Aphid BCR4 Structure and Activity Uncover a New Defensin Peptide Superfamily
Abstract
1. Introduction
2. Results
2.1. Total Synthesis of BCR4
2.2. Antimicrobial Activities of BCR4
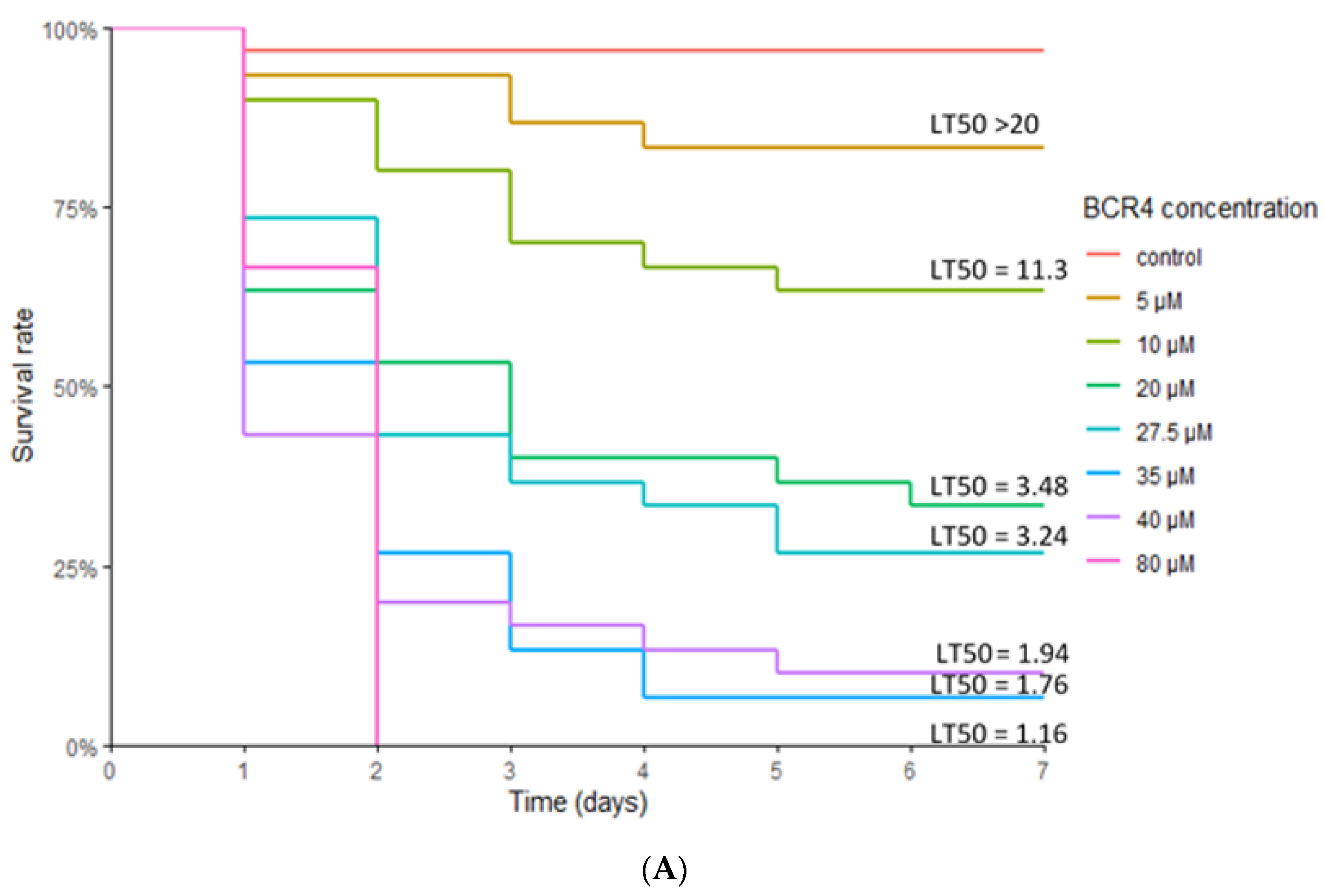
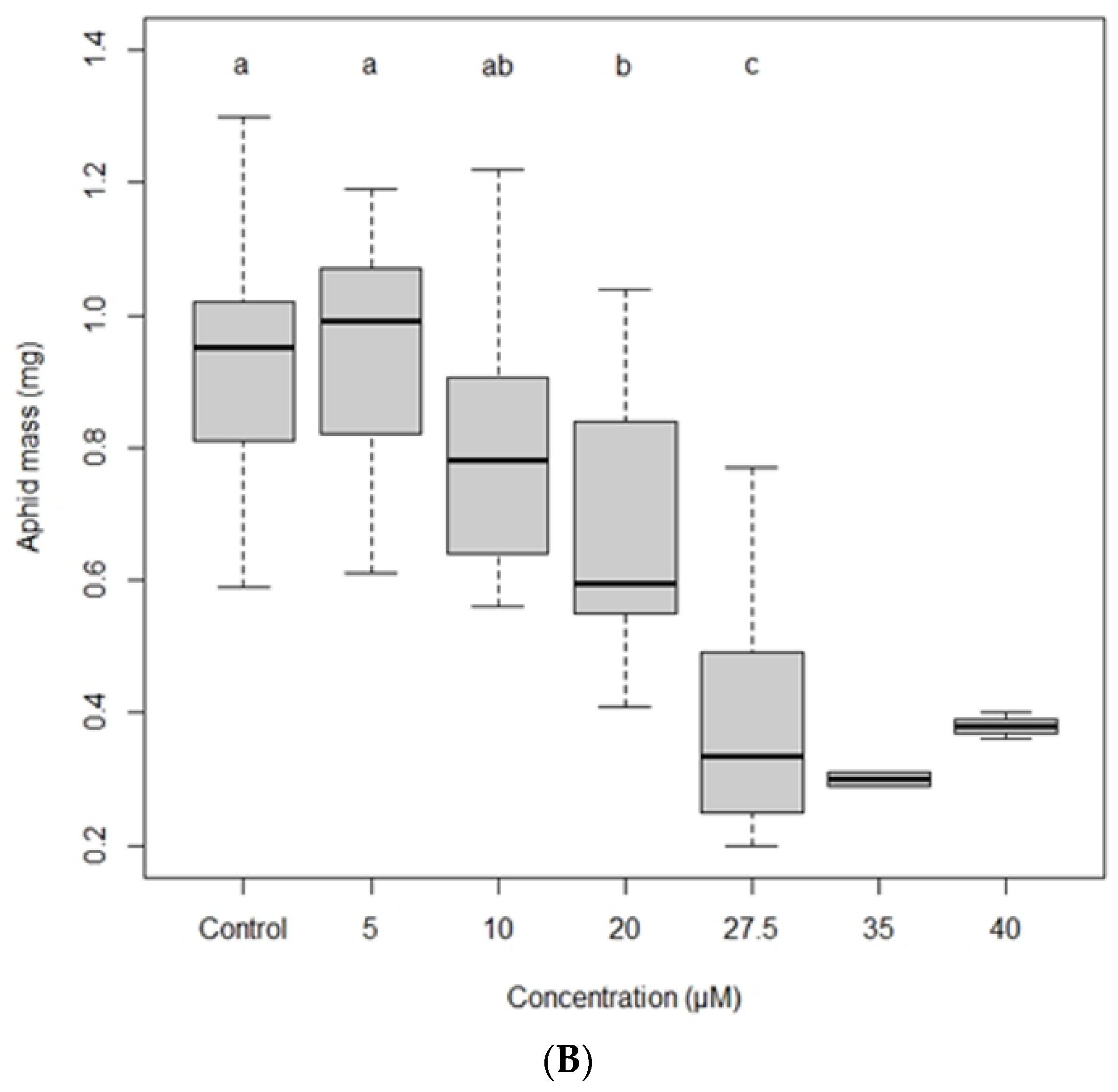
2.3. Insect Bioassays
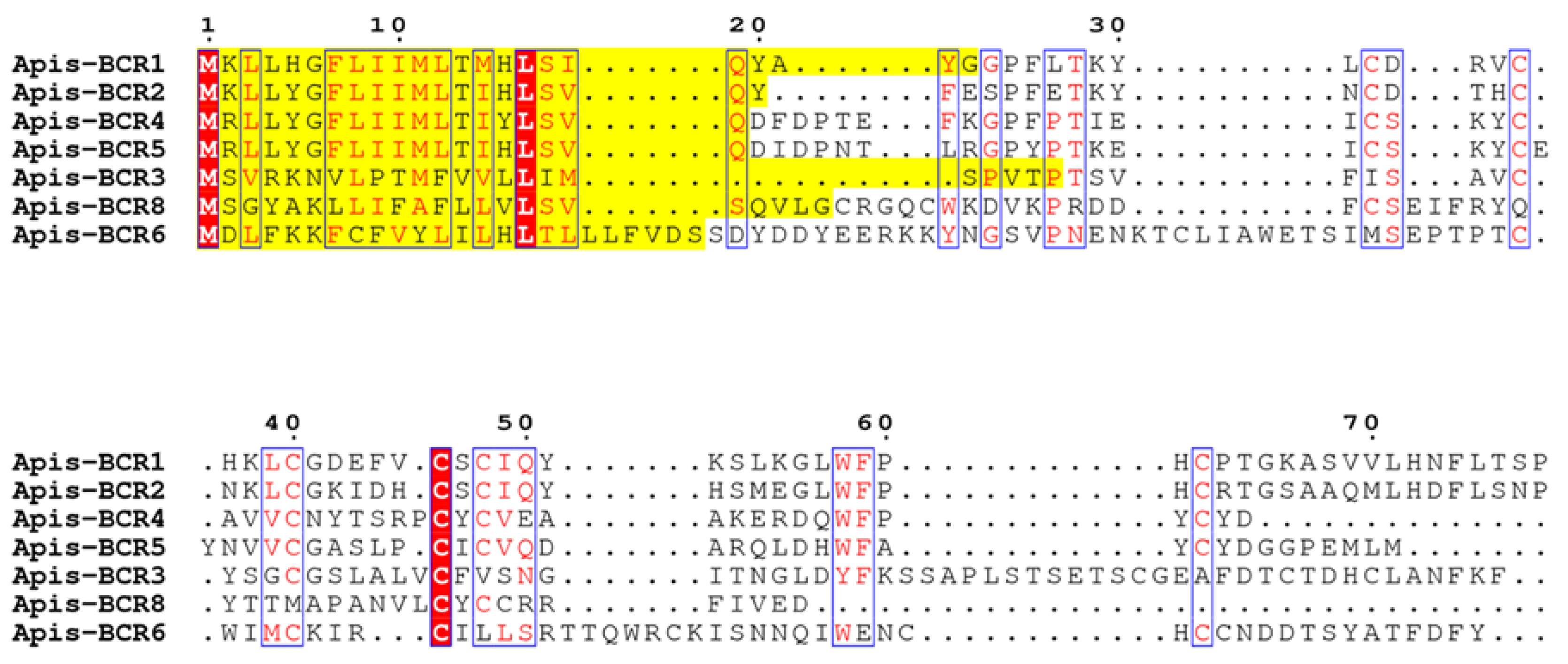
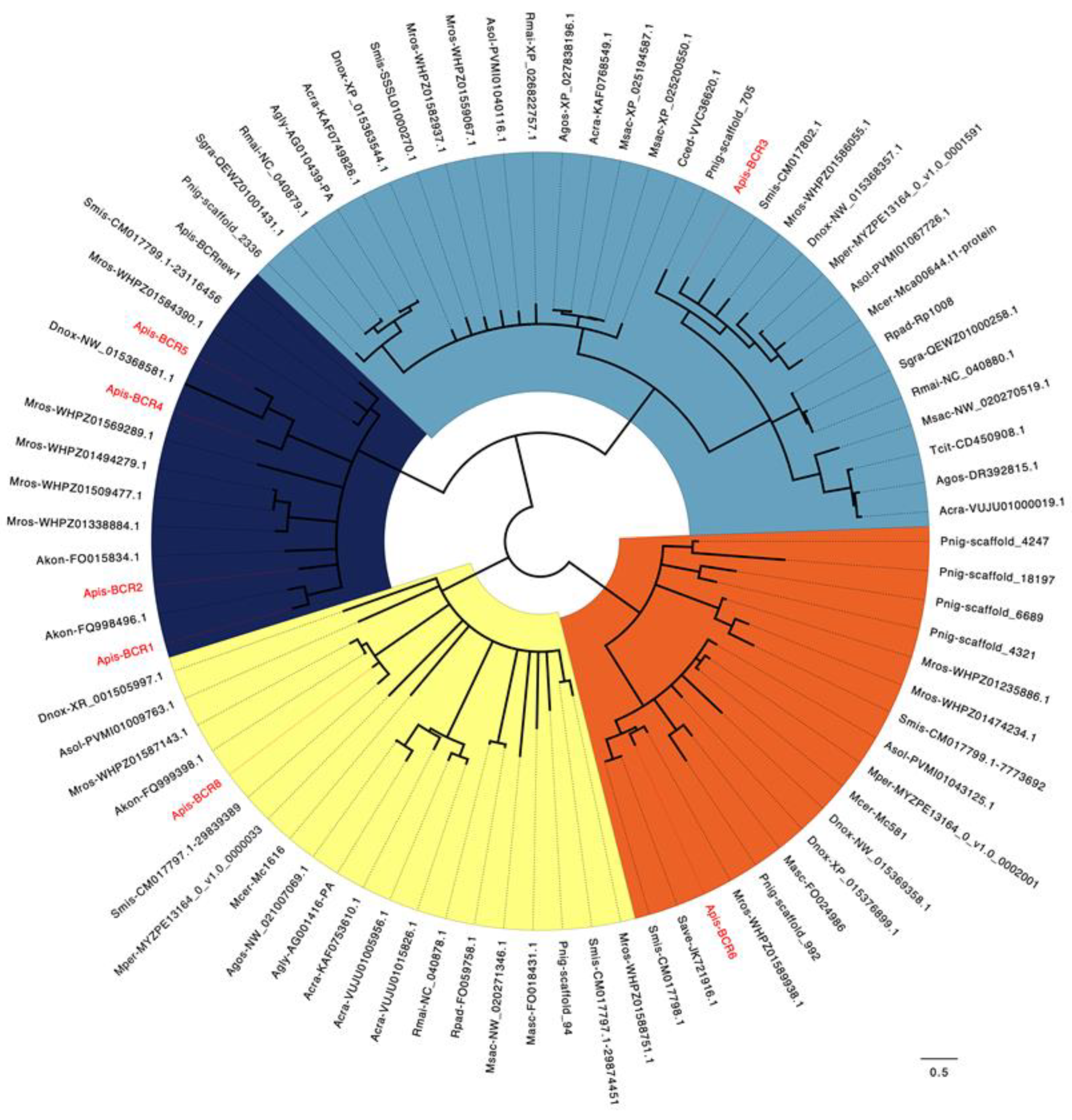
2.4. Phylogeny of the BCR Family
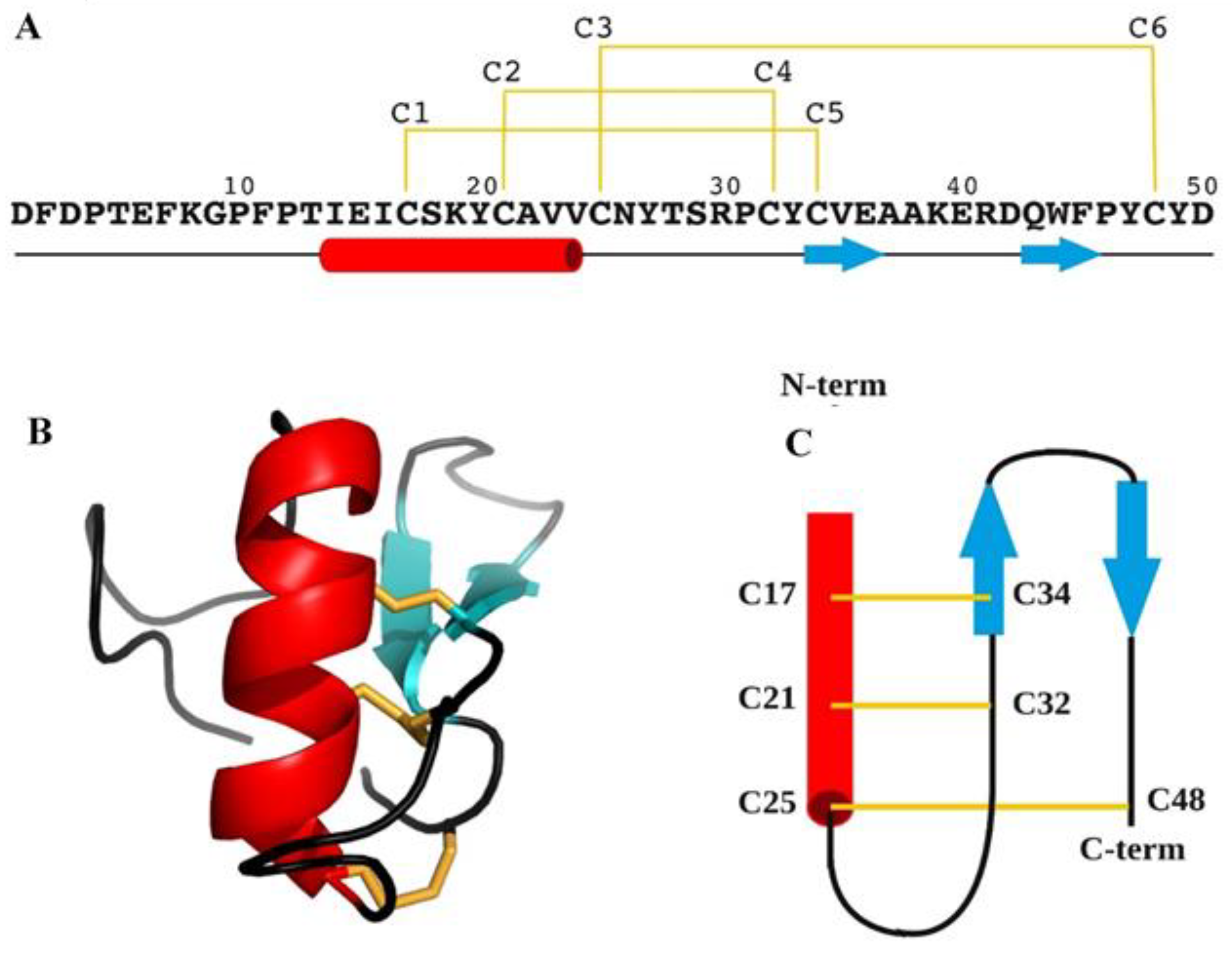
2.5. BCR4 Solution Structure
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. Antimicrobial Assays
4.3. Insects and Insect Assays
4.4. Sequence Analysis and Phylogeny
4.5. NMR Experiments
4.6. Structure Calculations
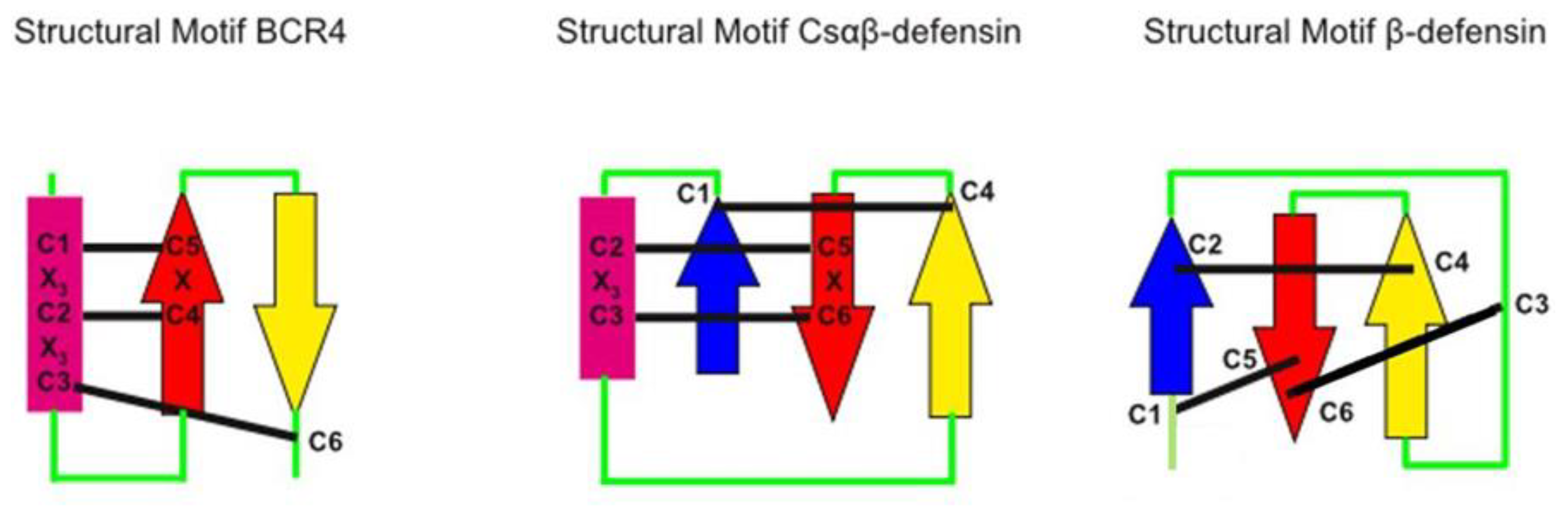
4.7. Structure Relationship of BCR4 Structure
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bradshaw, C.J.A.; Leroy, B.; Bellard, C.; Roiz, D.; Albert, C.; Fournier, A.; Barbet-Massin, M.; Salles, J.-M.; Simard, F.; Courchamp, F. Massive yet grossly underestimated global costs of invasive insects. Nat. Commun. 2016, 7, 12986. [Google Scholar] [CrossRef] [PubMed]
- Diagne, C.; Leroy, B.; Gozlan, R.E.; Vaissiere, A.C.; Assailly, C.; Nuninger, L.; Roiz, D.; Jourdain, F.; Jaric, I.; Courchamp, F. InvaCost, a public database of the economic costs of biological invasions worldwide. Sci. Data 2020, 7, 277. [Google Scholar] [CrossRef] [PubMed]
