An Original and Efficient Antibiotic Adjuvant Strategy to Enhance the Activity of Macrolide Antibiotics against Gram-Negative Resistant Strains
Abstract
1. Introduction
2. Results
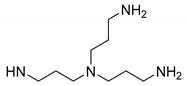
2.1. Synthesis of Polyaminoisoprenyl Derivatives 3–6
2.2. Antimicrobial Activity of Polyaminoisoprenyl Derivatives 3–6 against Gram-Negative Bacteria
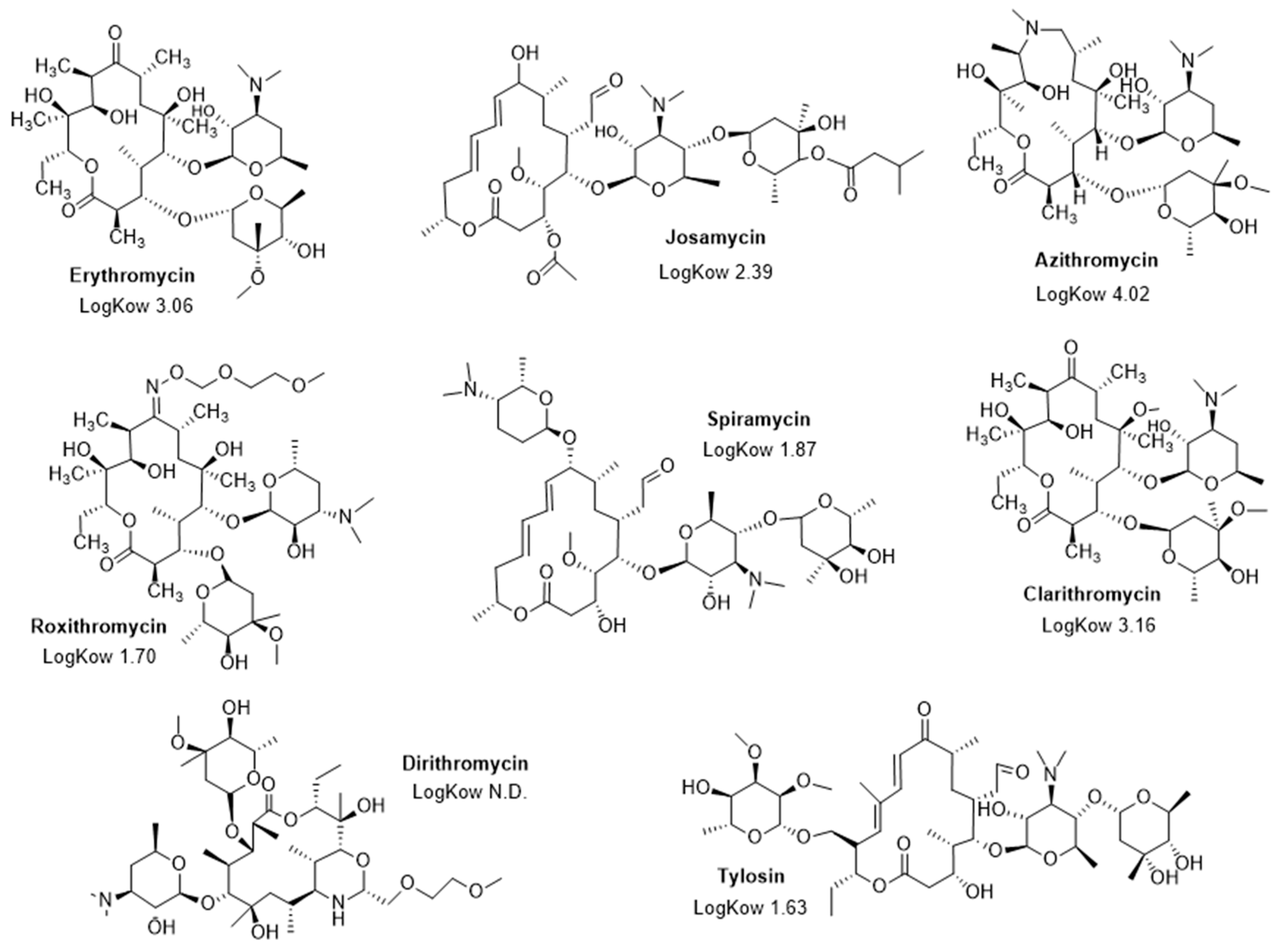
2.3. MICs of the Different Macrolides Tested against Various Gram-Negative Bacteria
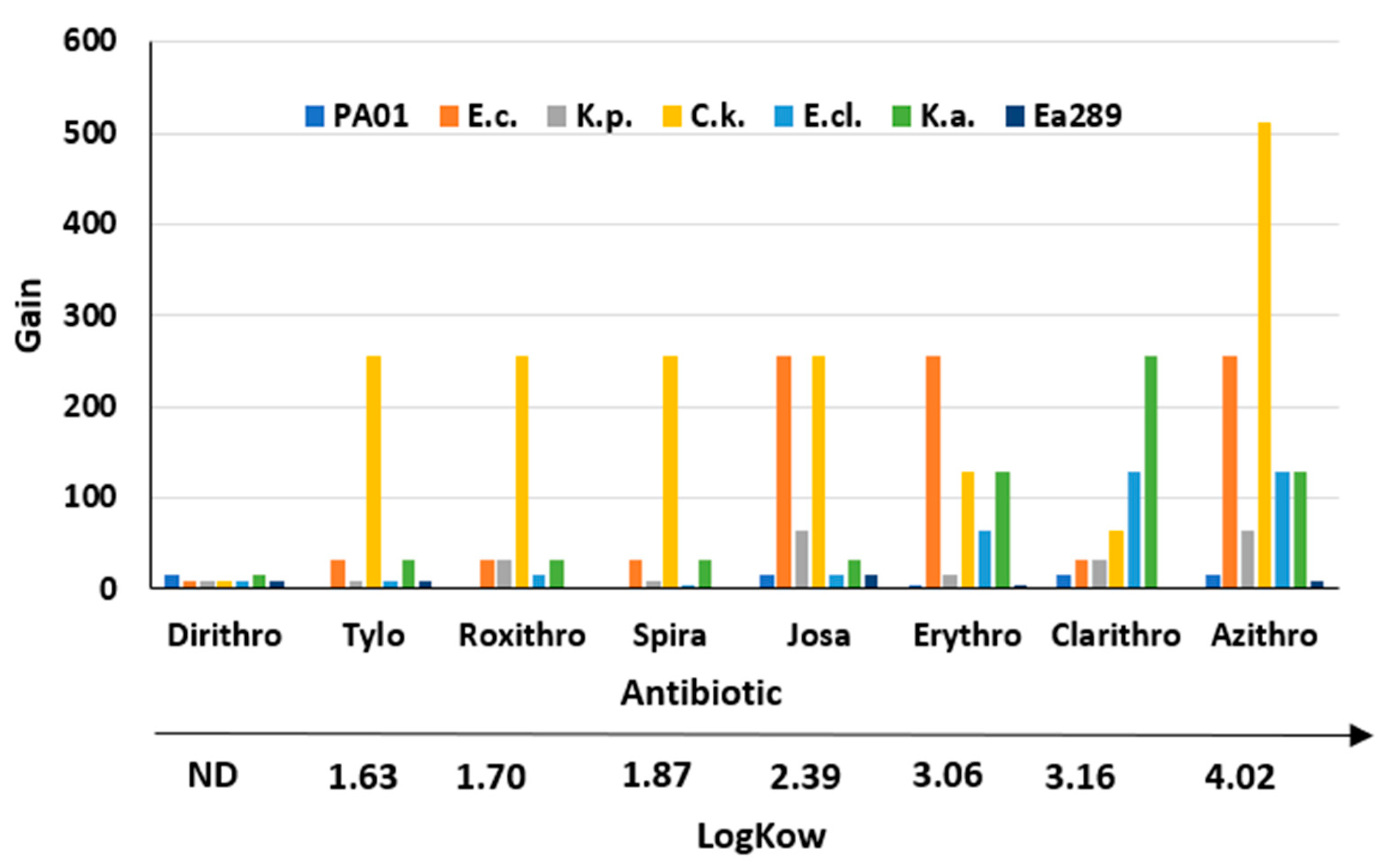
2.4. Restoration of Macrolides Activity against Various Gram-Negative Bacteria in Combination with Derivatives 3–6
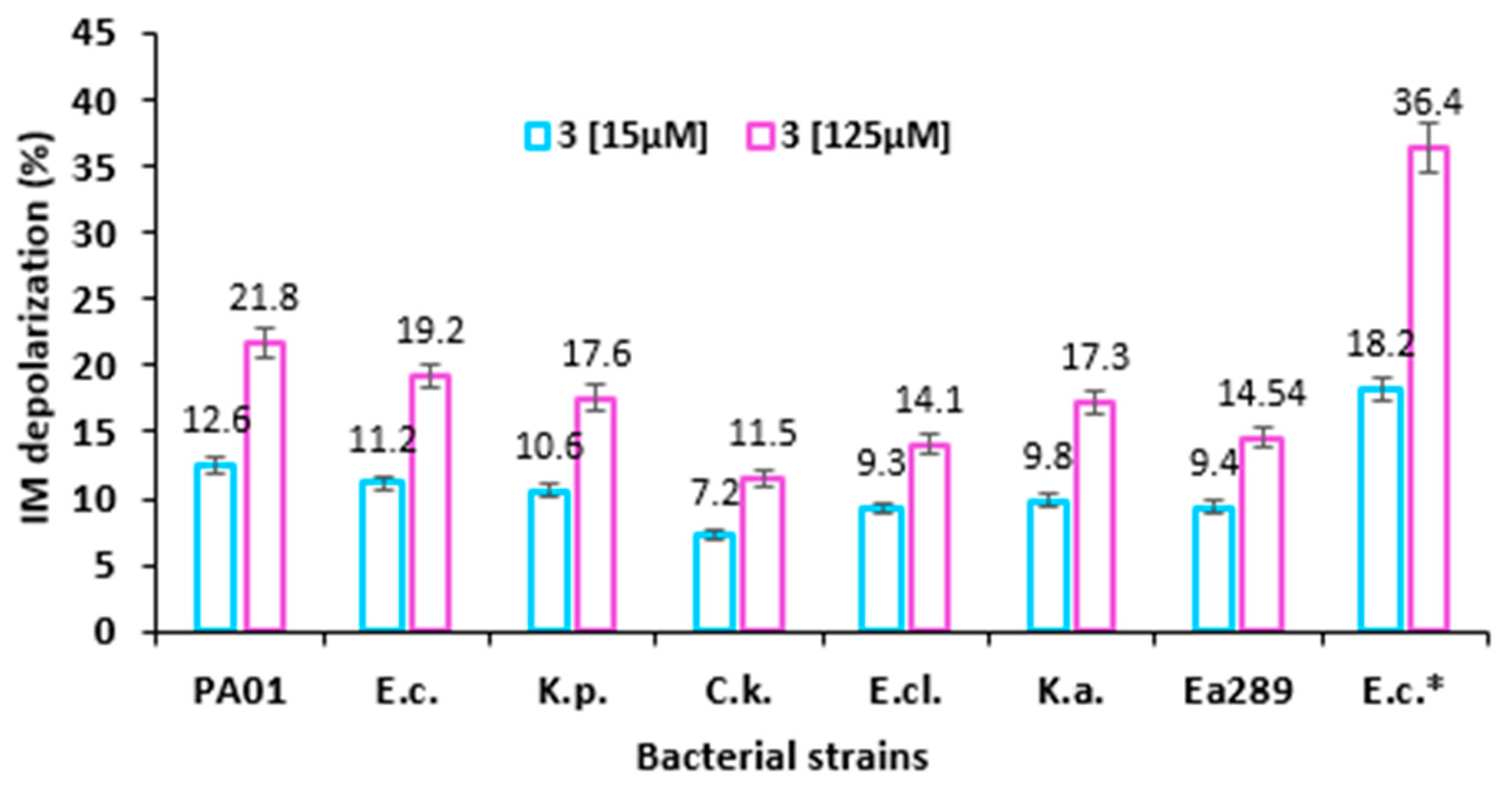
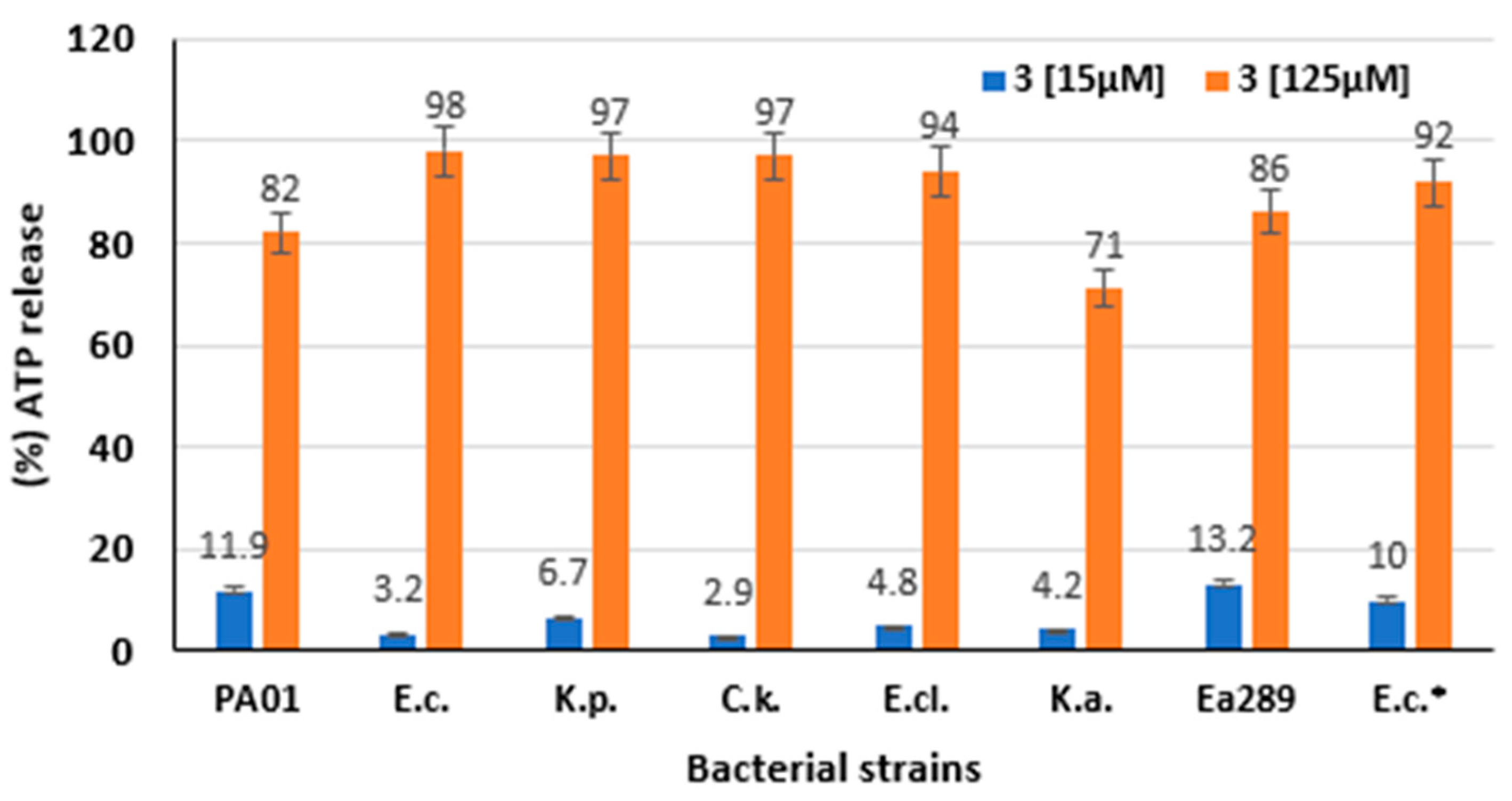
2.5. Mechanism of Action of Compound 3
Inner Membrane Depolarization Assay
2.6. ATP Efflux Measurement
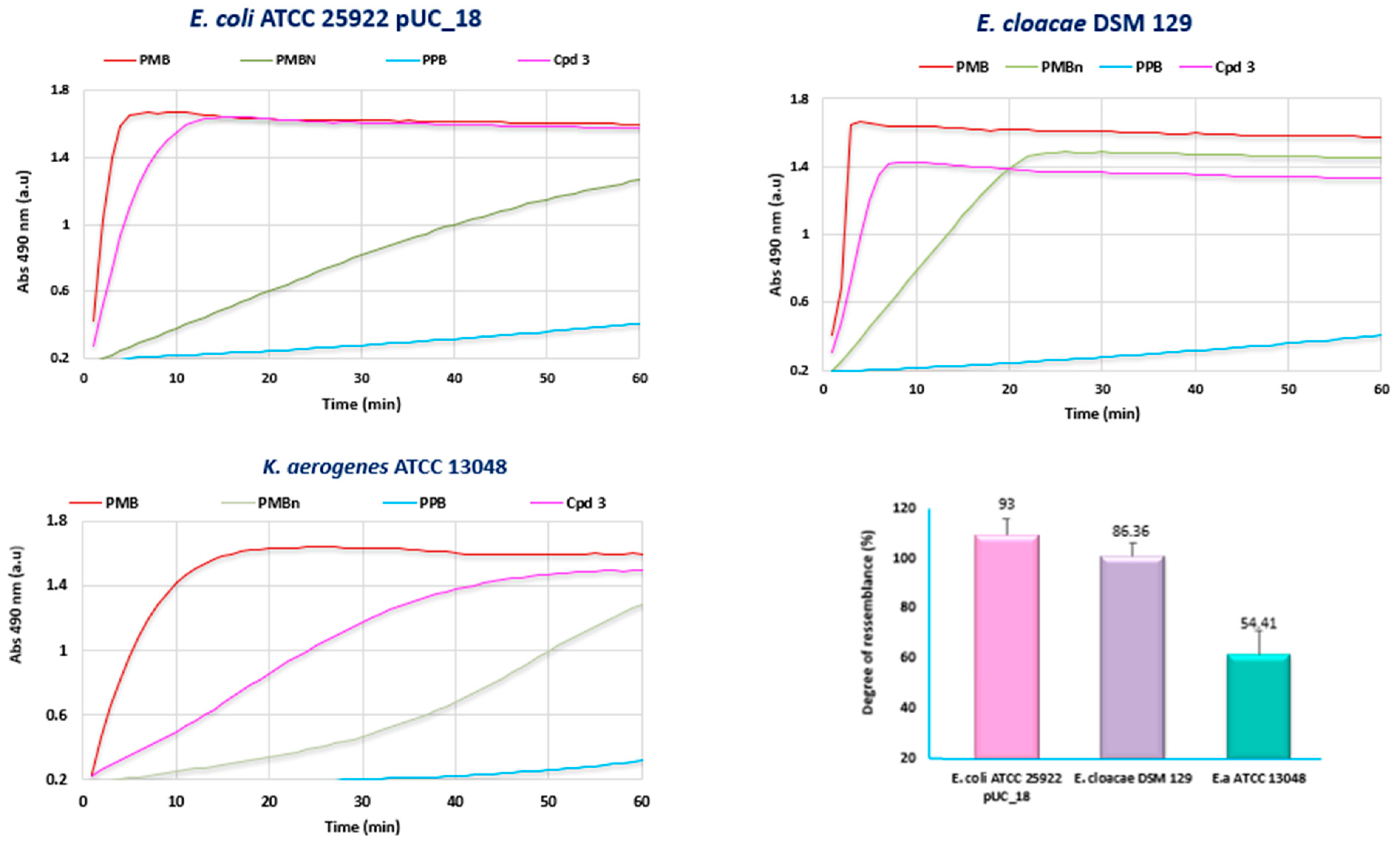
2.7. Outer Membrane Permeabilization
3. Discussion
4. Methods and Materials
4.1. Procedure for the Synthesis of Polyaminoisoprenyl Derivatives 3–6
Synthesis of Compound 3
4.2. Bacterial Strains
4.3. Transfer of Plasmid pUC-18 into Escherichia coli Strains
4.4. Antibiotics
4.5. MIC Determination of Macrolides and Polyaminoisoprenyl Derivatives
4.6. Determination of MICs of Macrolides in the Presence of Synergizing Compounds 3–6
4.7. Chequerboard Assay/Fractional Inhibitory Concentration Index (FICI)
4.8. Outer Membrane Permeabilization Assay
4.9. Membrane Depolarization Assay
4.10. ATP Efflux Measurement
4.11. Cytoxicity Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Livermore, D.M. The need for new antibiotics. Clin. Microbiol. Infect. 2004, 10 (Suppl. 4), 1–9. [Google Scholar] [CrossRef]
