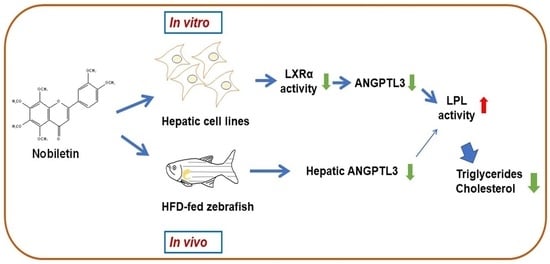
The Citrus Flavonoid Nobiletin Downregulates Angiopoietin-like Protein 3 (ANGPTL3) Expression and Exhibits Lipid-Modulating Effects in Hepatic Cells and Adult Zebrafish Models
Abstract
1. Introduction
2. Results
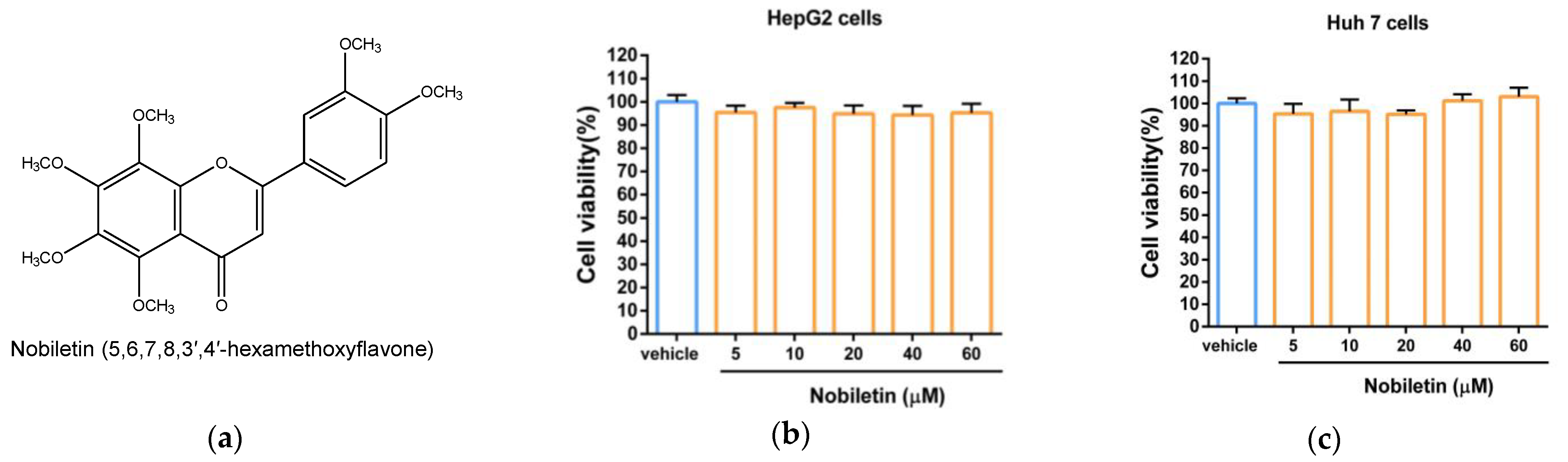
2.1. The Cytotoxic Effect of Nobiletin in Hepatic Cell Lines
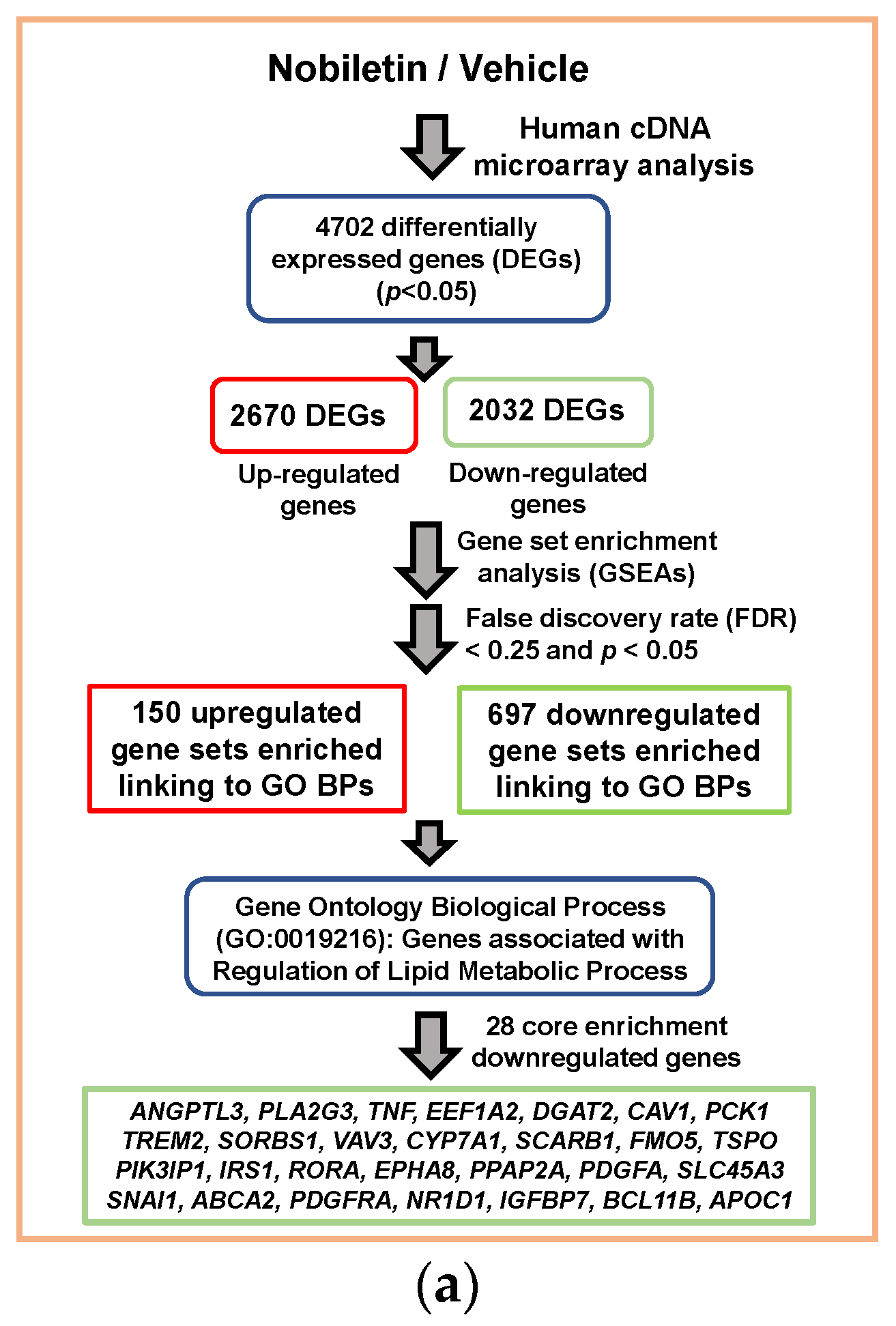
2.2. Analysis of Nobiletin-Modulated Differentially Expressed genes (DEGs) in HepG2 Cells
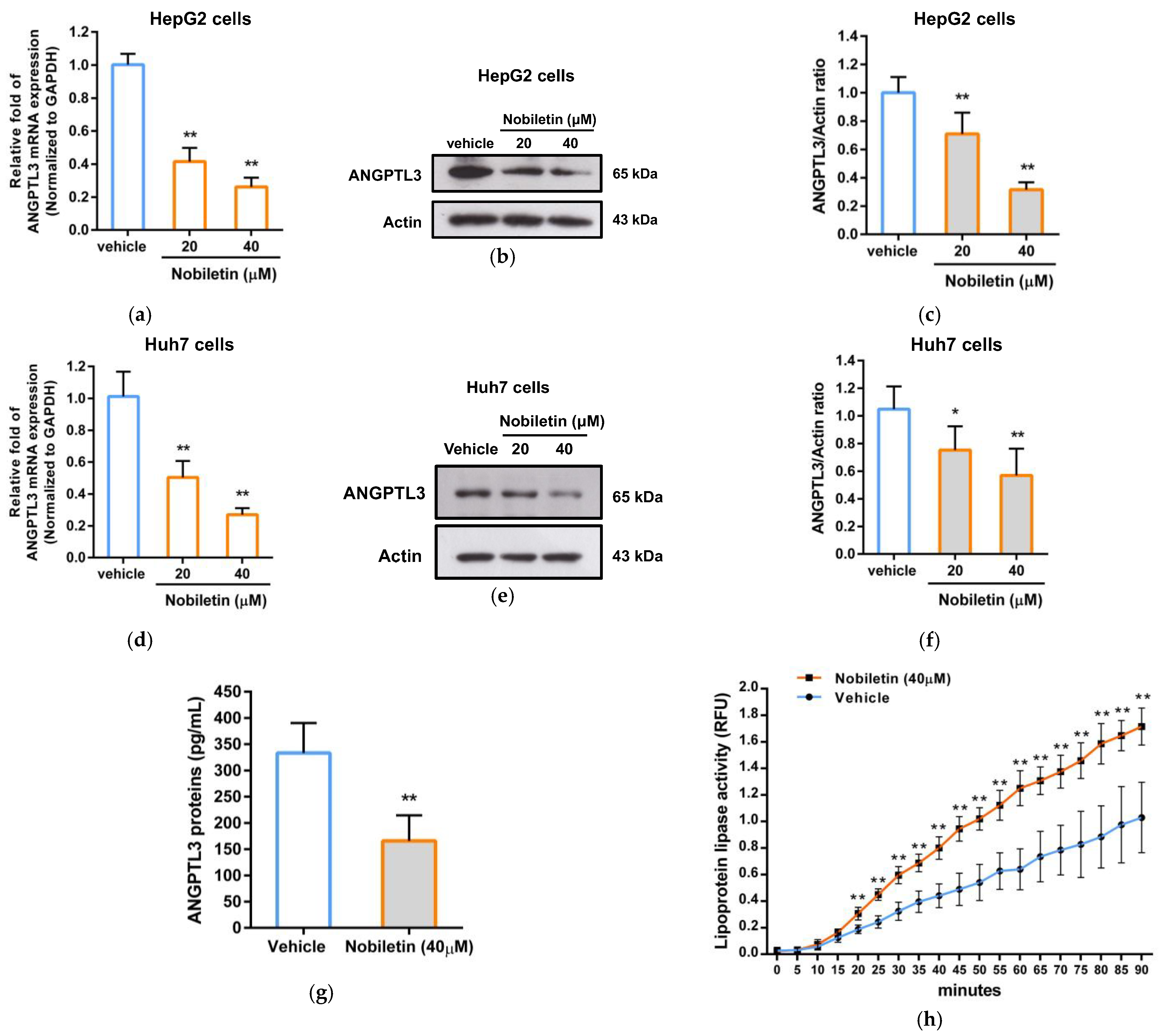
2.3. The Effect of Nobiletin on ANGPTL3 Gene Expression in Hepatic Cells
2.4. Nobiletin Suppresses the Transcriptional Activity of the ANGPTL3 Promoter in HepG2 Cells
2.5. Nobiletin Inhibits ANGPTL3 Expression by Counteracting LXRα Activity
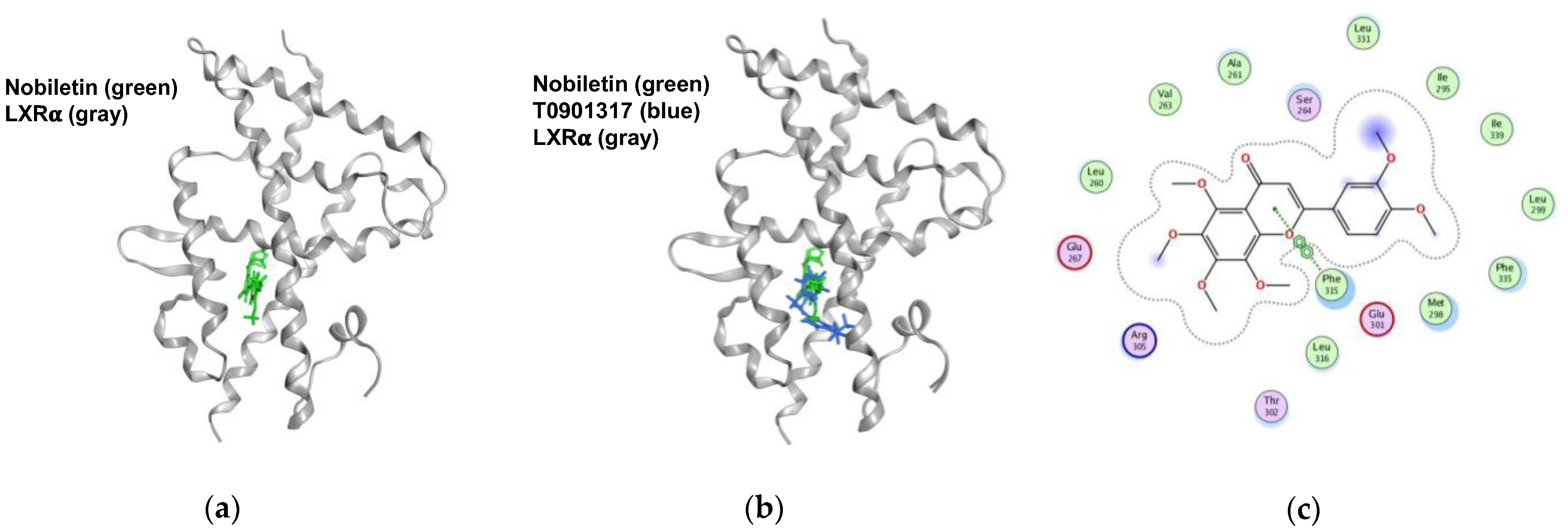
2.6. Nobiletin Docks into the Ligand-Binding Domain (LBD) of LXRα
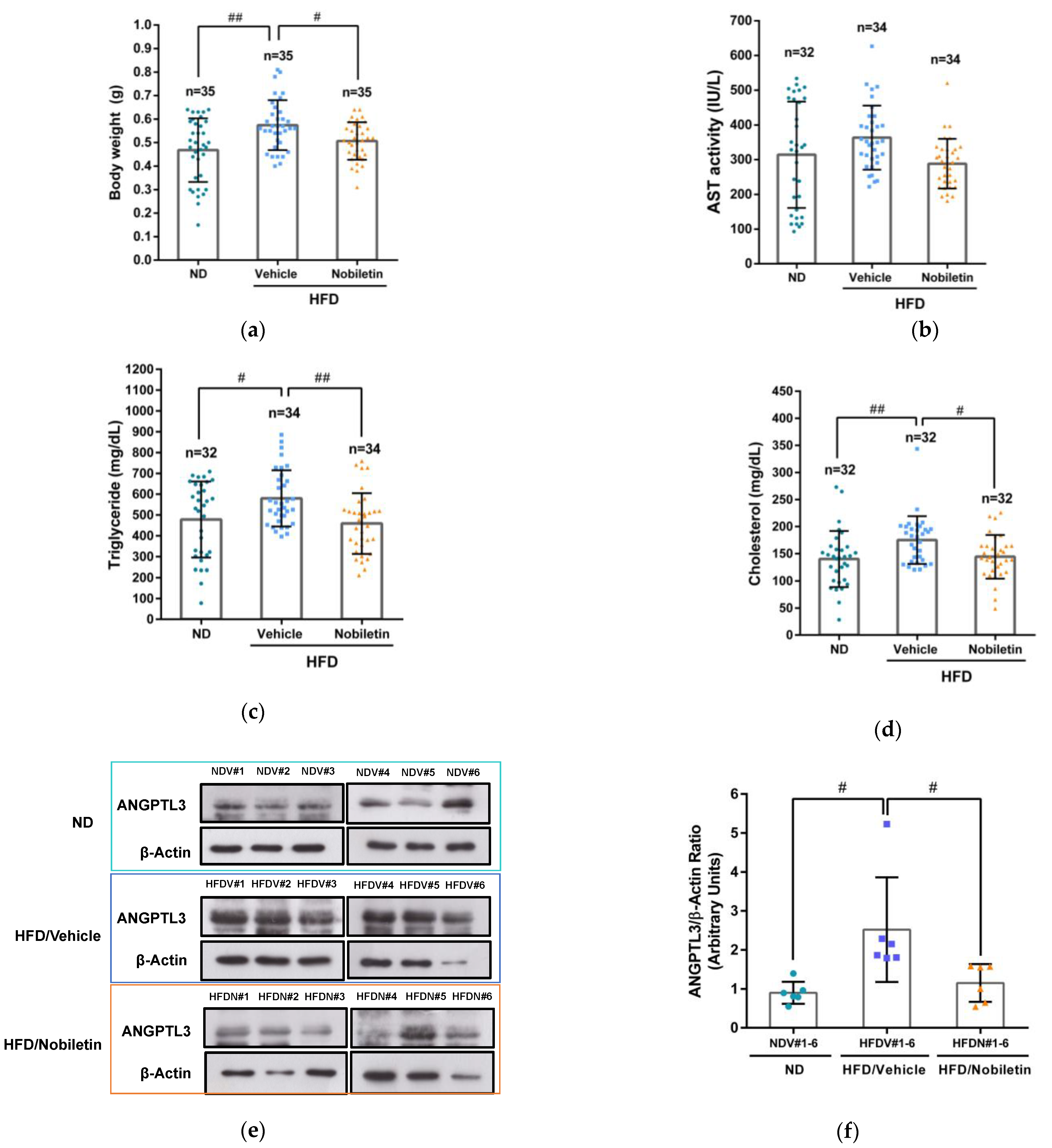
2.7. Nobiletin Modulates the Levels of Plasma Lipids and Hepatic ANGPTL3 Expression in Adult Zebrafish
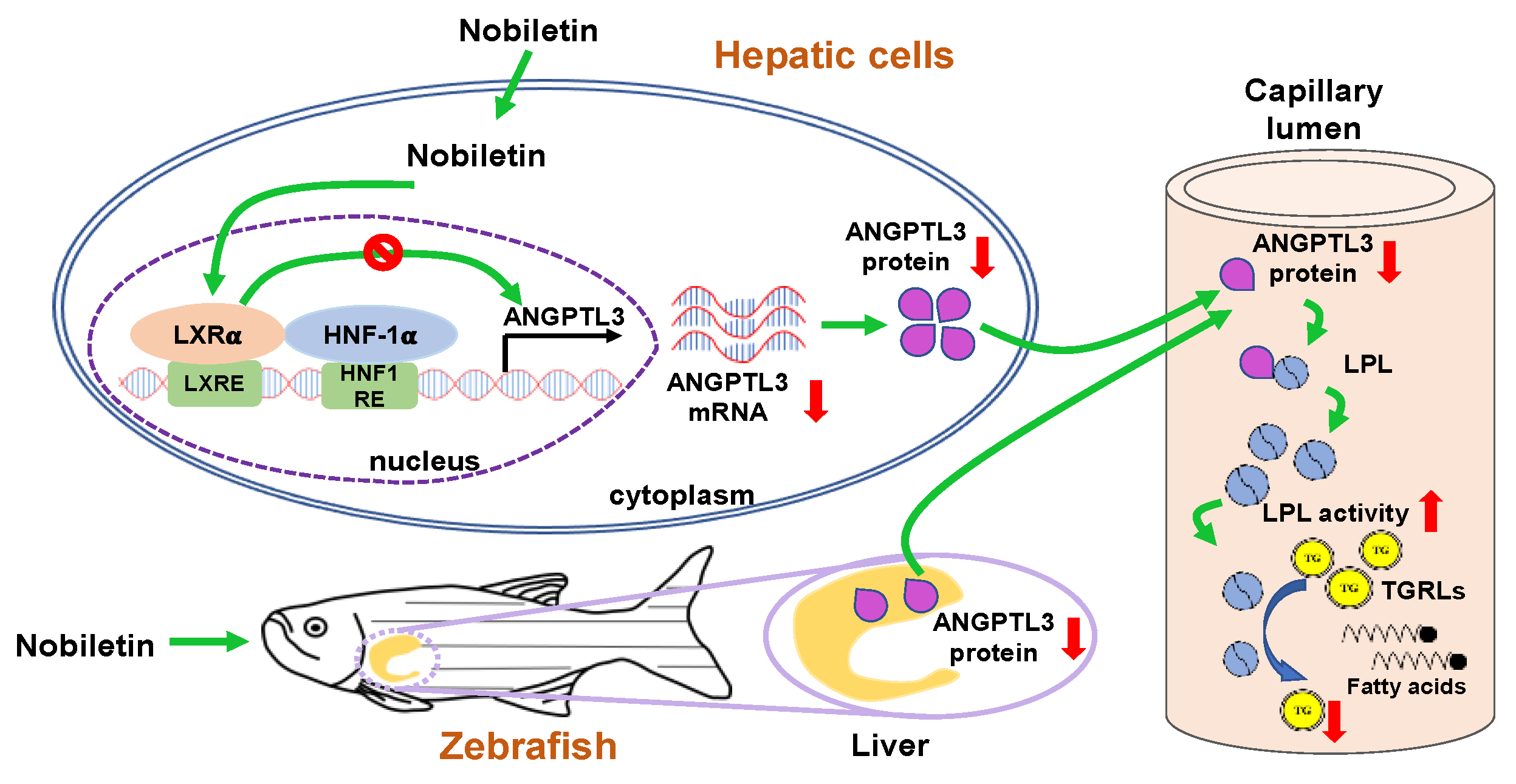
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture and Compounds Treatment
4.3. Measurement of Cell Viability
4.4. RNA Preparation and Transcriptome Analysis
4.5. Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA)
4.6. Reverse-Transcription Quantitative PCR (RT–qPCR) Analysis
4.7. Western Blot Analysis
4.8. Measurement of Extracellular ANGPTL3 Proteins
4.9. Analysis of Lipoprotein Lipase (LPL) Activity
4.10. Plasmid Transfection and Measurement of ANGPTL3 Promoter Activity
4.11. Molecular Docking of Nobiletin to the LXRα Protein
4.12. Zebrafish Experiments and Compound Administration
4.13. Biochemical Analysis of Plasma Lipids in Zebrafish
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hussain, A.; Ballantyne, C.M.; Saeed, A.; Virani, S.S. Triglycerides and ASCVD Risk Reduction: Recent Insights and Future Directions. Curr. Atheroscler. Rep. 2020, 22, 25. [Google Scholar] [CrossRef] [PubMed]
- Nordestgaard, B.G. Triglyceride-Rich Lipoproteins and Atherosclerotic Cardiovascular Disease: New Insights from Epidemiology, Genetics, and Biology. Circ. Res. 2016, 118, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Tall, A.R.; Thomas, D.G.; Gonzalez-Cabodevilla, A.G.; Goldberg, I.J. Addressing dyslipidemic risk beyond LDL-cholesterol. J. Clin. Investig. 2022, 132, 148559. [Google Scholar] [CrossRef] [PubMed]
- Sandesara, P.B.; Virani, S.S.; Fazio, S.; Shapiro, M.D. The Forgotten Lipids: Triglycerides, Remnant Cholesterol, and Atherosclerotic Cardiovascular Disease Risk. Endocr. Rev. 2019, 40, 537–557. [Google Scholar] [CrossRef]
- Olivecrona, G. Role of lipoprotein lipase in lipid metabolism. Curr. Opin. Lipidol. 2016, 27, 233–241. [Google Scholar] [CrossRef]