- Oerke, E.C.; Dehne, H.W. Safeguarding production—Losses in major crops and the role of crop protection. Crop Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- Calevro, F.; Tagu, D.; Callaerts, P. Acyrthosiphon pisum. Trends Genet. TIG 2019, 35, 781–782. [Google Scholar] [CrossRef]
- Dedryver, C.A.; Le Ralec, A.; Fabre, F. The conflicting relationships between aphids and men: A review of aphid damage and control strategies. Comptes. Rendus. Biol. 2010, 333, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D. Call to restrict neonicotinoids. Science 2018, 360, 973. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Du, Q.; Craik, D.J. Cyclotides: Disulfide-Rich peptide toxins in plants. Toxicon 2019, 172, 33–44. [Google Scholar] [CrossRef] [PubMed]
- King, G.F. Tying pest insects in knots: The deployment of spider-venom-derived knottins as bioinsecticides. Pest. Manag. Sci. 2019, 75, 2437–2445. [Google Scholar] [CrossRef] [PubMed]
- Gressent, F.; Da Silva, P.; Eyraud, V.; Karaki, L.; Royer, C. Pea Albumin 1 subunit b (PA1b), a promising bioinsecticide of plant origin. Toxins 2011, 3, 1502–1517. [Google Scholar] [CrossRef]
- Shafee, T.M.; Lay, F.T.; Phan, T.K.; Anderson, M.A.; Hulett, M.D. Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci. 2017, 74, 663–682. [Google Scholar] [CrossRef]
- Gonzalez-Castro, R.; Gomez-Lim, M.A.; Plisson, F. Cysteine-Rich Peptides: Hyperstable Scaffolds for Protein Engineering. Chembiochem. A Eur. J. Chem. Biol. 2021, 22, 961–973. [Google Scholar] [CrossRef]
