The Corticosterone–Glucocorticoid Receptor–AP1/CREB Axis Inhibits the Luteinizing Hormone Receptor Expression in Mouse Granulosa Cells
Abstract
:1. Introduction
2. Results
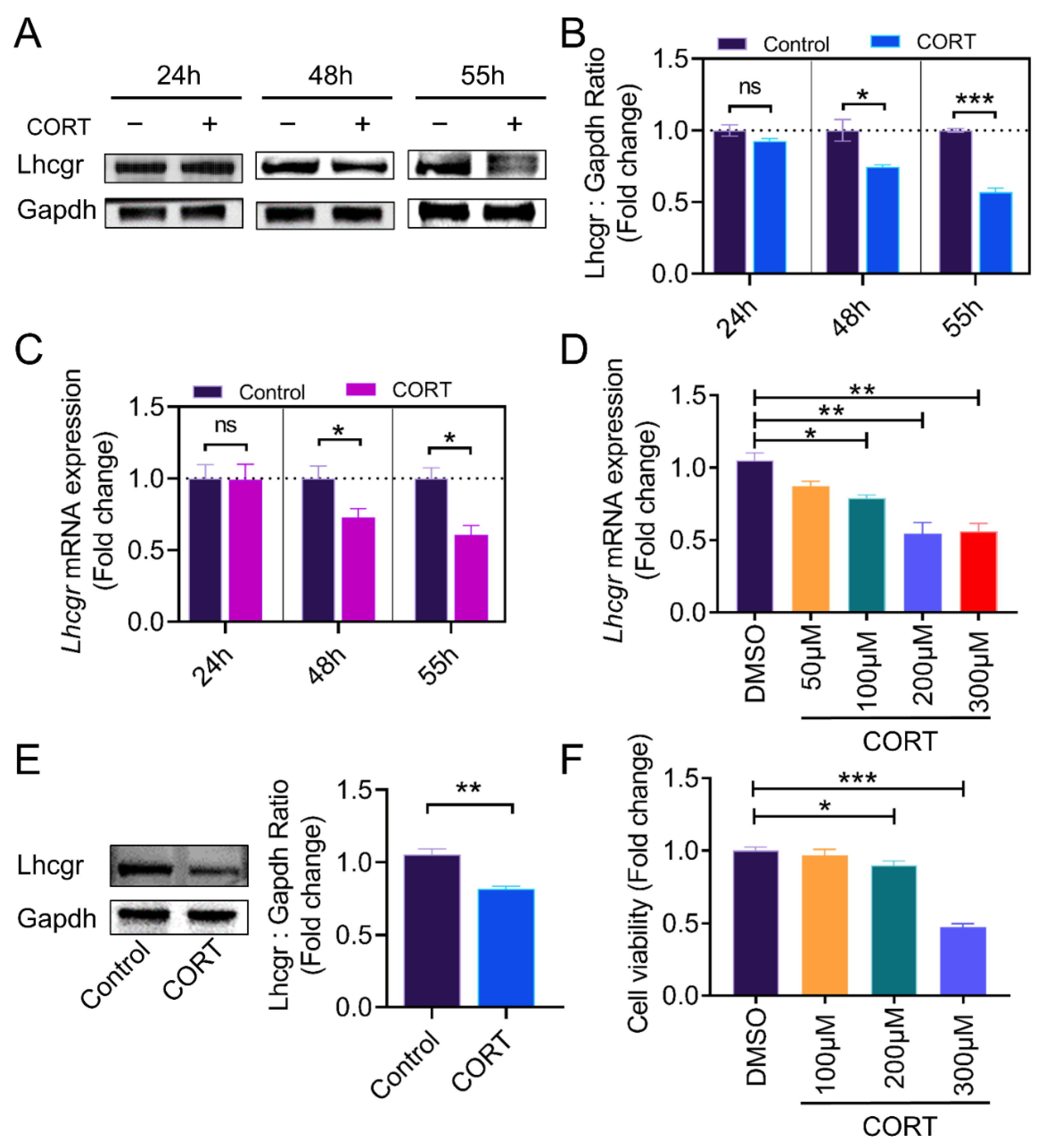
2.1. CORT Inhibits LH Receptor Expression in the Ovarian Granulosa Cells of Mice
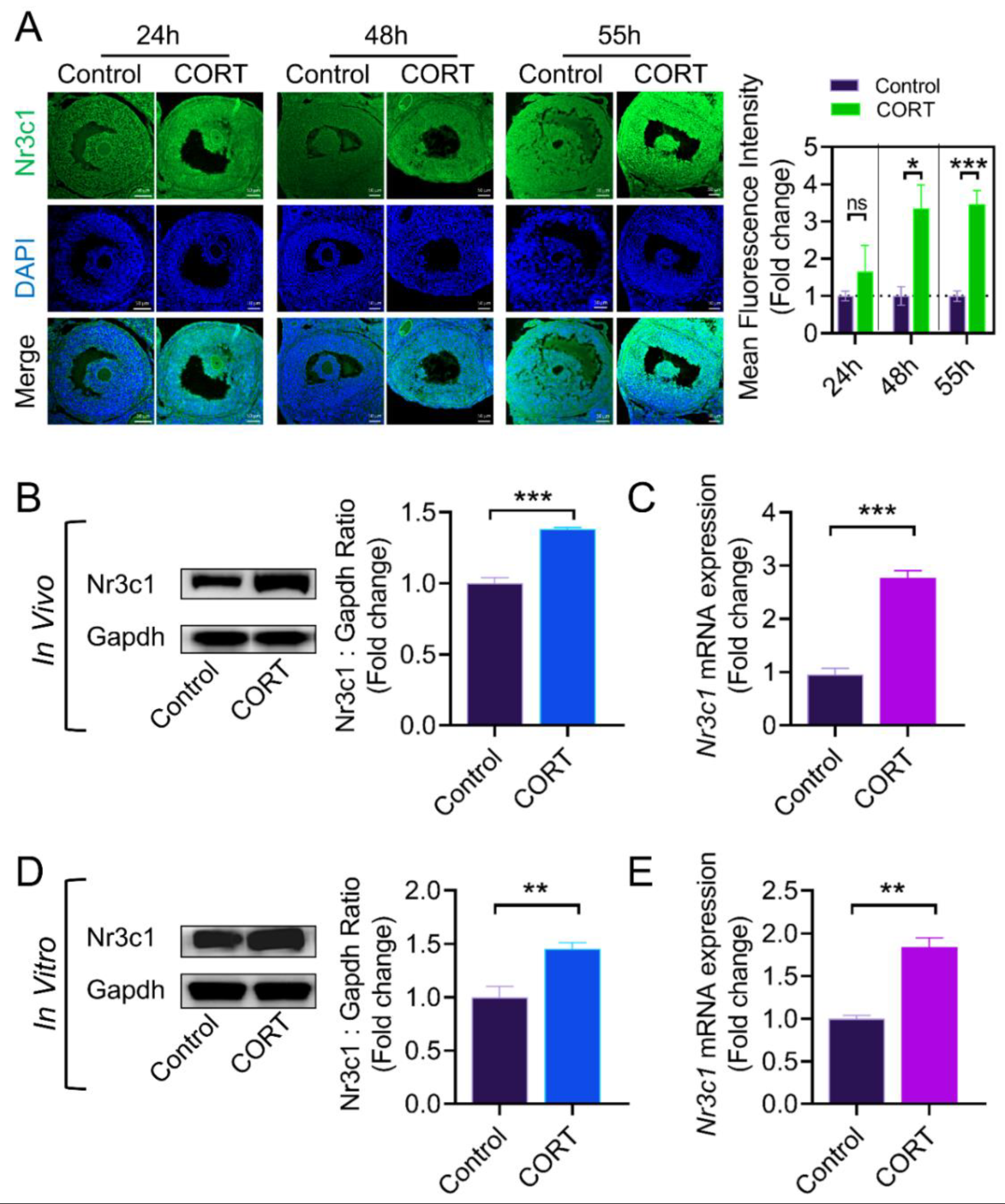
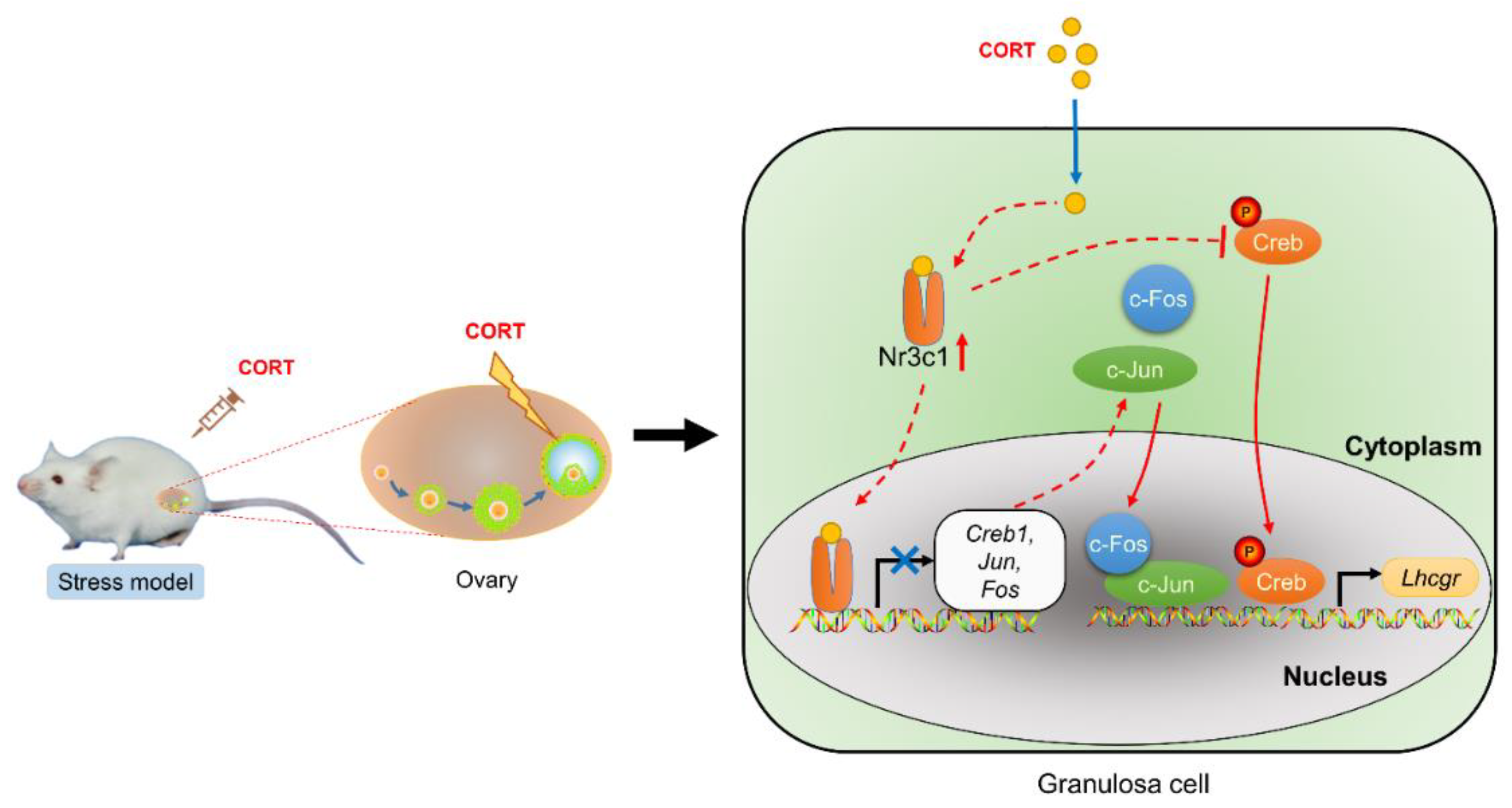
2.2. The CORT-Mediated Inhibition of Lhcgr Expression Is Dependent on the Activation of the Glucocorticoid Receptor (Nr3c1) in Mouse Ovarian GCs
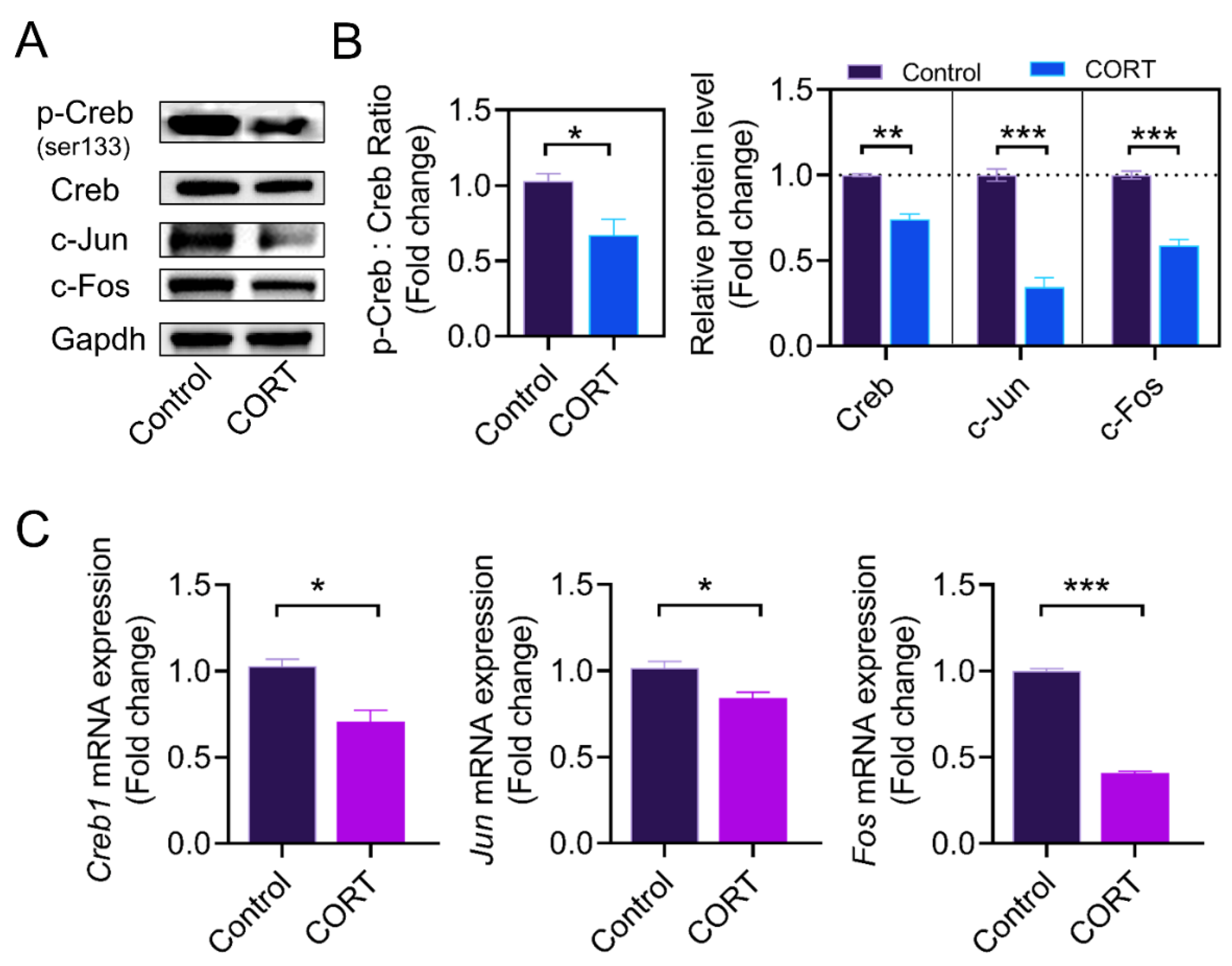
2.3. CORT Inhibits the Transcriptional Activity of Lhcgr Transcription Factors in GCs
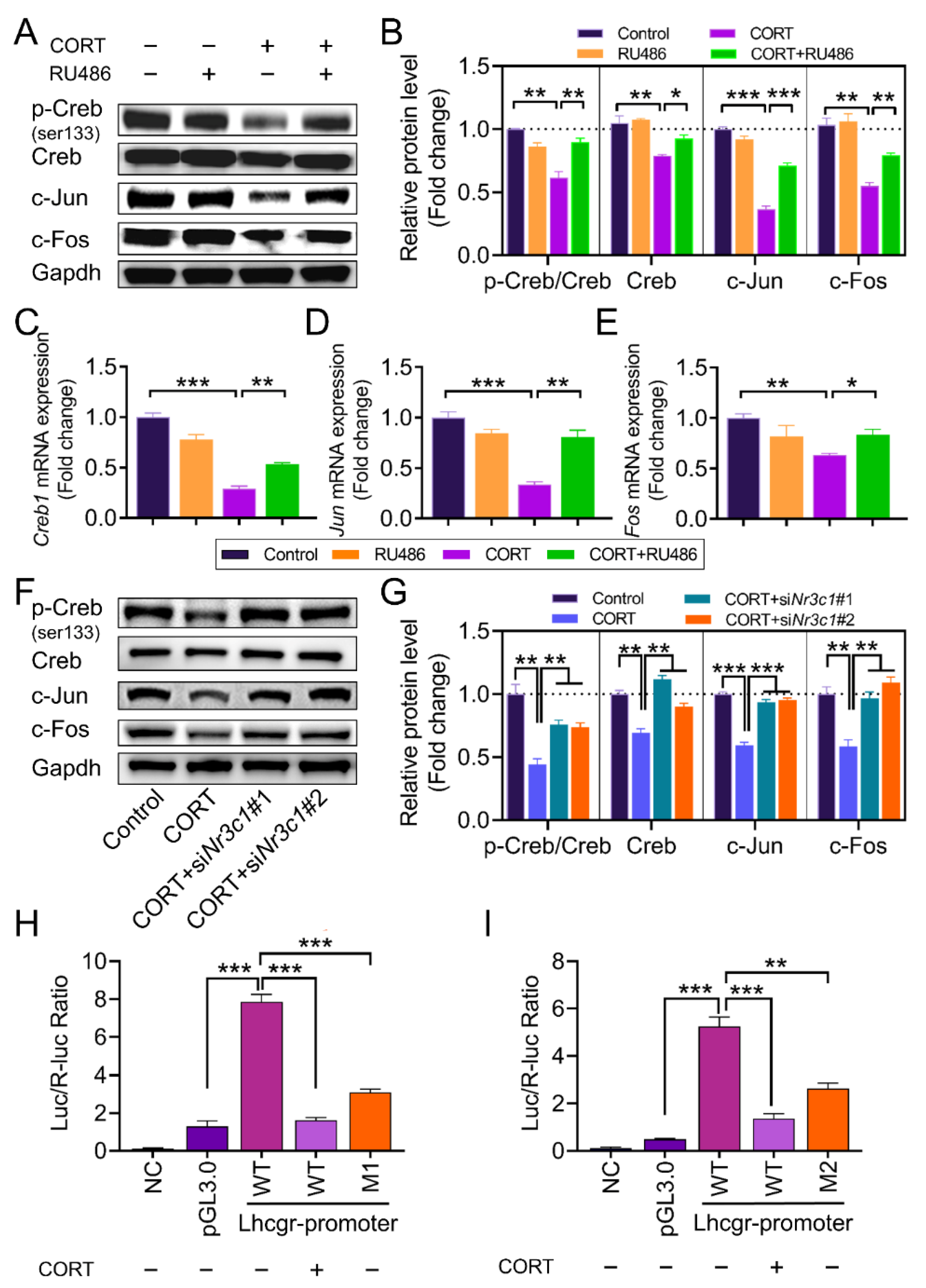
2.4. The Expression of AP1 and Creb Is Regulated by Nr3c1
3. Discussion
3.1. Impact of CORT on Lhcgr Expression
3.2. Lhcgr Transcriptional Regulation
3.3. Effect of Nr3c1 on Lhcgr Transcription
4. Materials and Methods
4.1. Chemicals and Antibodies
4.2. Animals and Ethics
4.3. Cell Culture and Treatments
4.4. Cell Viability Assay
4.5. Western Blotting
4.6. Quantitative RT-PCR (qRT-PCR)
4.7. Immunofluorescence
4.8. RNA Interference
4.9. Luciferase Reporter Gene Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Approval and Consent to Participate
Abbreviations
References
- Oke, O.E.; Uyanga, V.A.; Iyasere, O.S.; Oke, F.O.; Majekodunmi, B.C.; Logunleko, M.O.; Abiona, J.A.; Nwosu, E.U.; Abioja, M.O.; Daramola, J.O.; et al. Environmental stress and livestock productivity in hot-humid tropics: Alleviation and future perspectives. J. Biol. 2021, 100, 103077. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, J.C.; Sapolsky, R.M. Reproduction and resistance to stress: When and how. J. Neuroendocr. 2003, 15, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Ciborowska, P.; Michalczuk, M.; Bien, D. The Effect of Music on Livestock: Cattle, Poultry and Pigs. Animals 2021, 11, 3572. [Google Scholar] [CrossRef]