- Tsutsumi, K.; Inoue, Y.; Shima, A.; Iwasaki, K.; Kawamura, M.; Murase, T. The novel compound NO-1886 increases lipoprotein lipase activity with resulting elevation of high density lipoprotein cholesterol, and long-term administration inhibits atherogenesis in the coronary arteries of rats with experimental atherosclerosis. J. Clin. Investig. 1993, 92, 411–417. [Google Scholar] [CrossRef]
- Shimada, M.; Shimano, H.; Gotoda, T.; Yamamoto, K.; Kawamura, M.; Inaba, T.; Yazaki, Y.; Yamada, N. Overexpression of human lipoprotein lipase in transgenic mice. Resistance to diet-induced hypertriglyceridemia and hypercholesterolemia. J. Biol. Chem. 1993, 268, 17924–17929. [Google Scholar] [CrossRef]
- Gaziano, J.M.; Hennekens, C.H.; O‘Donnell, C.J.; Breslow, J.L.; Buring, J.E. Fasting triglycerides, high-density lipoprotein, and risk of myocardial infarction. Circulation 1997, 96, 2520–2525. [Google Scholar] [CrossRef]
- Koishi, R.; Ando, Y.; Ono, M.; Shimamura, M.; Yasumo, H.; Fujiwara, T.; Horikoshi, H.; Furukawa, H. Angptl3 regulates lipid metabolism in mice. Nat. Genet. 2002, 30, 151–157. [Google Scholar]
- Kersten, S. Angiopoietin-like 3 in lipoprotein metabolism. Nat. Rev. Endocrinol. 2017, 13, 731–739. [Google Scholar] [CrossRef]
- Chen, P.Y.; Gao, W.Y.; Liou, J.W.; Lin, C.Y.; Wu, M.J.; Yen, J.H. Angiopoietin-Like Protein 3 (ANGPTL3) Modulates Lipoprotein Metabolism and Dyslipidemia. Int. J. Mol. Sci. 2021, 22, 730. [Google Scholar] [CrossRef] [PubMed]
- Ruscica, M.; Zimetti, F.; Adorni, M.P.; Sirtori, C.R.; Lupo, M.G.; Ferri, N. Pharmacological aspects of ANGPTL3 and ANGPTL4 inhibitors: New therapeutic approaches for the treatment of atherogenic dyslipidemia. Pharmacol. Res. 2020, 153, 104653. [Google Scholar] [CrossRef] [PubMed]
- Conklin, D.; Gilbertson, D.; Taft, D.W.; Maurer, M.F.; Whitmore, T.E.; Smith, D.L.; Walker, K.M.; Chen, L.H.; Wattler, S.; Nehls, M.; et al. Identification of a mammalian angiopoietin-related protein expressed specifically in liver. Genomics 1999, 62, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Biterova, E.; Esmaeeli, M.; Alanen, H.I.; Saaranen, M.; Ruddock, L.W. Structures of Angptl3 and Angptl4, modulators of triglyceride levels and coronary artery disease. Sci. Rep. 2018, 8, 6752. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.C.; Mintah, I.J.; Alexa-Braun, C.A.; Shihanian, L.M.; Lee, J.S.; Banerjee, P.; Hamon, S.C.; Kim, H.I.; Cohen, J.C.; Hobbs, H.H.; et al. Angiopoietin-like protein 3 governs LDL-cholesterol levels through endothelial lipase-dependent VLDL clearance. J. Lipid Res. 2020, 61, 1271–1286. [Google Scholar] [CrossRef]
- Quagliarini, F.; Wang, Y.; Kozlitina, J.; Grishin, N.V.; Hyde, R.; Boerwinkle, E.; Valenzuela, D.M.; Murphy, A.J.; Cohen, J.C.; Hobbs, H.H. Atypical angiopoietin-like protein that regulates ANGPTL3. Proc. Natl. Acad. Sci. USA 2012, 109, 19751–19756. [Google Scholar] [CrossRef]
- Kaplan, R.; Zhang, T.; Hernandez, M.; Gan, F.X.; Wright, S.D.; Waters, M.G.; Cai, T.Q. Regulation of the angiopoietin-like protein 3 gene by LXR. J. Lipid Res. 2003, 44, 136–143. [Google Scholar] [CrossRef]