- Rahioui, I.; Eyraud, V.; Karaki, L.; Sasse, F.; Carre-Pierrat, M.; Qin, A.; Zheng, M.H.; Toepfer, S.; Sivignon, C.; Royer, C.; et al. Host range of the potential biopesticide Pea Albumin 1b (PA1b) is limited to insects. Toxicon 2014, 89, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.; Sukiran, N.A.; Walsh, S.; Fitches, E.C. The insecticidal activity of recombinant nemertide toxin alpha-1 from Lineus longissimus towards pests and beneficial species. Toxicon 2021, 197, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Delobel, B.; Grenier, A.M.; Gueguen, J.; Ferrasson, E.; Mbaiguinam, M. Utilisation d’un polypeptide dérivé d’une albumine PA1b de légumineuse comme insecticide (Brevet-98/05877). French Patent 1998, 98, 11. [Google Scholar]
- Shigenobu, S.; Stern, D.L. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc. Biol. Sci. 2013, 280, 20121952. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp APS. Nature 2000, 407, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Brinza, L.; Viñuelas, J.; Cottret, L.; Calevro, F.; Rahbé, Y.; Febvay, G.; Duport, G.; Colella, S.; Rabatel, A.; Gautier, C.; et al. Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. Comptes. Rendus. Biol. 2009, 332, 1034–1049. [Google Scholar] [CrossRef] [PubMed]