- Payne, D.J. Microbiology. Desperately seeking new antibiotics. Science 2008, 321, 1644–1645. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Nazir, M.; Ali, M.S.; Hussain, H.; Lee, Y.S.; Riaz, N.; Jabbar, A. Antimicrobial natural products: An update on future antibiotic drug candidates. Nat. Prod. Rep. 2010, 27, 238–254. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogen. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Vivas, R.; Barbosa, A.A.T.; Dolabela, S.S.; Jain, S. Multidrugresistant bacteria and alternative methods to control them: An overview. Microb. Drug Resist. 2019, 25, 890–908. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, G.; D’Inzeo, T.; Fiori, B.; Spanu, T.; Sganga, G. Burden of antibiotic resistant gram negative bacterial infections: Evidence and limits. J. Med. Microb. Diagn. 2014, 3, 1000132. [Google Scholar]
- Oduro-Mensah, D.; Obeng-Nkrumah, N.; Bonney, E.Y.; Oduro-Mensah, E.; Twum-Danso, K.; Osei, Y.D.; Sackey, S.T. Genetic characterization of TEM-type ESBL-associated antibacterial resistance in Enterobacteriaceae in a tertiary hospital in Ghana. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wagenlehner, F.M.E.; Dittmar, F. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Eur. Urol. 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Douafer, H.; Andrieu, V.; Phanstiel, O.; Brunel, J.M. Antibiotic Adjuvants: Make Antibiotics Great Again! J. Med. Chem. 2019, 62, 8665–8681. [Google Scholar] [CrossRef]
- Troudi, A.; Pages, J.M.; Brunel, J.M. Chemical Highlights Supporting the Role of Lipid A in Efficient Biological Adaptation of Gram-Negative Bacteria to External Stresses. J. Med. Chem. 2021, 64, 1816–1834. [Google Scholar] [CrossRef]