- Fugier, C.; Tousaint, J.J.; Prieur, X.; Plateroti, M.; Samarut, J.; Delerive, P. The lipoprotein lipase inhibitor ANGPTL3 is negatively regulated by thyroid hormone. J. Biol. Chem. 2006, 281, 11553–11559. [Google Scholar] [CrossRef]
- Inaba, T.; Matsuda, M.; Shimamura, M.; Takei, N.; Terasaka, N.; Ando, Y.; Yasumo, H.; Koishi, R.; Makishima, M.; Shimomura, I. Angiopoietin-like protein 3 mediates hypertriglyceridemia induced by the liver X receptor. J. Biol. Chem. 2003, 278, 21344–21351. [Google Scholar] [CrossRef]
- Dewey, F.E.; Gusarova, V.; Dunbar, R.L.; O‘Dushlaine, C.; Schurmann, C.; Gottesman, O.; McCarthy, S.; Van Hout, C.V.; Bruse, S.; Dansky, H.M.; et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 211–221. [Google Scholar] [CrossRef]
- Stitziel, N.O.; Khera, A.V.; Wang, X.; Bierhals, A.J.; Vourakis, A.C.; Sperry, A.E.; Natarajan, P.; Klarin, D.; Emdin, C.A.; Zekavat, S.M.; et al. ANGPTL3 Deficiency and Protection Against Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 2054–2063. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.; Qamar, A.; Qu, L.; Qasim, A.N.; Mehta, N.N.; Reilly, M.P.; Rader, D.J. Differential association of plasma angiopoietin-like proteins 3 and 4 with lipid and metabolic traits. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Kathiresan, S.; Melander, O.; Guiducci, C.; Surti, A.; Burtt, N.P.; Rieder, M.J.; Cooper, G.M.; Roos, C.; Voight, B.F.; Havulinna, A.S.; et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008, 40, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Sanna, S.; Jackson, A.U.; Scuteri, A.; Bonnycastle, L.L.; Clarke, R.; Heath, S.C.; Timpson, N.J.; Najjar, S.S.; Stringham, H.M.; et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008, 40, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Akoumianakis, I.; Zvintzou, E.; Kypreos, K.; Filippatos, T.D. ANGPTL3 and Apolipoprotein C-III as Novel Lipid-Lowering Targets. Curr. Atheroscler. Rep. 2021, 23, 20. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.J.; Lee, R.G.; Brandt, T.A.; Tai, L.J.; Fu, W.; Peralta, R.; Yu, R.; Hurh, E.; Paz, E.; McEvoy, B.W.; et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N. Engl. J. Med. 2017, 377, 222–232. [Google Scholar] [CrossRef]
- Mullard, A. FDA approves first anti-ANGPTL3 antibody, for rare cardiovascular indication. Nat. Rev. Drug Discov. 2021, 20, 251. [Google Scholar] [CrossRef]
- Ahmad, Z.; Banerjee, P.; Hamon, S.; Chan, K.C.; Bouzelmat, A.; Sasiela, W.J.; Pordy, R.; Mellis, S.; Dansky, H.; Gipe, D.A.; et al. Inhibition of Angiopoietin-Like Protein 3 with a Monoclonal Antibody Reduces Triglycerides in Hypertriglyceridemia. Circulation 2019, 140, 470–486. [Google Scholar] [CrossRef]
- Mulvihill, E.E.; Burke, A.C.; Huff, M.W. Citrus Flavonoids as Regulators of Lipoprotein Metabolism and Atherosclerosis. Annu. Rev. Nutr. 2016, 36, 275–299. [Google Scholar] [CrossRef]