- Simonet, P.; Gaget, K.; Balmand, S.; Ribeiro Lopes, M.; Parisot, N.; Buhler, K.; Duport, G.; Vulsteke, V.; Febvay, G.; Heddi, A.; et al. Bacteriocyte cell death in the pea aphid/Buchnera symbiotic system. Proc. Natl. Acad. Sci. USA 2018, 115, E1819–E1828. [Google Scholar] [CrossRef] [PubMed]
- Login, F.H.; Balmand, S.; Vallier, A.; Vincent-Monegat, C.; Vigneron, A.; Weiss-Gayet, M.; Rochat, D.; Heddi, A. Antimicrobial peptides keep insect endosymbionts under control. Science 2011, 334, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Uchi, N.; Fukudome, M.; Nozaki, N.; Suzuki, M.; Osuki, K.I.; Shigenobu, S.; Uchiumi, T. Antimicrobial Activities of Cysteine-rich Peptides Specific to Bacteriocytes of the Pea Aphid Acyrthosiphon pisum. Microbes Environ. 2019, 34, 155–160. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Shafee, T.M.; Lay, F.T.; Hulett, M.D.; Anderson, M.A. The Defensins Consist of Two Independent, Convergent Protein Superfamilies. Mol. Biol. Evol. 2016, 33, 2345–2356. [Google Scholar] [CrossRef]
- Huygens, C.; Ribeiro Lopes, M.; Gaget, K.; Duport, G.; Peignier, S.; De Groef, S.; Parisot, N.; Calevro, F.; Callaerts, P. Evolutionary diversification of insulin-related peptides (IRPs) in aphids and spatiotemporal distribution in Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 2022, 141, 103670. [Google Scholar] [CrossRef]
