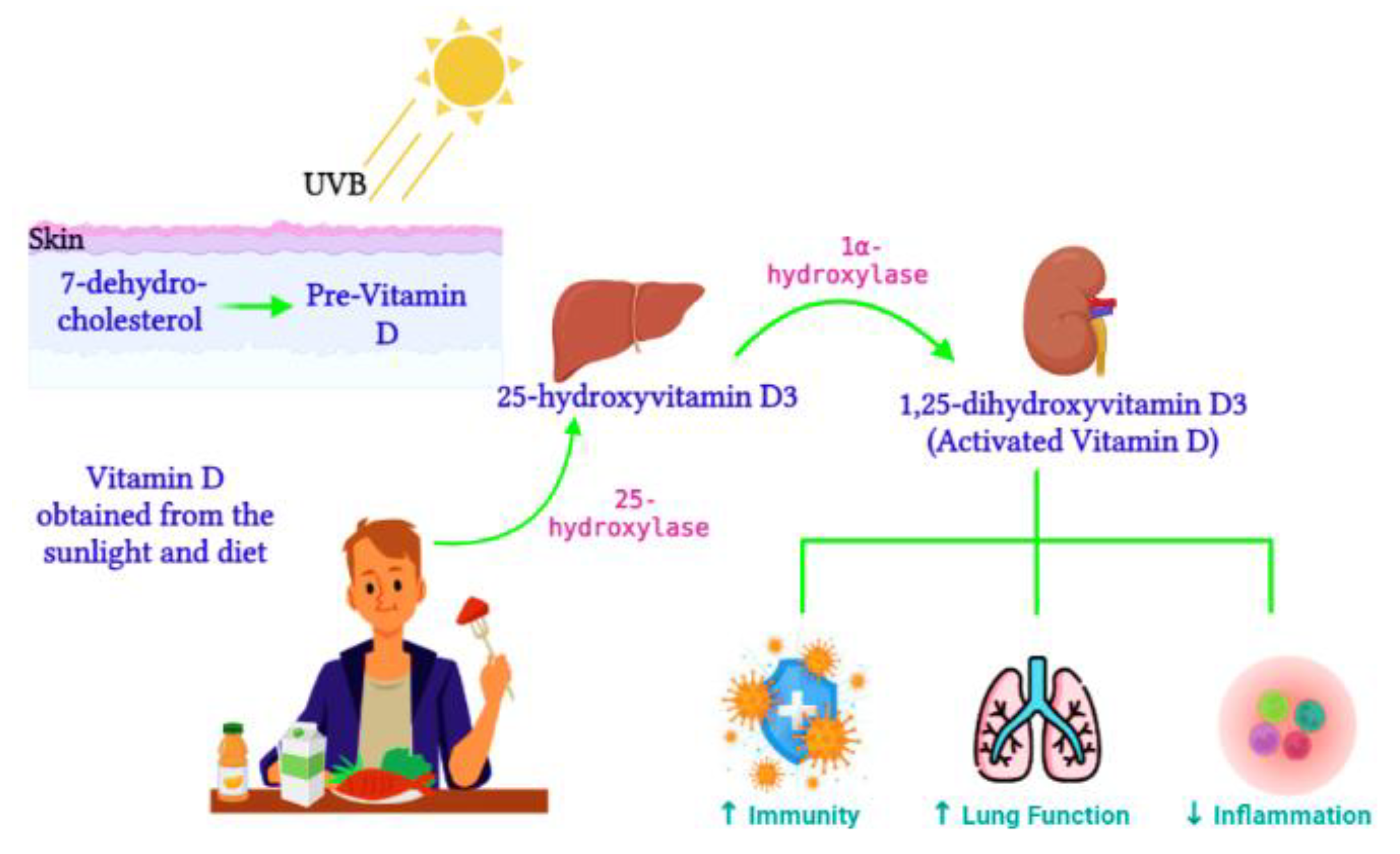
4. Vitamin D Regulation
4.1. The Impact of Vitamin D on Epigenetics and Gene Expression
4.2. The Role of Vitamin D in COVID-19 Infections
5. Potential COVID-19 Therapeutic Targets
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Sen, R.; Garbati, M.; Bryant, K.; Lu, Y. Epigenetic mechanisms influencing COVID-19. Genome 2021, 64, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Liu, Y. DNA methylation in human diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Surguchov, A. New putative epigenetic mechanism of epilepsy. Front. Neurol. 2017, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieberman, P.M. Epigenetics and Genetics of Viral Latency. Cell Host Microbe 2016, 19, 619–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019, 19, 417–432. [Google Scholar] [CrossRef]
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2021, 16, 263–270. [Google Scholar] [CrossRef]
- Holmes, L., Jr.; Lim, A.; Comeaux, C.R.; Dabney, K.W.; Okundaye, O. DNA Methylation of Candidate Genes (ACE II, IFN-γ, AGTR 1, CKG, ADD1, SCNN1B and TLR2) in Essential Hypertension: A Systematic Review and Quantitative Evidence Synthesis. Int. J. Environ. Res. Public Health 2019, 16, 4829. [Google Scholar] [CrossRef] [Green Version]
- Pruimboom, L. Methylation Pathways and SARS-CoV-2 Lung Infiltration and Cell Membrane-Virus Fusion Are Both Subject to Epigenetics. Front. Cell Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Ramakrishnan, V. Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 83–112. [Google Scholar] [CrossRef] [Green Version]
- Sodagar, H.; Alipour, S.; Hassani, S.; Aziz, S.G.; Ansari, M.H.K.; Asghari, R. The role of microRNAs in COVID-19 with a focus on miR-200c. J. Circ. Biomark 2022, 11, 14–23. [Google Scholar] [CrossRef]
- AbdelHamid, S.G.; Refaat, A.A.; Benjamin, A.M.; Elmawardy, L.A.; Elgendy, L.A.; Manolly, M.M.; Elmaksoud, N.A.; Sherif, N.; Hamdy, N.M. Deciphering epigenetic(s) role in modulating susceptibility to and severity of COVID-19 infection and/or outcome: A systematic rapid review. Environ. Sci. Pollut. Res. Int. 2021, 28, 54209–54221. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Benítez, I.D.; Pinilla, L.; Carratalá, A.; Moncusí-Moix, A.; Gort-Paniello, C.; Molinero, M.; González, J.; Torres, G.; Bernal, M.; et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021, 236, 147–159. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [Green Version]
- Amini-Farsani, Z.; Yadollahi-Farsani, M.; Arab, S.; Forouzanfar, F.; Yadollahi, M.; Asgharzade, S. Prediction and analysis of microRNAs involved in COVID-19 inflammatory processes associated with the NF-kB and JAK/STAT signaling pathways. Int. Immunopharmacol. 2021, 100, 108071. [Google Scholar] [CrossRef]
- Abedi, F.; Rezaee, R.; Hayes, A.W.; Nasiripour, S.; Karimi, G. MicroRNAs and SARS-CoV-2 life cycle, pathogenesis, and mutations: Biomarkers or therapeutic agents? Cell Cycle 2021, 20, 143–153. [Google Scholar] [CrossRef]
- Bouillon, R.; Carmeliet, G.; Verlinden, L.; van Etten, E.; Verstuyf, A.; Luderer, H.F.; Lieben, L.; Mathieu, C.; Demay, M. Vitamin D and human health: Lessons from vitamin D receptor null mice. Endocr. Rev. 2008, 29, 726–776. [Google Scholar]
- Azrielant, S.; Shoenfeld, Y. Vitamin D and the Immune System. Isr. Med. Assoc. J. 2017, 19, 510–511. [Google Scholar] [PubMed]
- Baeke, F.; Takiishi, T.; Korf, H.; Gysemans, C.; Mathieu, C. Vitamin D: Modulator of the immune system. Curr. Opin. Pharmacol. 2010, 10, 482–496. [Google Scholar] [CrossRef]