- Gandhi, G.R.; Vasconcelos, A.B.S.; Wu, D.T.; Li, H.B.; Antony, P.J.; Li, H.; Geng, F.; Gurgel, R.Q.; Narain, N.; Gan, R.Y. Citrus Flavonoids as Promising Phytochemicals Targeting Diabetes and Related Complications: A Systematic Review of In Vitro and In Vivo Studies. Nutrients 2020, 12, 2907. [Google Scholar] [CrossRef]
- Vajdi, M.; Farhangi, M.A. Citrus peel derived’ Poly-Methoxylated Flavones (PMF). Int. J. Vitam. Nutr. Res. 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Sato, T.; Takayama, Y.; Mimaki, Y.; Sashida, Y.; Yano, M.; Ito, A. Novel anti-inflammatory actions of nobiletin, a citrus polymethoxy flavonoid, on human synovial fibroblasts and mouse macrophages. Biochem. Pharmacol. 2003, 65, 2065–2071. [Google Scholar] [CrossRef]
- Huang, H.; Li, L.; Shi, W.; Liu, H.; Yang, J.; Yuan, X.; Wu, L. The Multifunctional Effects of Nobiletin and Its Metabolites In Vivo and In Vitro. Evid. Based Complement. Alternat. Med. 2016, 2016, 2918796. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.H.; Alex, D.; Lam, I.K.; Tsui, S.K.; Yang, Z.F.; Lee, S.M. Nobiletin, a polymethoxylated flavonoid from citrus, shows anti-angiogenic activity in a zebrafish in vivo model and HUVEC in vitro model. J. Cell. Biochem. 2011, 112, 3313–3321. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.H.; Lin, C.Y.; Chuang, C.H.; Chin, H.K.; Wu, M.J.; Chen, P.Y. Nobiletin Promotes Megakaryocytic Differentiation through the MAPK/ERK-Dependent EGR1 Expression and Exerts Anti-Leukemic Effects in Human Chronic Myeloid Leukemia (CML) K562 Cells. Cells 2020, 9, 877. [Google Scholar] [CrossRef]
- Nagase, H.; Yamakuni, T.; Matsuzaki, K.; Maruyama, Y.; Kasahara, J.; Hinohara, Y.; Kondo, S.; Mimaki, Y.; Sashida, Y.; Tank, A.W.; et al. Mechanism of neurotrophic action of nobiletin in PC12D cells. Biochemistry 2005, 44, 13683–13691. [Google Scholar] [CrossRef]
- Whitman, S.C.; Kurowska, E.M.; Manthey, J.A.; Daugherty, A. Nobiletin, a citrus flavonoid isolated from tangerines, selectively inhibits class A scavenger receptor-mediated metabolism of acetylated LDL by mouse macrophages. Atherosclerosis 2005, 178, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Kim, Y.; Ham, H.; Park, Y.; Jeong, H.S.; Lee, J. Nobiletin suppresses adipogenesis by regulating the expression of adipogenic transcription factors and the activation of AMP-activated protein kinase (AMPK). J. Agric. Food Chem. 2011, 59, 12843–12849. [Google Scholar] [CrossRef]
- Lin, Y.; Vermeer, M.A.; Bos, W.; van Buren, L.; Schuurbiers, E.; Miret-Catalan, S.; Trautwein, E.A. Molecular structures of citrus flavonoids determine their effects on lipid metabolism in HepG2 cells by primarily suppressing apoB secretion. J. Agric. Food Chem. 2011, 59, 4496–4503. [Google Scholar] [CrossRef]