- Ribeiro Lopes, M.; Parisot, N.; Gaget, K.; Huygens, C.; Peignier, S.; Duport, G.; Orlans, J.; Charles, H.; Baatsen, P.; Jousselin, E.; et al. Evolutionary novelty in the apoptotic pathway of aphids. Proc. Natl. Acad. Sci. USA 2020, 117, 32545–32556. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Lelievre, D.; Terrier, V.P.; Delmas, A.F.; Aucagne, V. Native Chemical Ligation Strategy to Overcome Side Reactions during Fmoc-Based Synthesis of C-Terminal Cysteine-Containing Peptides. Org. Lett. 2016, 18, 920–923. [Google Scholar] [CrossRef] [PubMed]
- Terrier, V.P.; Adihou, H.; Arnould, M.; Delmas, A.F.; Aucagne, V. A straightforward method for automated Fmoc-based synthesis of bio-inspired peptide crypto-thioesters. Chem. Sci. 2016, 7, 339–345. [Google Scholar] [CrossRef]
- Terrier, V.P.; Delmas, A.F.; Aucagne, V. Efficient synthesis of cysteine-rich cyclic peptides through intramolecular native chemical ligation of N-Hnb-Cys peptide crypto-thioesters. Org. Biomol. Chem. 2017, 15, 316–319. [Google Scholar] [CrossRef]
- Abboud, S.A.; Amoura, M.; Madinier, J.B.; Renoux, B.; Papot, S.; Piller, V.; Aucagne, V. Enzyme-Cleavable Linkers for Protein Chemical Synthesis through Solid-Phase Ligations. Angew. Chem. 2021, 60, 18612–18618. [Google Scholar] [CrossRef]
- Abboud, S.A.; Aucagne, V. An optimized protocol for the synthesis of N-2-hydroxybenzyl-cysteine peptide crypto-thioesters. Org. Biomol. Chem. 2020, 18, 8199–8208. [Google Scholar] [CrossRef] [PubMed]