- Greiller, C.L.; Martineau, A.R. Modulation of the immune response to respiratory viruses by vitamin D. Nutrients 2015, 7, 4240–4270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniel, C.; Sartory, N.A.; Zahn, N.; Radeke, H.H.; Stein, J.M. Immune Modulatory Treatment of Trinitrobenzene Sulfonic Acid Colitis with Calcitriol Is Associated with a Change of a T Helper (Th) 1/Th17 to a Th2 and Regulatory T Cell Profile. J. Pharmacol. Exp. Ther. 2008, 324, 23. [Google Scholar] [CrossRef] [Green Version]
- Arboleda, J.F.; Urcuqui-Inchima, S. Vitamin D Supplementation: A Potential Approach for Coronavirus/COVID-19 Therapeutics? Front. Immunol. 2020, 11, 1523. [Google Scholar] [CrossRef] [PubMed]
- Karlic, H.; Varga, F. Impact of vitamin D metabolism on clinical epigenetics. Clin. Epigenetics 2011, 2, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Kooraki, S.; Hosseiny, M.; Myers, L.; Gholamrezanezhad, A. Coronavirus (COVID-19) Outbreak: What the Department of Radiology Should Know. J. Am. Coll. Radiol. 2020, 17, 447–451. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Kandeel, M.; Ibrahim, A.; Fayez, M.; Al-Nazawi, M. From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol. 2020, 92, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Atlante, S.; Mongelli, A.; Barbi, V.; Martelli, F.; Farsetti, A.; Gaetano, C. The epigenetic implication in coronavirus infection and therapy. Clin. Epigenetics 2020, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Cao, Y.; Liu, W.; Li, J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses 2021, 13, 1115. [Google Scholar] [CrossRef]
- Aronson, J.K.; Ferner, R.E. Drugs and the renin-angiotensin system in COVID-19. BMJ 2020, 369, m1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Dupressoir, A.; Lavialle, C.; Heidmann, T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta 2012, 33, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Obata, Y.; Furusawa, Y.; Hase, K. Epigenetic modifications of the immune system in health and disease. Immunol. Cell. Biol. 2015, 93, 226–232. [Google Scholar] [CrossRef] [Green Version]
- Vavougios, G.D. A data-driven hypothesis on the epigenetic dysregulation of host metabolism by SARS coronaviral infection: Potential implications for the SARS-CoV-2 modus operandi. Med. Hypotheses 2020, 140, 109759. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Schäfer, A.; Burnum-Johnson, K.E.; Mitchell, H.D.; Eisfeld, A.J.; Walters, K.B.; Nicora, C.D.; Purvine, S.O.; Casey, C.P.; Monroe, M.E.; et al. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. USA 2018, 115, E1012–E1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, A.L.; McNamara, M.S.; Sinclair, D.A. Why does COVID-19 disproportionately affect older people? Aging 2020, 12, 9959–9981. [Google Scholar] [CrossRef] [PubMed]
- Holt, N.R.; Neumann, J.T.; McNeil, J.J.; Cheng, A.C. Implications of COVID-19 for an ageing population. Med. J. Aust. 2020, 213, 342–344.e341. [Google Scholar] [CrossRef]
- Zill, P.; Baghai, T.C.; Schüle, C.; Born, C.; Früstück, C.; Büttner, A.; Eisenmenger, W.; Varallo-Bedarida, G.; Rupprecht, R.; Möller, H.J.; et al. DNA methylation analysis of the angiotensin converting enzyme (ACE) gene in major depression. PLoS ONE 2012, 7, e40479. [Google Scholar] [CrossRef] [Green Version]
- Corley, M.J.; Ndhlovu, L.C. DNA methylation analysis of the COVID-19 host cell receptor, angiotensin I converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences. Preprints 2020, 2020030295. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Schatzmann Peron, J.P.; Nakaya, H.I. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Joubert, B.R.; Felix, J.F.; Yousefi, P.; Bakulski, K.M.; Just, A.C.; Breton, C.; Reese, S.E.; Markunas, C.A.; Richmond, R.C.; Xu, C.J.; et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016, 98, 680–696. [Google Scholar] [CrossRef] [Green Version]
- Cai, H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir. Med. 2020, 8, e20. [Google Scholar] [CrossRef]