- Wu, T.C.; Chen, H.T.; Chang, H.Y.; Yang, C.Y.; Hsiao, M.C.; Cheng, M.L.; Chen, J.C. Mineralocorticoid receptor antagonist spironolactone prevents chronic corticosterone induced depression-like behavior. Psychoneuroendocrinology 2013, 38, 871–883. [Google Scholar] [CrossRef]
- Richards, J.S.; Pangas, S.A. The ovary: Basic biology and clinical implications. J. Clin. Investig. 2010, 120, 963–972. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Liu, X.; Segaloff, D.L. A novel cyclic adenosine 3’,5’-monophosphate-responsive element involved in the transcriptional regulation of the lutropin receptor gene in granulosa cells. Mol. Endocrinol. 2000, 14, 1498–1508. [Google Scholar] [PubMed] [Green Version]
- Menon, K.M.; Clouser, C.L.; Nair, A.K. Gonadotropin receptors: Role of post-translational modifications and post-transcriptional regulation. Endocrine 2005, 26, 249–257. [Google Scholar] [CrossRef]
- Ubuka, T.; Tsutsui, K. Reproductive neuroendocrinology of mammalian gonadotropin-inhibitory hormone. Reprod. Med. Biol. 2019, 18, 225–233. [Google Scholar] [CrossRef]
- Zeleznik, A.J.; Schuler, H.M.; Reichert, L.E., Jr. Gonadotropin-binding sites in the rhesus monkey ovary: Role of the vasculature in the selective distribution of human chorionic gonadotropin to the preovulatory follicle. Endocrinology 1981, 109, 356–362. [Google Scholar] [CrossRef]
- Sugino, N.; Hirosawa-Takamori, M.; Zhong, L.; Telleria, C.M.; Shiota, K.; Gibori, G. Hormonal regulation of copper-zinc superoxide dismutase and manganese superoxide dismutase messenger ribonucleic acid in the rat corpus luteum: Induction by prolactin and placental lactogens. Biol. Reprod. 1998, 59, 599–605. [Google Scholar] [CrossRef]
- Casarini, L.; Riccetti, L.; De Pascali, F.; Gilioli, L.; Marino, M.; Vecchi, E.; Morini, D.; Nicoli, A.; La Sala, G.B.; Simoni, M. Estrogen Modulates Specific Life and Death Signals Induced by LH and hCG in Human Primary Granulosa Cells In Vitro. Int. J. Mol. Sci. 2017, 18, 926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sher, N.; Yivgi-Ohana, N.; Orly, J. Transcriptional regulation of the