- Clifton, L.A.; Skoda, M.W.; Le Brun, A.P.; Ciesielski, F.; Kuzmenko, I.; Holt, S.A.; Lakey, J.H. Effect of divalent cation removal on the structure of Gram-negative bacterial outer membrane models. Langmuir 2015, 31, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Decad, G.M.; Nikaido, H. Outer membrane of Gram-negative bacteria. XII. Molecular-sieving function of cell wall. J. Bacteriol. 1976, 128, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Outer membrane of Salmonella typhimurium transmembrane diffusion of some hydrophobic substances. Biochim. Biophys. Acta Biomembr. 1976, 433, 118–132. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, R.; Moser, H.E. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J. Med. Chem. 2008, 51, 2871–2878. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Klobucar, K.; French, S.; Côté, J.P.; Howes, J.R.; Brown, E.D. Genetic and chemical-genetic interactions map biogenesis and permeability determinants of the outer membrane of Escherichia coli. mBio 2020, 11, e00161-20. [Google Scholar] [CrossRef] [PubMed]
- Barker, C.A.; Farha, M.A.; Brown, E.D. Chemical genomic approaches to study model microbes. Chem. Biol. 2010, 17, 624–632. [Google Scholar] [CrossRef][Green Version]
- Specht, K.M.; Shokat, K.M. The emerging power of chemical genetics. Curr. Opin. Cell Biol. 2002, 14, 155–159. [Google Scholar] [CrossRef]
- Repaske, R. Lysis of Gram-negative organisms and the role of versene. Biochim. Biophys. Acta 1958, 30, 225–232. [Google Scholar] [CrossRef]
- Vaara, M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992, 56, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Corbett, D.; Wise, A.; Langley, T.; Skinner, K.; Trimby, E.; Birchall, S.; Dorali, A.; Sandiford, S.; Williams, J.; Warn, P.; et al. Potentiation of antibiotic activity by a novel cationic peptide: Potency and spectrum of activity of SPR741. Antimicrob. Agents Chemother. 2017, 61, e00200-17. [Google Scholar] [CrossRef] [PubMed]