- Leung, J.M.; Yang, C.X.; Tam, A.; Shaipanich, T.; Hackett, T.L.; Singhera, G.K.; Dorscheid, D.R.; Sin, D.D. ACE-2 expression in the small airway epithelia of smokers and COPD patients: Implications for COVID-19. Eur. Respir. J. 2020, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawalha, A.H.; Zhao, M.; Coit, P.; Lu, Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020, 215, 108410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 202, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Mao, S.Q.; Gu, T.L.; Zhong, F.D.; Gong, M.L.; Hao, L.M.; Yin, F.Y.; Dong, C.Z.; Zhang, L.N. Preliminary analysis of the association between methylation of the ACE2 promoter and essential hypertension. Mol. Med. Rep. 2017, 15, 3905–3911. [Google Scholar] [CrossRef]
- Kuderer, N.M.; Choueiri, T.K.; Shah, D.P.; Shyr, Y.; Rubinstein, S.M.; Rivera, D.R.; Shete, S.; Hsu, C.-Y.; Desai, A.; de Lima Lopes, G., Jr.; et al. Clinical impact of COVID-19 on patients with cancer (CCC19): A cohort study. Lancet 2020, 395, 1907–1918. [Google Scholar] [CrossRef]
- Chai, P.; Yu, J.; Ge, S.; Jia, R.; Fan, X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: A pan-cancer analysis. J. Hematol. Oncol. 2020, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Walden, M.; Tian, L.; Ross, R.L.; Sykora, U.M.; Byrne, D.P.; Hesketh, E.L.; Masandi, S.K.; Cassel, J.; George, R.; Ault, J.R.; et al. Metabolic control of BRISC-SHMT2 assembly regulates immune signalling. Nature 2019, 570, 194–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Zhang, J.; Xu, J.; Zhang, F.; Li, P.; He, Y.; Wu, J.; Wang, C.; Wang, X.; Zhang, W.; et al. An epigenome-wide DNA methylation study of patients with COVID-19. Ann. Hum. Genet. 2021, 85, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.J.; Pang, A.P.S.; Dody, K.; Mudd, P.A.; Patterson, B.K.; Seethamraju, H.; Bram, Y.; Peluso, M.J.; Torres, L.; Iyer, N.S.; et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J. Leukoc. Biol. 2021, 110, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Balnis, J.; Madrid, A.; Hogan, K.J.; Drake, L.A.; Chieng, H.C.; Tiwari, A.; Vincent, C.E.; Chopra, A.; Vincent, P.A.; Robek, M.D.; et al. Blood DNA methylation and COVID-19 outcomes. Clin. Epigenetics 2021, 13, 118. [Google Scholar] [CrossRef]
- Konigsberg, I.R.; Barnes, B.; Campbell, M.; Davidson, E.; Zhen, Y.; Pallisard, O.; Boorgula, M.P.; Cox, C.; Nandy, D.; Seal, S.; et al. Host methylation predicts SARS-CoV-2 infection and clinical outcome. Commun. Med. 2021, 1, 42. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, A.; Baric, R.S. Epigenetic Landscape during Coronavirus Infection. Pathogens 2017, 6, 8. [Google Scholar] [CrossRef]
- Lim, Y.X.; Ng, Y.L.; Tam, J.P.; Liu, D.X. Human Coronaviruses: A Review of Virus-Host Interactions. Diseases 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Li, H.; Zhou, L. EZH2-mediated H3K27me3 inhibits ACE2 expression. Biochem. Biophys. Res. Commun. 2020, 526, 947–952. [Google Scholar] [CrossRef]
- Huckriede, J.; de Vries, F.; Hultström, M.; Wichapong, K.; Reutelingsperger, C.; Lipcsey, M.; Garcia de Frutos, P.; Frithiof, R.; Nicolaes, G.A.F. Histone H3 Cleavage in Severe COVID-19 ICU Patients. Front. Cell. Infect. Microbiol. 2021, 11, 694186. [Google Scholar] [CrossRef]
- Bouchard, B.A.; Colovos, C.; Lawson, M.A.; Osborn, Z.T.; Sackheim, A.M.; Mould, K.J.; Janssen, W.J.; Cohen, M.J.; Majumdar, D.; Freeman, K. Increased histone-DNA complexes and endothelial-dependent thrombin generation in severe COVID-19. Vasc. Pharmacol. 2022, 142, 106950. [Google Scholar] [CrossRef] [PubMed]