cholesterol side chain cleavage cytochrome P450 gene (CYP11A1) revisited: Binding of GATA, cyclic adenosine 3’,5’-monophosphate response element-binding protein and activating protein (AP)-1 proteins to a distal novel cluster of cis-regulatory elements potentiates AP-2 and steroidogenic factor-1-dependent gene expression in the rodent placenta and ovary. Mol. Endocrinol. 2007, 21, 948–962. [Google Scholar] [PubMed] [Green Version]
- Zhang, F.P.; Poutanen, M.; Wilbertz, J.; Huhtaniemi, I. Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LuRKO) mice. Mol. Endocrinol. 2001, 15, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.M.; Mishra, S.; Zou, W.; Xu, B.; Foltz, M.; Li, X.; Rao, C.V. Targeted disruption of luteinizing hormone/human chorionic gonadotropin receptor gene. Mol. Endocrinol. 2001, 15, 184–200. [Google Scholar] [CrossRef]
- Mizuki, D.; Matsumoto, K.; Tanaka, K.; Thi Le, X.; Fujiwara, H.; Ishikawa, T.; Higuchi, Y. Antidepressant-like effect of Butea superba in mice exposed to chronic mild stress and its possible mechanism of action. J. Ethnopharmacol. 2014, 156, 16–25. [Google Scholar] [CrossRef]
- Yan, T.; Xu, M.; Wan, S.; Wang, M.; Wu, B.; Xiao, F.; Bi, K.; Jia, Y. Schisandra chinensis produces the antidepressant-like effects in repeated corticosterone-induced mice via the BDNF/TrkB/CREB signaling pathway. Psychiatry Res. 2016, 243, 135–142. [Google Scholar] [CrossRef]
- Valsamakis, G.; Chrousos, G.; Mastorakos, G. Stress, female reproduction and pregnancy. Psychoneuroendocrinology 2019, 100, 48–57. [Google Scholar] [CrossRef]
- Wagenmaker, E.R.; Moenter, S.M. Exposure to Acute Psychosocial Stress Disrupts the Luteinizing Hormone Surge Independent of Estrous Cycle Alterations in Female Mice. Endocrinology 2017, 158, 2593–2602. [Google Scholar] [CrossRef] [Green Version]
- Whirledge, S.; Cidlowski, J.A. Glucocorticoids, stress, and fertility. Minerva. Endocrinol. 2010, 35, 109–125. [Google Scholar]