- French, S.; Farha, M.A.; Ellis, M.J.; Sameer, Z.; Côté, J.-P.; Cotroneo, N.; Lister, T.; Rubio, A.; Brown, E.D. Polymyxin B analog SPR741 potentiates antibiotics against Gram-negative bacteria and uniquely perturbs the outer membrane. ACS Infect. Dis. 2020, 6, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Schindler, M.; Osborn, M.J. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry 1979, 18, 4425–4430. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M.; Viljanen, P. Binding of polymyxin B nonapeptide to Gram-negative bacteria. Antimicrob. Agents Chemother. 1985, 27, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.; Pais, J.P.; Nunes, R.; Blázquez-Sánchez, M.T.; Marquês, J.T.; Almeida, A.F.; Serra, P.; Xavier, N.M.; Vila-Viçosa, D.; Machuqueiro, M.; et al. Sugar-based bactericides targeting phosphatidylethanolamine-enriched membranes. Nat. Commun. 2018, 19, 4857. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Laslop, N.; Mankin, A.S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef]
- Gomes, C.; Martínez-Puchol, S.; Palma, N.; Horna, G.; Ruiz-Roldán, L.; Pons, M.J.; Ruiz, J. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit. Rev. Microbiol. 2017, 43, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Arsic, B.; Barber, J.; Čikoš, A.; Mladenovic, M.; Stankovic, N.; Novak, P. 16-membered macrolide antibiotics: A review. Int. J. Antimicrob. Agents 2018, 51, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Borselli, D.; Lieutaud, A.; Thefenne, H.; Garnotel, E.; Pagès, J.-M.; Brunel, J.M.; Bolla, J.-M. Polyamino-isoprenic derivatives block intrinsic resistance of P. aeruginosa to doxycycline and chloramphenicol in vitro. PLoS ONE 2016, 11, e0154490. [Google Scholar] [CrossRef] [PubMed]