- Christophorou, M.A.; Castelo-Branco, G.; Halley-Stott, R.P.; Oliveira, C.S.; Loos, R.; Radzisheuskaya, A.; Mowen, K.A.; Bertone, P.; Silva, J.C.R.; Zernicka-Goetz, M.; et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 2014, 507, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Leppkes, M.; Knopf, J.; Naschberger, E.; Lindemann, A.; Singh, J.; Herrmann, I.; Stürzl, M.; Staats, L.; Mahajan, A.; Schauer, C.; et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine 2020, 58, 102925. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Stadler, S.; Correll, S.; Li, P.; Wang, D.; Hayama, R.; Leonelli, L.; Han, H.; Grigoryev, S.A.; et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009, 184, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.-j.; Ni, Z.-y.; Hu, Y.; Liang, W.-h.; Ou, C.-q.; He, J.-x.; Liu, L.; Shan, H.; Lei, C.-l.; Hui, D.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Veras, F.P.; Pontelli, M.C.; Silva, C.M.; Toller-Kawahisa, J.E.; de Lima, M.; Nascimento, D.C.; Schneider, A.H.; Caetité, D.; Tavares, L.A.; Paiva, I.M.; et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.Y.; Park, H.H.; Park, W.; Kim, H.; Jang, J.G.; Hong, K.S.; Lee, J.Y.; Seo, H.S.; Na, D.H.; Kim, T.H.; et al. Long-acting nanoparticulate DNase-1 for effective suppression of SARS-CoV-2-mediated neutrophil activities and cytokine storm. Biomaterials 2021, 267, 120389. [Google Scholar] [CrossRef] [PubMed]
- de Gonzalo-Calvo, D.; Vea, A.; Bär, C.; Fiedler, J.; Couch, L.S.; Brotons, C.; Llorente-Cortes, V.; Thum, T. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: A novel tool for personalized medicine? Eur. Heart J. 2019, 40, 1643–1650. [Google Scholar] [CrossRef] [PubMed]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef]
- Arisan, E.D.; Dart, A.; Grant, G.H.; Arisan, S.; Cuhadaroglu, S.; Lange, S.; Uysal-Onganer, P. The Prediction of miRNAs in SARS-CoV-2 Genomes: hsa-miR Databases Identify 7 Key miRs Linked to Host Responses and Virus Pathogenicity-Related KEGG Pathways Significant for Comorbidities. Viruses 2020, 12, 614. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J.-h. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef]
- Fulzele, S.; Sahay, B.; Yusufu, I.; Lee, T.J.; Sharma, A.; Kolhe, R.; Isales, C.M. COVID-19 Virulence in Aged Patients Might Be Impacted by the Host Cellular MicroRNAs Abundance/Profile. Aging Dis. 2020, 11, 509–522. [Google Scholar] [CrossRef]
- Roganović, J. Downregulation of microRNA-146a in diabetes, obesity and hypertension may contribute to severe COVID-19. Med. Hypotheses 2021, 146, 110448. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Svegliati Baroni, S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef]
- Bertolazzi, G.; Cipollina, C.; Benos, P.V.; Tumminello, M.; Coronnello, C. miR-1207-5p can contribute to dysregulation of inflammatory response in COVID-19 via targeting SARS-CoV-2 RNA. Front. Cell. Infect. Microbiol. 2020, 10, 586592. [Google Scholar] [CrossRef]
- Balmeh, N.; Mahmoudi, S.; Mohammadi, N.; Karabedianhajiabadi, A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Inform. Med. Unlocked 2020, 20, 100407. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.T.-A.; Khalid, Z.; Zahid, H.; Yousaf, M.A.; Shakoori, A.R. A computational and bioinformatic analysis of ACE2: An elucidation of its dual role in COVID-19 pathology and finding its associated partners as potential therapeutic targets. J. Biomol. Struct. Dyn. 2022, 40, 1813–1829. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Chatterjee, S.; Xiao, K.; Riedel, I.; Wang, Y.; Foo, R.; Bär, C.; Thum, T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020, 148, 46–49. [Google Scholar] [CrossRef]
- Chen, L.J.; Xu, R.; Yu, H.M.; Chang, Q.; Zhong, J.C. The ACE2/Apelin Signaling, MicroRNAs, and Hypertension. Int. J. Hypertens. 2015, 2015, 896861. [Google Scholar] [CrossRef] [Green Version]