- Abboud, S.A.; Cisse, E.H.; Doudeau, M.; Benedetti, H.; Aucagne, V. A straightforward methodology to overcome solubility challenges for N-terminal cysteinyl peptide segments used in native chemical ligation. Chem. Sci. 2021, 12, 3194–3201. [Google Scholar] [CrossRef] [PubMed]
- Derache, C.; Meudal, H.; Aucagne, V.; Mark, K.J.; Cadene, M.; Delmas, A.F.; Lalmanach, A.C.; Landon, C. Initial insights into structure-activity relationships of avian defensins. J. Biol. Chem. 2012, 287, 7746–7755. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Hograindleur, J.P.; Voisin, S.; Abi Nahed, R.; Abd El Aziz, T.M.; Escoffier, J.; Bessonnat, J.; Fovet, C.M.; De Waard, M.; Hennebicq, S.; et al. Spermaurin, an La1-like peptide from the venom of the scorpion Scorpio Maurus palmatus, improves sperm motility and fertilization in different mammalian species. Mol. Hum. Reprod. 2016, 23, 116–131. [Google Scholar] [CrossRef]
- Legeai, F.; Shigenobu, S.; Gauthier, J.P.; Colbourne, J.; Rispe, C.; Collin, O.; Richards, S.; Wilson, A.C.; Murphy, T.; Tagu, D. AphidBase: A centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol. Biol. 2010, 19, 5–12. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 390–401. [Google Scholar] [CrossRef]
- Hodcroft, E. TreeCollapseCL 4. Available online: http://emmahodcroft.com/treecollapsecl.html (accessed on 22 July 2022).