- Yuk, T.; Kim, Y.; Yang, J.; Sung, J.; Jeong, H.S.; Lee, J. Nobiletin Inhibits Hepatic Lipogenesis via Activation of AMP-Activated Protein Kinase. Evid. Based Complement. Alternat. Med. 2018, 2018, 7420265. [Google Scholar] [CrossRef]
- Lee, Y.S.; Cha, B.Y.; Choi, S.S.; Choi, B.K.; Yonezawa, T.; Teruya, T.; Nagai, K.; Woo, J.T. Nobiletin improves obesity and insulin resistance in high-fat diet-induced obese mice. J. Nutr. Biochem. 2013, 24, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Bunbupha, S.; Pakdeechote, P.; Maneesai, P.; Prasarttong, P. Nobiletin alleviates high-fat diet-induced nonalcoholic fatty liver disease by modulating AdipoR1 and gp91(phox) expression in rats. J. Nutr. Biochem. 2021, 87, 108526. [Google Scholar] [CrossRef] [PubMed]
- Morrow, N.M.; Burke, A.C.; Samsoondar, J.P.; Seigel, K.E.; Wang, A.; Telford, D.E.; Sutherland, B.G.; O’Dwyer, C.; Steinberg, G.R.; Fullerton, M.D.; et al. The citrus flavonoid nobiletin confers protection from metabolic dysregulation in high-fat-fed mice independent of AMPK. J. Lipid Res. 2020, 61, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Yoon, D.S.; Jung, U.J. Efficacy of nobiletin in improving hypercholesterolemia and nonalcoholic fatty liver disease in high-cholesterol diet-fed mice. Nutr. Res. Pract. 2021, 15, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Morrow, N.M.; Trzaskalski, N.A.; Hanson, A.A.; Fadzeyeva, E.; Telford, D.E.; Chhoker, S.S.; Sutherland, B.G.; Edwards, J.Y.; Huff, M.W.; Mulvihill, E.E. Nobiletin Prevents High-Fat Diet-Induced Dysregulation of Intestinal Lipid Metabolism and Attenuates Postprandial Lipemia. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 127–144. [Google Scholar] [CrossRef]
- Morelli, M.B.; Chavez, C.; Santulli, G. Angiopoietin-like proteins as therapeutic targets for cardiovascular disease: Focus on lipid disorders. Expert Opin. Ther. Targets 2020, 24, 79–88. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, S.; Yang, W.; Huang, Q.; Ho, C.T. The biological fate and bioefficacy of citrus flavonoids: Bioavailability, biotransformation, and delivery systems. Food Funct. 2021, 12, 3307–3323. [Google Scholar] [CrossRef]
- Liao, W.; Liu, Z.; Zhang, T.; Sun, S.; Ye, J.; Li, Z.; Mao, L.; Ren, J. Enhancement of Anti-Inflammatory Properties of Nobiletin in Macrophages by a Nano-Emulsion Preparation. J. Agric. Food Chem. 2018, 66, 91–98. [Google Scholar] [CrossRef]
- Tung, Y.C.; Li, S.; Huang, Q.; Hung, W.L.; Ho, C.T.; Wei, G.J.; Pan, M.H. 5-Demethylnobiletin and 5-Acetoxy-6,7,8,3′,4′-pentamethoxyflavone Suppress Lipid Accumulation by Activating the LKB1-AMPK Pathway in 3T3-L1 Preadipocytes and High Fat Diet-Fed C57BL/6 Mice. J. Agric. Food Chem. 2016, 64, 3196–3205. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, C.; Chen, J.; Tian, G.; McClements, D.J.; Xiao, H.; Zheng, J. Encapsulation of Polymethoxyflavones in Citrus Oil Emulsion-Based Delivery Systems. J. Agric. Food Chem. 2017, 65, 1732–1739. [Google Scholar] [CrossRef]