- Lv, F.; Fan, G.; Wan, Y.; Chen, Y.; Ni, Y.; Huang, J.; Xu, D.; Zhang, W.; Wang, H. Intrauterine endogenous high glucocorticoids program ovarian dysfunction in female offspring secondary to prenatal caffeine exposure. Sci. Total Environ. 2021, 789, 147691. [Google Scholar] [CrossRef]
- Rabin, D.S.; Johnson, E.O.; Brandon, D.D.; Liapi, C.; Chrousos, G.P. Glucocorticoids inhibit estradiol-mediated uterine growth: Possible role of the uterine estradiol receptor. Biol. Reprod. 1990, 42, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Whirledge, S.; Cidlowski, J.A. Glucocorticoids and Reproduction: Traffic Control on the Road to Reproduction. Trends. Endocrinol. Metab. 2017, 28, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, W.; Meng, X.; Zhang, L.; Shen, M.; Liu, H. Corticosterone Injection Impairs Follicular Development, Ovulation and Steroidogenesis Capacity in Mice Ovary. Animals 2019, 9, 1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breen, K.M.; Billings, H.J.; Wagenmaker, E.R.; Wessinger, E.W.; Karsch, F.J. Endocrine basis for disruptive effects of cortisol on preovulatory events. Endocrinology 2005, 146, 2107–2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.J.; Han, X.; He, N.; Wang, G.L.; Gong, S.; Lin, J.; Gao, M.; Tan, J.H. Glucocorticoids impair oocyte developmental potential by triggering apoptosis of ovarian cells via activating the Fas system. Sci. Rep. 2016, 6, 24036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caradonna, S.G.; Einhorn, N.R.; Saudagar, V.; Khalil, H.; Petty, G.H.; Lihagen, A.; LeFloch, C.; Lee, F.S.; Akil, H.; Guidotti, A.; et al. Corticosterone induces discrete epigenetic signatures in the dorsal and ventral hippocampus that depend upon sex and genotype: Focus on methylated Nr3c1 gene. Transl. Psychiatry 2022, 12, 109. [Google Scholar] [CrossRef]
- Li, J.; Hlavka-Zhang, J.; Shrimp, J.H.; Piper, C.; Dupere-Richer, D.; Roth, J.S.; Jing, D.; Casellas Roman, H.L.; Troche, C.; Swaroop, A.; et al. PRC2 Inhibitors Overcome Glucocorticoid Resistance Driven by NSD2 Mutation in Pediatric Acute Lymphoblastic Leukemia. Cancer Discov. 2022, 12, 186–203. [Google Scholar] [CrossRef]