- Widiasta, A.; Sribudiani, Y.; Nugrahapraja, H.; Hilmanto, D.; Sekarwana, N.; Rachmadi, D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-coding RNA Res. 2020, 5, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Bilezikian, J.P.; Bikle, D.; Hewison, M.; Lazaretti-Castro, M.; Formenti, A.M.; Gupta, A.; Madhavan, M.V.; Nair, N.; Babalyan, V.; Hutchings, N. Mechanisms in endocrinology: Vitamin D and COVID-19. Eur. J. Endocrinol. 2020, 183, R133–R147. [Google Scholar] [CrossRef]
- Hewison, M. Vitamin D and the intracrinology of innate immunity. Mol. Cell. Endocrinol. 2010, 321, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginde, A.A.; Mansbach, J.M.; Camargo, C.A. Vitamin D, respiratory infections, and asthma. Curr. Allergy Asthma Rep. 2009, 9, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.; Sadoughi, A.; Kohn, N.; Miller, R.; Chandak, T.; Talwar, A. Vitamin D deficiency in advanced lung disease. In A102. Interstitial Lung Disease: Exploring the Pathobiology of Interstitial Lung Disease: What Can We Learn from Histopathology, Genomics, and Biomarkers? American Thoracic Society: New York, NY, USA, 2011; p. A2346. [Google Scholar]
- Dusso, A.S. Vitamin D receptor: Mechanisms for vitamin D resistance in renal failure. Kidney Int. 2003, 63 (Suppl. 85), S6–S9. [Google Scholar] [CrossRef] [Green Version]
- Herr, C.; Greulich, T.; Koczulla, R.A.; Meyer, S.; Zakharkina, T.; Branscheidt, M.; Eschmann, R.; Bals, R. The role of vitamin D in pulmonary disease: COPD, asthma, infection, and cancer. Respir. Res. 2011, 12, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.-T.; Dabbas, B.; Laperriere, D.; Bitton, A.J.; Soualhine, H.; Tavera-Mendoza, L.E.; Dionne, S.; Servant, M.J.; Bitton, A.; Seidman, E.G. Direct and indirect induction by 1, 25-dihydroxyvitamin D3 of the NOD2/CARD15-defensin β2 innate immune pathway defective in Crohn disease. J. Biol. Chem. 2010, 285, 2227–2231. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. Vitamin D deficiency. N. Engl. J. Med. 2007, 357, 266–281. [Google Scholar] [CrossRef]
- Hansdottir, S.; Monick, M.M.; Lovan, N.; Powers, L.S.; Hunninghake, G.W. Smoking disrupts vitamin D metabolism in the lungs. In A36. Inflammation and the Airway Epithelium; American Thoracic Society: New York, NY, USA, 2010; p. A1425. [Google Scholar]
- Hughes, D.; Norton, R. Vitamin D and respiratory health. Clin. Exp. Immunol. 2009, 158, 20–25. [Google Scholar] [CrossRef]
- Meeran, S.M.; Ahmed, A.; Tollefsbol, T.O. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin. Epigenetics 2010, 1, 101–116. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.S.; Chung, I.; Trump, D.L. Epigenetic silencing of CYP24 in the tumor microenvironment. J. Steroid. Biochem. Mol. Biol. 2010, 121, 338–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishikawa, J.; Kitaura, M.; Matsumoto, M.; Imagawa, M.; Nishihara, T. Difference and similarity of DNA sequence recognized by VDR homodimer and VDR/RXR heterodimer. Nucleic Acids Res. 1994, 22, 2902–2907. [Google Scholar] [CrossRef] [Green Version]
- Fujiki, R.; Kim, M.-s.; Sasaki, Y.; Yoshimura, K.; Kitagawa, H.; Kato, S. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 2005, 24, 3881–3894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marik, R.; Fackler, M.; Gabrielson, E.; Zeiger, M.A.; Sukumar, S.; Stearns, V.; Umbricht, C.B. DNA methylation-related vitamin D receptor insensitivity in breast cancer. Cancer Biol. Ther. 2010, 10, 44–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essa, S.; Denzer, N.; Mahlknecht, U.; Klein, R.; Collnot, E.M.; Tilgen, W.; Reichrath, J. VDR microRNA expression and epigenetic silencing of vitamin D signaling in melanoma cells. J. Steroid. Biochem. Mol. Biol. 2010, 121, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Thorne, J.L.; Maguire, O.; Doig, C.L.; Battaglia, S.; Fehr, L.; Sucheston, L.E.; Heinaniemi, M.; O’Neill, L.P.; McCabe, C.J.; Turner, B.M.; et al. Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells. Nucleic Acids Res. 2011, 39, 2045–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seuter, S.; Pehkonen, P.; Heikkinen, S.; Carlberg, C. Dynamics of 1α,25-dihydroxyvitamin D3-dependent chromatin accessibility of early vitamin D receptor target genes. Biochim. Biophys. Acta 2013, 1829, 1266–1275. [Google Scholar] [CrossRef]