- Verbist, B.; Klambauer, G.; Vervoort, L.; Talloen, W.; Consortium, Q.; Shkedy, Z.; Thas, O.; Bender, A.; Gohlmann, H.W.; Hochreiter, S. Using transcriptomics to guide lead optimization in drug discovery projects: Lessons learned from the QSTAR project. Drug Discov. Today 2015, 20, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Jia, J.; Wang, X.; Zhang, R.; Niu, S.; Ni, L.; Di, X.; Liu, C. Differential roles of Scavenger receptor class B type I: A protective molecule and a facilitator of atherosclerosis (Review). Mol. Med. Rep. 2020, 22, 2599–2604. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sethi, A.; Yanek, L.R.; Knapper, C.; Nordestgaard, B.G.; Tybjaerg-Hansen, A.; Becker, D.M.; Mathias, R.A.; Remaley, A.T.; Becker, L.C. SCARB1 Gene Variants Are Associated With the Phenotype of Combined High High-Density Lipoprotein Cholesterol and High Lipoprotein (a). Circ. Cardiovasc. Genet. 2016, 9, 408–418. [Google Scholar] [CrossRef]
- Rinninger, F.; Brundert, M.; Brosch, I.; Donarski, N.; Budzinski, R.M.; Greten, H. Lipoprotein lipase mediates an increase in selective uptake of HDL-associated cholesteryl esters by cells in culture independent of scavenger receptor BI. J. Lipid Res. 2001, 42, 1740–1751. [Google Scholar] [CrossRef]
- Brundert, M.; Ewert, A.; Heeren, J.; Behrendt, B.; Ramakrishnan, R.; Greten, H.; Merkel, M.; Rinninger, F. Scavenger receptor class B type I mediates the selective uptake of high-density lipoprotein-associated cholesteryl ester by the liver in mice. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, D.; Karwatowska-Prokopczuk, E.; Baum, S.J.; Hurh, E.; Kingsbury, J.; Bartlett, V.J.; Figueroa, A.L.; Piscitelli, P.; Singleton, W.; Witztum, J.L.; et al. Vupanorsen, an N-acetyl galactosamine-conjugated antisense drug to ANGPTL3 mRNA, lowers triglycerides and atherogenic lipoproteins in patients with diabetes, hepatic steatosis, and hypertriglyceridaemia. Eur. Heart J. 2020, 41, 3936–3945. [Google Scholar] [CrossRef]
- Xiao, H.B.; Sun, Z.L.; Zhou, N. 1,3,5,8-tetrahydroxyxanthone regulates ANGPTL3-LPL pathway to lessen the ketosis in mice. Eur. J. Pharm. Sci. 2012, 46, 26–31. [Google Scholar] [CrossRef]
- Chen, P.Y.; Chao, T.Y.; Hsu, H.J.; Wang, C.Y.; Lin, C.Y.; Gao, W.Y.; Wu, M.J.; Yen, J.H. The Lipid-Modulating Effect of Tangeretin on the Inhibition of Angiopoietin-like 3 (ANGPTL3) Gene Expression through Regulation of LXRalpha Activation in Hepatic Cells. Int. J. Mol. Sci. 2021, 22, 9853. [Google Scholar] [CrossRef]
- Wang, B.; Tontonoz, P. Liver X receptors in lipid signalling and membrane homeostasis. Nat. Rev. Endocrinol. 2018, 14, 452–463. [Google Scholar] [CrossRef]
- Jiang, S.; Qiu, G.H.; Zhu, N.; Hu, Z.Y.; Liao, D.F.; Qin, L. ANGPTL3: A novel biomarker and promising therapeutic target. J. Drug Target 2019, 27, 876–884. [Google Scholar] [CrossRef]