- Van der Auwera, S.; Klinger-Konig, J.; Wittfeld, K.; Terock, J.; Hannemann, A.; Bulow, R.; Nauck, M.; Volker, U.; Volzke, H.; Grabe, H.J. The interplay between genetic variation and gene expression of the glucocorticoid receptor gene NR3C1 and blood cortisol levels on verbal memory and hippocampal volumes. Eur. Arch. Psychiatry Clin. Neurosci. 2022, 1–12. [Google Scholar] [CrossRef]
- Garabedian, M.J.; Harris, C.A.; Jeanneteau, F. Glucocorticoid receptor action in metabolic and neuronal function. F1000Research 2017, 6, 1208. [Google Scholar] [CrossRef] [Green Version]
- Spencer, R.L.; Deak, T. A users guide to HPA axis research. Physiol. Behav. 2017, 178, 43–65. [Google Scholar] [CrossRef]
- Miranda, T.B.; Morris, S.A.; Hager, G.L. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol. Cell. Endocrinol. 2013, 380, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.T.; Fu, T.; Zhang, H.Y.; Yang, Z.S.; Zheng, Z.H.; Yang, Z.M. Progesterone-regulated Hsd11b2 as a barrier to balance mouse uterine corticosterone. J. Endocrinol. 2020, 244, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Joels, M.; Baram, T.Z. The neuro-symphony of stress. Nat. Rev. Neurosci. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, A.C.; Dunn, H.; Ferguson, S.S. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br. J. Pharm. 2012, 165, 1717–1736. [Google Scholar] [CrossRef] [Green Version]
- Miller, B.H.; Takahashi, J.S. Central circadian control of female reproductive function. Front. Endocrinol 2013, 4, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einarsson, S.; Brandt, Y.; Lundeheim, N.; Madej, A. Stress and its influence on reproduction in pigs: A review. Acta Vet. Scand. 2008, 50, 48. [Google Scholar] [CrossRef] [Green Version]
- De Kloet, E.R.; Vreugdenhil, E.; Oitzl, M.S.; Joels, M. Brain corticosteroid receptor balance in health and disease. Endocr. Rev. 1998, 19, 269–301. [Google Scholar]