- Lieutaud, A.; Pieri, C.; Bolla, J.M.; Brunel, J.M. New Polyaminoisoprenyl Antibiotics Enhancers against Two Multidrug-Resistant Gram-Negative Bacteria from Enterobacter and Salmonella Species. J. Med. Chem. 2020, 63, 10496–10508. [Google Scholar] [CrossRef] [PubMed]
- Troudi, A.; Fethi, M.; El Asli, M.S.; Bolla, J.M.; Klibi, N.; Brunel, J.M. Efficiency of a tetracycline-adjuvant combination against multidrug resistant Pseudomonas aeruginosa Tunisian clinical isolates. Antibiotics 2020, 9, 919. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Brunel, J.-M.; Bolla, J.-M.; Van Bambeke, F. The polyaminoisoprenyl potentiator NV716 revives old disused antibiotics against intracellular forms of infection by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2021, 65, e02028. [Google Scholar] [CrossRef] [PubMed]
- Bisacchi, G.S.; Manchester, J.I. A New-Class Antibacterial—Almost. Lessons in Drug Discovery and Development: A Critical Analysis of More than 50 Years of Effort toward ATPase Inhibitors of DNA Gyrase and Topoisomerase IV. ACS Infect. Dis. 2015, 1, 4–41. [Google Scholar] [CrossRef]
- te Winkel, J.D.; Gray, D.A.; Seistrup, K.H.; Hamoen, L.W.; Strahl, H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front. Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Borselli, D.; Blanchet, M.; Bolla, J.-M.; Muth, A.; Skruber, K.; Phanstiel, O.I.V.; Brunel, J.M. Motuporamine Derivatives as Antimicrobial Agents and Antibiotic Enhancers against Resistant Gram-Negative Bacteria. Chembiochem 2017, 18, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Naseem, R.; Wann, K.T.; Holland, I.B.; Campbell, A.K. ATP regulates calcium efflux and growth in E. coli. J. Mol. Biol. 2009, 391, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Committee, S.A. Comité de l’Antibiogramme de la Société Française de Microbiologie report 2003. Int. J. Antimicrob. Agents 2003, 21, 364–391. [Google Scholar]