- Kahn, B.B.; Alquier, T.; Carling, D.; Hardie, D.G. AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005, 1, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hong, S.W.; Park, S.E.; Rhee, E.J.; Park, C.Y.; Oh, K.W.; Park, S.W.; Lee, W.Y. AMP-activated protein kinase suppresses the expression of LXR/SREBP-1 signaling-induced ANGPTL8 in HepG2 cells. Mol. Cell. Endocrinol. 2015, 414, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Ka, J.; Jin, S.W. Zebrafish as an Emerging Model for Dyslipidemia and Associated Diseases. J. Lipid Atheroscler. 2021, 10, 42–56. [Google Scholar] [CrossRef]
- Tang, D.; Geng, F.; Yu, C.; Zhang, R. Recent Application of Zebrafish Models in Atherosclerosis Research. Front. Cell Dev. Biol. 2021, 9, 643697. [Google Scholar] [CrossRef] [PubMed]
- Ka, J.; Pak, B.; Han, O.; Lee, S.; Jin, S.W. Comparison of transcriptomic changes between zebrafish and mice upon high fat diet reveals evolutionary convergence in lipid metabolism. Biochem. Biophys. Res. Commun. 2020, 530, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; So, J.H.; Kim, H.T.; Choi, J.H.; Lee, M.S.; Choi, S.Y.; Kim, C.H.; Kim, M.J. Angiopoietin-like 3 regulates hepatocyte proliferation and lipid metabolism in zebrafish. Biochem. Biophys. Res. Commun. 2014, 446, 1237–1242. [Google Scholar] [CrossRef]
- Chen, P.Y.; Wang, C.Y.; Tsao, E.C.; Chen, Y.T.; Wu, M.J.; Ho, C.T.; Yen, J.H. 5-Demethylnobiletin Inhibits Cell Proliferation, Downregulates ID1 Expression, Modulates the NF-kappaB/TNF-alpha Pathway and Exerts Antileukemic Effects in AML Cells. Int. J. Mol. Sci. 2022, 23, 7392. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell. Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Chen, S.F.; Chen, P.Y.; Hsu, H.J.; Wu, M.J.; Yen, J.H. Xanthohumol Suppresses Mylip/Idol Gene Expression and Modulates LDLR Abundance and Activity in HepG2 Cells. J. Agric. Food Chem. 2017, 65, 7908–7918. [Google Scholar] [CrossRef]
- Velasco-Santamaria, Y.M.; Korsgaard, B.; Madsen, S.S.; Bjerregaard, P. Bezafibrate, a lipid-lowering pharmaceutical, as a potential endocrine disruptor in male zebrafish (Danio rerio). Aquat. Toxicol. 2011, 105, 107–118. [Google Scholar]




| Gene Symbol | Description | Rank in Metric Score | Running ES | log2(Nobiletin/Vehicle) |
|---|---|---|---|---|
| ANGPTL3 | angiopoietin like 3 | −0.890 | 0.012 | −0.883 |
| PLA2G3 | phospholipase A2 group III | −0.826 | −0.016 | −0.817 |
| TNF | tumor necrosis factor | −0.739 | −0.034 | −0.738 |
| EEF1A2 | eukaryotic translation elongation factor 1 alpha 2 | −0.699 | −0.054 | −0.700 |
| DGAT2 | diacylglycerol O-acyltransferase 2 | −0.591 | −0.044 | −0.577 |
| CAV1 | caveolin 1 | −0.578 | −0.063 | −0.570 |
| PCK1 | phosphoenolpyruvate carboxy kinase 1 | −0.547 | −0.066 | −0.547 |
| TREM2 | triggering receptor expressed on myeloid cells 2 | −0.545 | −0.085 | −0.535 |
| SORBS1 | sorbin and SH3 domain containing 1 | −0.538 | −0.101 | −0.537 |