- Jeppesen, J.V.; Kristensen, S.G.; Nielsen, M.E.; Humaidan, P.; Dal Canto, M.; Fadini, R.; Schmidt, K.T.; Ernst, E.; Yding Andersen, C. LH-receptor gene expression in human granulosa and cumulus cells from antral and preovulatory follicles. J. Clin. Endocrinol. Metab. 2012, 97, E1524–E1531. [Google Scholar] [CrossRef]
- Breen, K.M.; Thackray, V.G.; Hsu, T.; Mak-McCully, R.A.; Coss, D.; Mellon, P.L. Stress levels of glucocorticoids inhibit LHbeta-subunit gene expression in gonadotrope cells. Mol. Endocrinol. 2012, 26, 1716–1731. [Google Scholar] [CrossRef] [Green Version]
- Convissar, S.; Winston, N.J.; Fierro, M.A.; Scoccia, H.; Zamah, A.M.; Stocco, C. Sp1 regulates steroidogenic genes and LHCGR expression in primary human luteinized granulosa cells. J. Steroid Biochem. Mol. Biol. 2019, 190, 183–192. [Google Scholar] [CrossRef]
- De Rensis, F.; Garcia-Ispierto, I.; Lopez-Gatius, F. Seasonal heat stress: Clinical implications and hormone treatments for the fertility of dairy cows. Theriogenology 2015, 84, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.S.; Qian, Z.; Liu, H.L.; Bao, E.D. ACTH-induced stress in weaned sows impairs LH receptor expression and steroidogenesis capacity in the ovary. Reprod. Biol. Endocrinol. 2016, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- He, P.J.; Fujimoto, Y.; Yamauchi, N.; Hattori, M.A. Real-time monitoring of cAMP response element binding protein signaling in porcine granulosa cells modulated by ovarian factors. Mol. Cell. Biochem. 2006, 290, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Peegel, H.; Towns, R.; Nair, A.; Menon, K.M. A novel mechanism for the modulation of luteinizing hormone receptor mRNA expression in the rat ovary. Mol. Cell. Endocrinol. 2005, 233, 65–72. [Google Scholar] [CrossRef] [PubMed]
- LaVoie, H.A. Transcriptional control of genes mediating ovarian follicular growth, differentiation, and steroidogenesis in pigs. Mol. Reprod. Dev. 2017, 84, 788–801. [Google Scholar] [CrossRef] [Green Version]