- Schanda, P.; Kupce, E.; Brutscher, B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 2005, 33, 199–211. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Holm, L. Dali server: Structural unification of protein families. Nucleic Acids Res. 2022, 50, W210–W215. [Google Scholar] [CrossRef]
- M’Barek, S.; Chagot, B.; Andreotti, N.; Visan, V.; Mansuelle, P.; Grissmer, S.; Marrakchi, M.; El Ayeb, M.; Sampieri, F.; Darbon, H.; et al. Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity. Proteins 2005, 60, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Gressent, F.; Duport, G.; Rahioui, I.; Pauchet, Y.; Bolland, P.; Specty, O.; Rahbe, Y. Biological activity and binding site characteristics of the PA1b entomotoxin on insects from different orders. J. Insect Sci. 2007, 7, 12. [Google Scholar] [CrossRef]
- Login, F.H.; Heddi, A. Insect immune system maintains long-term resident bacteria through a local response. J. Insect Physiol. 2013, 59, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, W.; Zehirov, G.; Szatmari, A.; Debreczeny, M.; Ishihara, H.; Kevei, Z.; Farkas, A.; Mikulass, K.; Nagy, A.; Tiricz, H.; et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 2010, 327, 1122–1126. [Google Scholar] [CrossRef]
- Carro, L.; Pujic, P.; Alloisio, N.; Fournier, P.; Boubakri, H.; Hay, A.E.; Poly, F.; Francois, P.; Hocher, V.; Mergaert, P.; et al. Alnus peptides modify membrane porosity and induce the release of nitrogen-rich metabolites from nitrogen-fixing Frankia. ISME J. 2015, 9, 1723–1733. [Google Scholar] [CrossRef]
- Mergaert, P.; Kikuchi, Y.; Shigenobu, S.; Nowack, E.C.M. Metabolic Integration of Bacterial Endosymbionts through Antimicrobial Peptides. Trends Microbiol. 2017, 25, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.; Strzepa, A.; Jouvensal, L.; Rahioui, I.; Gressent, F.; Delmas, A.F. A folded and functional synthetic PA1b, an interlocked entomotoxic miniprotein. Biopolymers 2009, 92, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Febvay, G.; Delobel, B.; Rahbe, Y. Influence of the Amino-Acid Balance on the Improvement of an Artificial Diet for a Biotype of Acyrthosiphon-Pisum (Homoptera, Aphididae). Can. J. Zool. 1988, 66, 2449–2453. [Google Scholar] [CrossRef]
- Simonet, P.; Gaget, K.; Parisot, N.; Duport, G.; Rey, M.; Febvay, G.; Charles, H.; Callaerts, P.; Colella, S.; Calevro, F. Disruption of phenylalanine hydroxylase reduces adult lifespan and fecundity, and impairs embryonic development in parthenogenetic pea aphids. Sci. Rep. 2016, 6, 34321. [Google Scholar] [CrossRef]
- Loth, K.; Costechareyre, D.; Effantin, G.; Rahbe, Y.; Condemine, G.; Landon, C.; da Silva, P. New Cyt-like delta-endotoxins from Dickeya dadantii: Structure and aphicidal activity. Sci. Rep. 2015, 5, 8791. [Google Scholar] [CrossRef]
- Rahbe, Y.; Febvay, G. Protein Toxicity to Aphids—An Invitro Test on Acyrthosiphon-Pisum. Entomol. Exp. Appl. 1993, 67, 149–160. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Schaffer, A.A.; Agarwala, R.; Altschul, S.F.; Lipman, D.J.; Madden, T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012, 7, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 19854763. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, P.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Brunger, A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007, 2, 2728–2733. [Google Scholar] [CrossRef]
- Brunger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. D 1998, 54, 905–921. [Google Scholar] [CrossRef]
- Rieping, W.; Habeck, M.; Bardiaux, B.; Bernard, A.; Malliavin, T.E.; Nilges, M. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 2007, 23, 381–382. [Google Scholar] [CrossRef]
- Cheung, M.S.; Maguire, M.L.; Stevens, T.J.; Broadhurst, R.W. DANGLE: A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 2010, 202, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.G.; Thornton, J.M. PROMOTIF—A program to identify and analyze structural motifs in proteins. Protein Sci. 1996, 5, 212–220. [Google Scholar] [CrossRef] [PubMed]
- De Lano, W.L. Pymol; DeLano Scientific: South San Francisco, CA, USA, 2002. [Google Scholar]