| RNH2 | Cpd-(Isolated Yield (%)) | IC50 (µM) CHO | ||
|---|---|---|---|---|
| 3–4 | 5–6 | |||
 | 3 (72) | 5 (64) | 142.79 | >150 |
 | 4 (49) | 6 (63) | >150 | 126.82 |
| MIC (µM) (µg/mL) | ||||
|---|---|---|---|---|
| Strains | Cpd 3 | Cpd 4 | Cpd 5 | Cpd 6 |
| P. aeruginosa PA01 | 25 (10) | 200 (78) | >400 (>135) | >400 (>130) |
| E. coli ATCC 25922 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| K. pneumoniae ATCC 13883 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| C. koseri IP8294 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| E. cloacae DSM 129 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| K. aerogenes ATCC 13048 | 100 (40) | >400 (>156) | >400 (>135) | >400 (>130) |
| K. aerogenes 289 | 100 (40) | >400 (>156) | >400 (>135) | >400 (>130) |
| AG100A_pUC18 | 12.5 (5) | 50 (19) | 100 (33) | >200 (>65) |
| MIC (µg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strains | Erythro | Josa | Azithro | Roxithro | Spira | Clarithro | Dirithro | Tylo |
| PA01 | 512 | >1024 | 128 | >1024 | >1024 | 512 | 1024 | >1024 |
| E. coli ATCC 25922 | 128 | 1024 | 8 | 512 | 512 | 128 | 64 | 512 |
| K. pneumoniae ATCC 13883 | 128 | 1024 | 16 | 512 | 512 | 128 | 64 | 1024 |
| C. koseri IP8294 | 256 | 1024 | 16 | 1024 | 1024 | 128 | 32 | 1024 |
| E. cloacae DSM 129 | >1024 | >1024 | 64 | >1024 | >1024 | 1024 | 128 | >1024 |
| K. aerogenes ATCC 13048 | 512 | 512 | 64 | 512 | 1024 | 256 | 64 | 1024 |
| K. aerogenes 289 | >1024 | >1024 | 64 | >1024 | 1024 | >1024 | 1024 | >1024 |
| AG100A_pUC18 | 8 | 32 | 2 | 32 | 128 | 64 | 16 | 256 |
| Antibiotic | Cpd | PA01 | E.c. a | K.p. b | C.k. c | E.cl. d | K.a. e | Ea289 f | E.c. g |
|---|---|---|---|---|---|---|---|---|---|
| Erythromycine | 3 | 128 | 0.5 | 8 | 2 | 16 | 4 | 256 | <0.0005 |