- Pan, L.; Matloob, A.F.; Du, J.; Pan, H.; Dong, Z.; Zhao, J.; Feng, Y.; Zhong, Y.; Huang, B.; Lu, J. Vitamin D stimulates apoptosis in gastric cancer cells in synergy with trichostatin A/sodium butyrate-induced and 5-aza-2′-deoxycytidine-induced PTEN upregulation. FEBS J. 2010, 277, 989–999. [Google Scholar] [CrossRef]
- Abedin, S.A.; Banwell, C.M.; Colston, K.W.; Carlberg, C.; Campbell, M.J. Epigenetic corruption of VDR signalling in malignancy. Anticancer Res. 2006, 26, 2557–2566. [Google Scholar]
- Khanim, F.L.; Gommersall, L.M.; Wood, V.H.; Smith, K.L.; Montalvo, L.; O’Neill, L.P.; Xu, Y.; Peehl, D.M.; Stewart, P.M.; Turner, B.M.; et al. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene 2004, 23, 6712–6725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X.; Lutz, W.; Schroeder, T.M.; Bachman, L.A.; Westendorf, J.J.; Kumar, R.; Griffin, M.D. Regulation of relB in dendritic cells by means of modulated association of vitamin D receptor and histone deacetylase 3 with the promoter. Proc. Natl. Acad. Sci. USA 2005, 102, 16007–16012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, B.-S.; Tavera-Mendoza, L.E.; Dimitrov, V.; Wang, X.; Calderon, M.R.; Wang, H.-J.; White, J.H. Stimulation of Sirt1-regulated FoxO protein function by the ligand-bound vitamin D receptor. Mol. Cell. Biol. 2010, 30, 4890–4900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavu, S.; Boss, O.; Elliott, P.J.; Lambert, P.D. Sirtuins—Novel therapeutic targets to treat age-associated diseases. Nat. Rev. Drug Discov. 2008, 7, 841–853. [Google Scholar] [CrossRef]
- Hansdottir, S.; Monick, M.M.; Hinde, S.L.; Lovan, N.; Look, D.C.; Hunninghake, G.W. Respiratory epithelial cells convert inactive vitamin D to its active form: Potential effects on host defense. J. Immunol. 2008, 181, 7090–7099. [Google Scholar] [CrossRef] [Green Version]
- Hansdottir, S.; Monick, M.M.; Lovan, N.; Powers, L.; Gerke, A.; Hunninghake, G.W. Vitamin D decreases respiratory syncytial virus induction of NF-κB–linked chemokines and cytokines in airway epithelium while maintaining the antiviral state. J. Immunol. 2010, 184, 965–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantorna, M.T.; Snyder, L.; Lin, Y.-D.; Yang, L. Vitamin D and 1, 25 (OH) 2D regulation of T cells. Nutrients 2015, 7, 3011–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human antimicrobial peptides as therapeutics for viral infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, J.; Lin, E.; He, L.; Yu, J.; Tan, P.; Zhou, Y. Autophagy and viral infection. In Autophagy Regulation of Innate Immunity; Springer: Singapore, 2019; pp. 55–78. [Google Scholar]
- Mushegian, A.A. Autophagy and vitamin D. Sci. Signal. 2017, 10, eaan2526. [Google Scholar] [CrossRef] [PubMed]
- Teymoori-Rad, M.; Shokri, F.; Salimi, V.; Marashi, S.M. The interplay between vitamin D and viral infections. Rev. Med. Virol. 2019, 29, e2032. [Google Scholar] [CrossRef]
- Lau, F.H.; Majumder, R.; Torabi, R.; Saeg, F.; Hoffman, R.; Cirillo, J.D.; Greiffenstein, P. Vitamin D insufficiency is prevalent in severe COVID-19. MedRxiv 2020. [Google Scholar] [CrossRef]
- Lanham-New, S.A.; Webb, A.R.; Cashman, K.D.; Buttriss, J.L.; Fallowfield, J.L.; Masud, T.; Hewison, M.; Mathers, J.C.; Kiely, M.; Welch, A.A. Vitamin D and SARS-CoV-2 virus/COVID-19 disease. BMJ Nutr. Prev. Health 2020, 3, 106. [Google Scholar] [CrossRef]