- Piszkiewicz, D.; Landon, M.; Smith, E.L. Anomalous cleavage of aspartyl-proline peptide bonds during amino acid sequence determinations. Biochem. Biophys. Res. Commun. 1970, 40, 1173–1178. [Google Scholar] [CrossRef]
- Segalas, I.; Thai, R.; Menez, R.; Vita, C. A particularly labile Asp–Pro bond in the green mamba muscarinic toxin Mtx2—Effect of protein conformation on the rate of cleavage. FEBS Lett. 1995, 371, 171–175. [Google Scholar] [CrossRef]


| MIC a (µM) | |
|---|---|
| Escherichia coli NM522 | 17.0 ± 2.4 |
| Micrococcus luteus | >80 |
| Concentration (µM) | |||||||
|---|---|---|---|---|---|---|---|
| BCR4 peptide | 80 | 40 | 35 | 27.5 | 20 | 10 | 5 |
| LT50 (days) | 1.16 [0.98–1.37] | 1.76 [1.27–2.42] | 1.94 [1.47–2.57] | 3.24 [2.22–4.72] | 3.48 [2.28–5.31] | 11.3 [4.16–30.8] | >20 |
| Number of Cysteines | Consensus Motif | Cysteine Bonding Pattern | Number of Orthologous Sequences in Aphid Species a | |
|---|---|---|---|---|
| BCR1-2-4-5 | 6 | CX3CX3-5CX4-6CXCX11-13C | 1:5, 2:4, 3:6 | Akon (2), Apis (5), Mros (4), Smis (1), Mros (1), Dnox (1) |
| BCR3 | 6 | CX3CX6CX21-25CX6CX3C | Unknown | Acra (3), Agly (1), Agos (2), Apis (1), Asol (2), Cced (1), Dnox (2), Mcer (1), Mper (1), Mros (3), Msac (3), Pnig (2), Rma (3), Rpad (1), Sgra (2), Smis (2), Tcit (1) |
| BCR6 | 8 | CX15CX3CX3CX10CX10CXCC | Unknown | Apis (1), Asol (1), Dnox (2), Masc (1), Mcer (1), Mper (1), Mros (3), Pnig (5), Save (1), Smis (2) |
| BCR8 | 6 | CX3-4CX10CX15-17CXCC | Unknown | Akon (1), Acra (3), Agly (1), Agos (1), Apis (1), Asol (1), Dnox (1), Masc (1), Mcer (1), Mper (1), Msac (1), Mros (2), Pnig (1), Rmai (1), Rpad (1), Smis (2) |
| CSαβ | 6 | CX6-15CX3CX9-10CX4-7CXC | 1:4, 2:5, 3:6 | insects |
| 8 | CX10CX5CX3CX9-10CX6-8CXCX3C | 1:8, 2:5, 3:6, 4:7 | plants | |
| α-defensin | 6 | CXCX3-5CX9CX6-10CC | 1:6, 2:4, 3:5 | primates |
| β-defensin | 6 | CX5-7CX3-5CX8-11CX4-6CC | 1:5, 2:4, 3:6 | vertebrates |
| NMR Restraints | |
|---|---|
| Distance Restraints | |
| Total NOE | 923 |
| Unambiguous | 763 |
| Ambiguous | 160 |
| Hydrogen bonds | 16 |
| Dihedral Angle Restraints | 88 |
| Disulfide bridges a | 3 |
| Structural Statistics (7PQW.pdb) | |
| Average Violations Per Structure | |
| NOEs ≥ 0.3Å | 0 |
| Hydrogen bonds ≥ 0.3Å | 0 |
| Dihedrals ≥ 15° | 3 |
| Dihedrals ≥ 10° | 8 |
| Average pairwise rmsd (Å) | backbone atoms 0.45 ± 0.14 heavy atoms 1.14 ± 0.15 |
| Ramachandran Analysis | |
| Most favored region and allowed region | 96.9 |
| Generously allowed | 0.7 |
| Disallowed | 2.4 |
| Energies (kcal·mol−1) b | |
| Electrostatic | −1631.48 ± 29.53 |
| Van der Waals | −334.18 ± 11.14 |
| Total energy | −1196.56 ± 31.80 |
| Residual NOE energy | 56.38 ± 5.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loth, K.; Parisot, N.; Paquet, F.; Terrasson, H.; Sivignon, C.; Rahioui, I.; Ribeiro Lopes, M.; Gaget, K.; Duport, G.; Delmas, A.F.; et al. Aphid BCR4 Structure and Activity Uncover a New Defensin Peptide Superfamily. Int. J. Mol. Sci. 2022, 23, 12480. https://doi.org/10.3390/ijms232012480
Loth K, Parisot N, Paquet F, Terrasson H, Sivignon C, Rahioui I, Ribeiro Lopes M, Gaget K, Duport G, Delmas AF, et al. Aphid BCR4 Structure and Activity Uncover a New Defensin Peptide Superfamily. International Journal of Molecular Sciences. 2022; 23(20):12480. https://doi.org/10.3390/ijms232012480
Chicago/Turabian StyleLoth, Karine, Nicolas Parisot, Françoise Paquet, Hugo Terrasson, Catherine Sivignon, Isabelle Rahioui, Mélanie Ribeiro Lopes, Karen Gaget, Gabrielle Duport, Agnès F. Delmas, and et al. 2022. "Aphid BCR4 Structure and Activity Uncover a New Defensin Peptide Superfamily" International Journal of Molecular Sciences 23, no. 20: 12480. https://doi.org/10.3390/ijms232012480
APA StyleLoth, K., Parisot, N., Paquet, F., Terrasson, H., Sivignon, C., Rahioui, I., Ribeiro Lopes, M., Gaget, K., Duport, G., Delmas, A. F., Aucagne, V., Heddi, A., Calevro, F., & da Silva, P. (2022). Aphid BCR4 Structure and Activity Uncover a New Defensin Peptide Superfamily. International Journal of Molecular Sciences, 23(20), 12480. https://doi.org/10.3390/ijms232012480