- Rusovici, R.; LaVoie, H.A. Expression and distribution of AP-1 transcription factors in the porcine ovary. Biol. Reprod. 2003, 69, 64–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sefton, C.; Harno, E.; Davies, A.; Small, H.; Allen, T.J.; Wray, J.R.; Lawrence, C.B.; Coll, A.P.; White, A. Elevated Hypothalamic Glucocorticoid Levels Are Associated With Obesity and Hyperphagia in Male Mice. Endocrinology 2016, 157, 4257–4265. [Google Scholar] [CrossRef] [Green Version]
- Chrousos, G.P.; Kino, T. Glucocorticoid signaling in the cell. Expanding clinical implications to complex human behavioral and somatic disorders. Ann. N. Y. Acad. Sci. 2009, 1179, 153–166. [Google Scholar] [CrossRef] [Green Version]
- Langlais, D.; Couture, C.; Balsalobre, A.; Drouin, J. The Stat3/GR interaction code: Predictive value of direct/indirect DNA recruitment for transcription outcome. Mol. Cell 2012, 47, 38–49. [Google Scholar] [CrossRef] [Green Version]
- Shyu, A.B.; Belasco, J.G.; Greenberg, M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991, 5, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Rao, N.A.; McCalman, M.T.; Moulos, P.; Francoijs, K.J.; Chatziioannou, A.; Kolisis, F.N.; Alexis, M.N.; Mitsiou, D.J.; Stunnenberg, H.G. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011, 21, 1404–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Kloet, E.R.; Meijer, O.C.; de Nicola, A.F.; de Rijk, R.H.; Joels, M. Importance of the brain corticosteroid receptor balance in metaplasticity, cognitive performance and neuro-inflammation. Front. Neuroendocr. 2018, 49, 124–145. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.H.; Madhana, R.M.; Athira, K.V.; Kasala, E.R.; Bodduluru, L.N.; Pitta, S.; Mahareddy, J.R.; Lahkar, M. Resveratrol ameliorates depressive-like behavior in repeated corticosterone-induced depression in mice. Steroids 2015, 101, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Mifsud, K.R.; Reul, J. Mineralocorticoid and glucocorticoid receptor-mediated control of genomic responses to stress in the brain. Stress 2018, 21, 389–402. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Jiang, Y.; Guan, Z.; Cao, Y.; Li, L.; Liu, H.; Sun, S.C. Protective mechanism of FSH against oxidative damage in mouse ovarian granulosa cells by repressing autophagy. Autophagy 2017, 13, 1364–1385. [Google Scholar] [CrossRef]
- Shen, M.; Lin, F.; Zhang, J.; Tang, Y.; Chen, W.K.; Liu, H. Involvement of the up-regulated FoxO1 expression in follicular granulosa cell apoptosis induced by oxidative stress. J. Biol. Chem. 2012, 287, 25727–25740. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wei, Y.; Li, X.; Li, C.; Zhang, L.; Liu, Z.; Cao, Y.; Li, W.; Zhang, X.; Zhang, J.; et al. The Corticosterone–Glucocorticoid Receptor–AP1/CREB Axis Inhibits the Luteinizing Hormone Receptor Expression in Mouse Granulosa Cells. Int. J. Mol. Sci. 2022, 23, 12454. https://doi.org/10.3390/ijms232012454
Zhang X, Wei Y, Li X, Li C, Zhang L, Liu Z, Cao Y, Li W, Zhang X, Zhang J, et al. The Corticosterone–Glucocorticoid Receptor–AP1/CREB Axis Inhibits the Luteinizing Hormone Receptor Expression in Mouse Granulosa Cells. International Journal of Molecular Sciences. 2022; 23(20):12454. https://doi.org/10.3390/ijms232012454
Chicago/Turabian StyleZhang, Xuan, Yinghui Wei, Xiaoxuan Li, Chengyu Li, Liangliang Zhang, Zhaojun Liu, Yan Cao, Weijian Li, Xiying Zhang, Jiaqing Zhang, and et al. 2022. "The Corticosterone–Glucocorticoid Receptor–AP1/CREB Axis Inhibits the Luteinizing Hormone Receptor Expression in Mouse Granulosa Cells" International Journal of Molecular Sciences 23, no. 20: 12454. https://doi.org/10.3390/ijms232012454
APA StyleZhang, X., Wei, Y., Li, X., Li, C., Zhang, L., Liu, Z., Cao, Y., Li, W., Zhang, X., Zhang, J., Shen, M., & Liu, H. (2022). The Corticosterone–Glucocorticoid Receptor–AP1/CREB Axis Inhibits the Luteinizing Hormone Receptor Expression in Mouse Granulosa Cells. International Journal of Molecular Sciences, 23(20), 12454. https://doi.org/10.3390/ijms232012454