- Daneshkhah, A.; Agrawal, V.; Eshein, A.; Subramanian, H.; Roy, H.K.; Backman, V. The possible role of vitamin D in suppressing cytokine storm and associated mortality in COVID-19 patients. MedRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Maghbooli, Z.; Sahraian, M.A.; Ebrahimi, M.; Pazoki, M.; Kafan, S.; Tabriz, H.M.; Hadadi, A.; Montazeri, M.; Nasiri, M.; Shirvani, A. Vitamin D sufficiency, a serum 25-hydroxyvitamin D at least 30 ng/mL reduced risk for adverse clinical outcomes in patients with COVID-19 infection. PLoS ONE 2020, 15, e0239799. [Google Scholar] [CrossRef] [PubMed]
- Alipio, M. Vitamin D supplementation could possibly improve clinical outcomes of patients infected with coronavirus-2019 (COVID-19). SSRN 2020, 3571484. [Google Scholar] [CrossRef]
- Dhar, A.; Mir, H.; Koul, P.A. Vitamin D Levels and Length of Hospitalization in Indian Patients With COVID-19: A Single-Center Prospective Study. Cureus 2022, 14, e26704. [Google Scholar] [CrossRef] [PubMed]
- Autier, P.; Mullie, P.; Macacu, A.; Dragomir, M.; Boniol, M.; Coppens, K.; Pizot, C.; Boniol, M. Effect of vitamin D supplementation on non-skeletal disorders: A systematic review of meta-analyses and randomised trials. Lancet Diabetes Endocrinol. 2017, 5, 986–1004. [Google Scholar] [CrossRef]
- Tan, C.W.; Ho, L.P.; Kalimuddin, S.; Cherng, B.P.Z.; Teh, Y.E.; Thien, S.Y.; Wong, H.M.; Tern, P.J.W.; Chandran, M.; Chay, J.W.M. A cohort study to evaluate the effect of combination Vitamin D, Magnesium and Vitamin B12 (DMB) on progression to severe outcome in older COVID-19 patients. MedRxiv 2020. [Google Scholar] [CrossRef]
- Zurita-Cruz, J.; Fonseca-Tenorio, J.; Villasís-Keever, M.; López-Alarcón, M.; Parra-Ortega, I.; López-Martínez, B.; Miranda-Novales, G. Efficacy and safety of vitamin D supplementation in hospitalized COVID-19 pediatric patients: A randomized controlled trial. Front. Pediatr. 2022, 10, 943529. [Google Scholar] [CrossRef]
- Raharusun, P.; Priambada, S.; Budiarti, C.; Agung, E.; Budi, C. Patterns of COVID-19 mortality and vitamin D: An Indonesian study. SSRN 2020, 3585561. [Google Scholar] [CrossRef]
- Gennari, L.; Campi, I.; Merlotti, D.; Mingiano, C.; Frosali, A.; Giovanelli, L.; Torlasco, C.; Pengo, M.; Heilbron, F.; Soranna, D. Vitamin D deficiency is independently associated with COVID-19 severity and mortality. J. Bone Miner. Res. 2020, 35, 7–8. [Google Scholar]
- Luo, X.; Liao, Q.; Shen, Y.; Li, H.; Cheng, L. Vitamin D Deficiency Is Associated with COVID-19 Incidence and Disease Severity in Chinese People. J. Nutr. 2020, 151, 98–103. [Google Scholar] [CrossRef]
- Abrishami, A.; Dalili, N.; Mohammadi Torbati, P.; Asgari, R.; Arab-Ahmadi, M.; Behnam, B.; Sanei-Taheri, M. Possible association of vitamin D status with lung involvement and outcome in patients with COVID-19: A retrospective study. Eur. J. Nutr. 2021, 60, 2249–2257. [Google Scholar] [CrossRef]
- Peng, D.; Huang, H.; Liu, Z.; Gao, Y.; Liu, Y. Vitamin D levels and clinical outcomes of SARS-CoV-2 Omicron subvariant BA.2 in children: A longitudinal cohort study. Front. Nutr. 2022, 9, 960859. [Google Scholar] [CrossRef]
- Hou, Y.-M.; Zhao, J.-Y.; Liu, H.-Y. Impact of serum 25-hydroxyvitamin D on cardiac prognosis in Chinese patients with heart failure. Br. J. Nutr. 2019, 122, 162–171. [Google Scholar] [CrossRef]
- Zhou, Y.-F.; Luo, B.-A.; Qin, L.-L. The association between vitamin D deficiency and community-acquired pneumonia: A meta-analysis of observational studies. Medicine 2019, 98, e17252. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e910. [Google Scholar] [CrossRef] [PubMed]
- Verdecchia, P.; Cavallini, C.; Spanevello, A.; Angeli, F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020, 76, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Krammer, F. SARS-CoV-2 vaccines: Status report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Herbein, G. Management of epigenomic networks entailed in coronavirus infections and COVID-19. Clin. Epigenetics 2020, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef] [Green Version]