| 4 | 256 | 4 | 16 | 32 | 64 | 32 | 512 | 0.031 | |
| 5 | 512 | 64 | 32 | 64 | 256 | 128 | >1024 | 4 | |
| 6 | 512 | 128 | 64 | 256 | 512 | 128 | >1024 | 2 | |
| Josamycine | 3 | 64 | 4 | 16 | 4 | 64 | 16 | 64 | 0.0019 |
| 4 | 512 | 16 | 32 | 64 | 128 | 64 | 128 | 1 | |
| 5 | 1024 | 128 | 64 | 256 | 256 | 128 | 512 | 8 | |
| 6 | >1024 | 512 | 128 | 512 | 1024 | 256 | 1024 | 16 | |
| Azithromycine | 3 | 8 | 0.031 | 0.25 | 0.031 | 0.5 | 0.5 | 8 | <0.0005 |
| 4 | 32 | 1 | 0.5 | 1 | 4 | 1 | 16 | 0.25 | |
| 5 | 64 | 4 | 4 | 4 | 16 | 4 | 32 | 1 | |
| 6 | 64 | 4 | 4 | 8 | 32 | 4 | 64 | 2 | |
| Roxithromycine | 3 | 512 | 16 | 16 | 4 | 64 | 16 | 512 | <0.0005 |
| 4 | 1024 | 16 | 64 | 32 | 256 | 64 | 1024 | 4 | |
| 5 | 1024 | 128 | 128 | 256 | 512 | 256 | >1024 | 16 | |
| 6 | 1024 | 512 | 256 | 256 | 1024 | 256 | >1024 | 16 | |
| Spiramycine | 3 | >1024 | 16 | 64 | 4 | 256 | 32 | 512 | <0.0005 |
| 4 | >1024 | 64 | 128 | 256 | 512 | 128 | 256 | 1 | |
| 5 | 1024 | 256 | 128 | 512 | 1024 | 512 | 512 | 1 | |
| 6 | >1024 | 256 | 256 | 1024 | 1024 | 1024 | 512 | 1 | |
| clarithromycine | 3 | 32 | 4 | 4 | 2 | 8 | 1 | 512 | 0.0039 |
| 4 | 128 | 8 | 8 | 4 | 64 | 8 | 1024 | 16 | |
| 5 | 128 | 16 | 32 | 32 | 256 | 64 | 1024 | 16 | |
| 6 | 128 | 32 | 32 | 64 | 512 | 64 | 1024 | 32 | |
| Dirithromycine | 3 | 64 | 8 | 8 | 4 | 16 | 4 | 128 | <0.0005 |
| 4 | 512 | 16 | 16 | 8 | 64 | 8 | 256 | 2 | |
| 5 | 512 | 16 | 16 | 16 | 64 | 32 | 512 | 4 | |
| 6 | 512 | 32 | 16 | 16 | 128 | 32 | 512 | 8 | |
| Tylosine | 3 | 512 | 16 | 128 | 4 | 128 | 32 | 128 | 4 |
| 4 | >1024 | 256 | 256 | 128 | 1024 | 256 | 256 | 32 | |
| 5 | >1024 | 512 | 512 | 512 | >1024 | 512 | 512 | 64 | |
| 6 | >1024 | 512 | 512 | 512 | >1024 | 512 | 1024 | 128 |
| Strain | FICI | |||||||
|---|---|---|---|---|---|---|---|---|