| VAV3 | vav guanine nucleotide exchange factor 3 | −0.478 | −0.083 | −0.462 |
| CYP7A1 | cytochrome P450 family 7 subfamily A member 1 | −0.462 | −0.088 | −0.447 |
| SCARB1 | scavenger receptor class B member 1 | −0.458 | −0.089 | −0.436 |
| FMO5 | flavin containing dimethylaniline monoxygenase 5 | −0.442 | −0.099 | −0.439 |
| TSPO | translocator protein | −0.432 | −0.094 | −0.431 |
| PIK3IP1 | phosphoinositide-3-kinase interacting protein 1 | −0.432 | −0.115 | −0.431 |
| IRS1 | insulin receptor substrate 1 | −0.425 | −0.100 | −0.424 |
| RORA | RAR related orphan receptor A | −0.410 | −0.122 | −0.398 |
| EPHA8 | EPH receptor A8 | −0.407 | −0.114 | −0.414 |
| PPAP2A | phosphatidic acid phosphatase type 2A | −0.402 | −0.127 | −0.409 |
| PDGFA | platelet derived growth factor subunit A | −0.396 | −0.135 | −0.388 |
| SLC45A3 | solute carrier family 45 member 3 | −0.395 | −0.142 | −0.380 |
| SNAI1 | snail family transcriptional repressor 1 | −0.389 | −0.156 | −0.384 |
| ABCA2 | ATP binding cassette subfamily A member 2 | −0.383 | −0.162 | −0.389 |
| PDGFRA | platelet derived growth factor receptor alpha | −0.383 | −0.180 | −0.383 |
| NR1D1 | nuclear receptor subfamily 1 group D member 1 | −0.371 | −0.167 | −0.359 |
| IGFBP7 | insulin like growth factor binding protein 7 | −0.368 | −0.179 | −0.364 |
| BCL11B | BAF chromatin remodeling complex subunit BCL11B | −0.368 | −0.203 | −0.366 |
| APOC1 | apolipoprotein C1 | −0.361 | −0.189 | −0.354 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-Y.; Chen, P.-Y.; Hsu, H.-J.; Gao, W.-Y.; Wu, M.-J.; Yen, J.-H. The Citrus Flavonoid Nobiletin Downregulates Angiopoietin-like Protein 3 (ANGPTL3) Expression and Exhibits Lipid-Modulating Effects in Hepatic Cells and Adult Zebrafish Models. Int. J. Mol. Sci. 2022, 23, 12485. https://doi.org/10.3390/ijms232012485
Lin C-Y, Chen P-Y, Hsu H-J, Gao W-Y, Wu M-J, Yen J-H. The Citrus Flavonoid Nobiletin Downregulates Angiopoietin-like Protein 3 (ANGPTL3) Expression and Exhibits Lipid-Modulating Effects in Hepatic Cells and Adult Zebrafish Models. International Journal of Molecular Sciences. 2022; 23(20):12485. https://doi.org/10.3390/ijms232012485
Chicago/Turabian StyleLin, Ching-Yen, Pei-Yi Chen, Hao-Jen Hsu, Wan-Yun Gao, Ming-Jiuan Wu, and Jui-Hung Yen. 2022. "The Citrus Flavonoid Nobiletin Downregulates Angiopoietin-like Protein 3 (ANGPTL3) Expression and Exhibits Lipid-Modulating Effects in Hepatic Cells and Adult Zebrafish Models" International Journal of Molecular Sciences 23, no. 20: 12485. https://doi.org/10.3390/ijms232012485
APA StyleLin, C.-Y., Chen, P.-Y., Hsu, H.-J., Gao, W.-Y., Wu, M.-J., & Yen, J.-H. (2022). The Citrus Flavonoid Nobiletin Downregulates Angiopoietin-like Protein 3 (ANGPTL3) Expression and Exhibits Lipid-Modulating Effects in Hepatic Cells and Adult Zebrafish Models. International Journal of Molecular Sciences, 23(20), 12485. https://doi.org/10.3390/ijms232012485