- Sfera, A.; Bullock, K.; Price, A.; Inderias, L.; Osorio, C. Ferrosenescence: The iron age of neurodegeneration? Mech. Ageing Dev. 2018, 174, 63–75. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Mehta, S.; Jeffrey, K.L. Beyond receptors and signaling: Epigenetic factors in the regulation of innate immunity. Immunol. Cell Biol. 2015, 93, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Zwergel, C.; Stazi, G.; Valente, S.; Mai, A. Histone Deacetylase Inhibitors: Updated Studies in Various Epigenetic-Related Diseases. J. Clin. Epigenetics 2016, 2, 7. [Google Scholar]
- Yang, T.; Li, S.; Zhang, X.; Pang, X.; Lin, Q.; Cao, J. Resveratrol, sirtuins, and viruses. Rev. Med. Virol. 2015, 25, 431–445. [Google Scholar] [CrossRef]
- Nehme, Z.; Pasquereau, S.; Herbein, G. Control of viral infections by epigenetic-targeted therapy. Clin. Epigenetics 2019, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Ayaz, S.; Crea, F. Targeting SARS-CoV-2 using polycomb inhibitors as antiviral agents. Epigenomics 2020, 12, 811–812. [Google Scholar] [CrossRef] [PubMed]
- Geller, A.; Yan, J. Could the Induction of Trained Immunity by β-Glucan Serve as a Defense Against COVID-19? Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Arghiani, N.; Nissan, T.; Matin, M.M. Role of microRNAs in COVID-19 with implications for therapeutics. Biomed. Pharmacother. 2021, 144, 112247. [Google Scholar] [CrossRef]
- Gallicano, G.I.; Casey, J.L.; Fu, J.; Mahapatra, S. Molecular targeting of vulnerable RNA sequences in SARS CoV-2: Identifying clinical feasibility. Gene Therapy 2022, 29, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.-A.-K.; Sany, M.R.U.; Islam, M.S.; Islam, A.B.M.M.K. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.K.; Zhou, Z.; Jiang, X.M.; Zheng, Y.; Chen, X.; Fu, Z.; Xiao, G.; Zhang, C.Y.; Zhang, L.K.; Yi, Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Glinsky, G. Genomics-guided molecular maps of coronavirus targets in human cells: A path toward the repurposing of existing drugs to mitigate the pandemic. arXiv 2020, arXiv:2003.13665. [Google Scholar]
- Ilie, P.C.; Stefanescu, S.; Smith, L. The role of vitamin D in the prevention of coronavirus disease 2019 infection and mortality. Aging Clin. Exp. Res. 2020, 32, 1195–1198. [Google Scholar] [CrossRef]
- Wang, T.-T.; Nestel, F.P.; Bourdeau, V.; Nagai, Y.; Wang, Q.; Liao, J.; Tavera-Mendoza, L.; Lin, R.; Hanrahan, J.W.; Mader, S. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 2004, 173, 2909–2912. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Yang, Y.L.; Jang, S.-H.; Jang, Y.-S. Human β-defensin 2 plays a regulatory role in innate antiviral immunity and is capable of potentiating the induction of antigen-specific immunity. Virol. J. 2018, 15, 1–12. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foolchand, A.; Mazaleni, S.; Ghazi, T.; Chuturgoon, A.A. A Review: Highlighting the Links between Epigenetics, COVID-19 Infection, and Vitamin D. Int. J. Mol. Sci. 2022, 23, 12292. https://doi.org/10.3390/ijms232012292
Foolchand A, Mazaleni S, Ghazi T, Chuturgoon AA. A Review: Highlighting the Links between Epigenetics, COVID-19 Infection, and Vitamin D. International Journal of Molecular Sciences. 2022; 23(20):12292. https://doi.org/10.3390/ijms232012292
Chicago/Turabian StyleFoolchand, Ashmika, Siyanda Mazaleni, Terisha Ghazi, and Anil A. Chuturgoon. 2022. "A Review: Highlighting the Links between Epigenetics, COVID-19 Infection, and Vitamin D" International Journal of Molecular Sciences 23, no. 20: 12292. https://doi.org/10.3390/ijms232012292