| Erythro | Josa | Azithro | Roxithro | Spira | Clarithro | Dirithro | Tylo | |
| Pa01 | 0.65 | 0.46 | 0.46 | 0.90 | 1.40 | 0.46 | 0.46 | 0.90 |
| E.c. a | 0.20 | 0.20 | 0.20 | 0.20 | 0.23 | 0.23 | 0.33 | 0.23 |
| K.p. b | 0.26 | 0.22 | 0.22 | 0.23 | 0.33 | 0.23 | 0.33 | 0.33 |
| C.k. c | 0.21 | 0.20 | 0.20 | 0.22 | 0.20 | 0.22 | 0.33 | 0.20 |
| E.cl. d | 0.22 | 0.26 | 0.21 | 0.26 | 0.45 | 0.21 | 0.33 | 0.33 |
| K.a. e | 0.11 | 0.13 | 0.11 | 0.13 | 0.13 | 0.10 | 0.16 | 0.13 |
| Ea289 f | 0.35 | 0.16 | 0.23 | 0.60 | 0.60 | 0.60 | 0.23 | 0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troudi, A.; Bolla, J.M.; Klibi, N.; Brunel, J.M. An Original and Efficient Antibiotic Adjuvant Strategy to Enhance the Activity of Macrolide Antibiotics against Gram-Negative Resistant Strains. Int. J. Mol. Sci. 2022, 23, 12457. https://doi.org/10.3390/ijms232012457
Troudi A, Bolla JM, Klibi N, Brunel JM. An Original and Efficient Antibiotic Adjuvant Strategy to Enhance the Activity of Macrolide Antibiotics against Gram-Negative Resistant Strains. International Journal of Molecular Sciences. 2022; 23(20):12457. https://doi.org/10.3390/ijms232012457
Chicago/Turabian StyleTroudi, Azza, Jean Michel Bolla, Naouel Klibi, and Jean Michel Brunel. 2022. "An Original and Efficient Antibiotic Adjuvant Strategy to Enhance the Activity of Macrolide Antibiotics against Gram-Negative Resistant Strains" International Journal of Molecular Sciences 23, no. 20: 12457. https://doi.org/10.3390/ijms232012457
APA StyleTroudi, A., Bolla, J. M., Klibi, N., & Brunel, J. M. (2022). An Original and Efficient Antibiotic Adjuvant Strategy to Enhance the Activity of Macrolide Antibiotics against Gram-Negative Resistant Strains. International Journal of Molecular Sciences, 23(20), 12457. https://doi.org/10.3390/ijms232012457






