Assessing and Evaluating the Scope and Constraints of Idylla Molecular Assays by Using Different Source Materials in Routine Diagnostic Settings
Abstract
:1. Introduction
2. Results
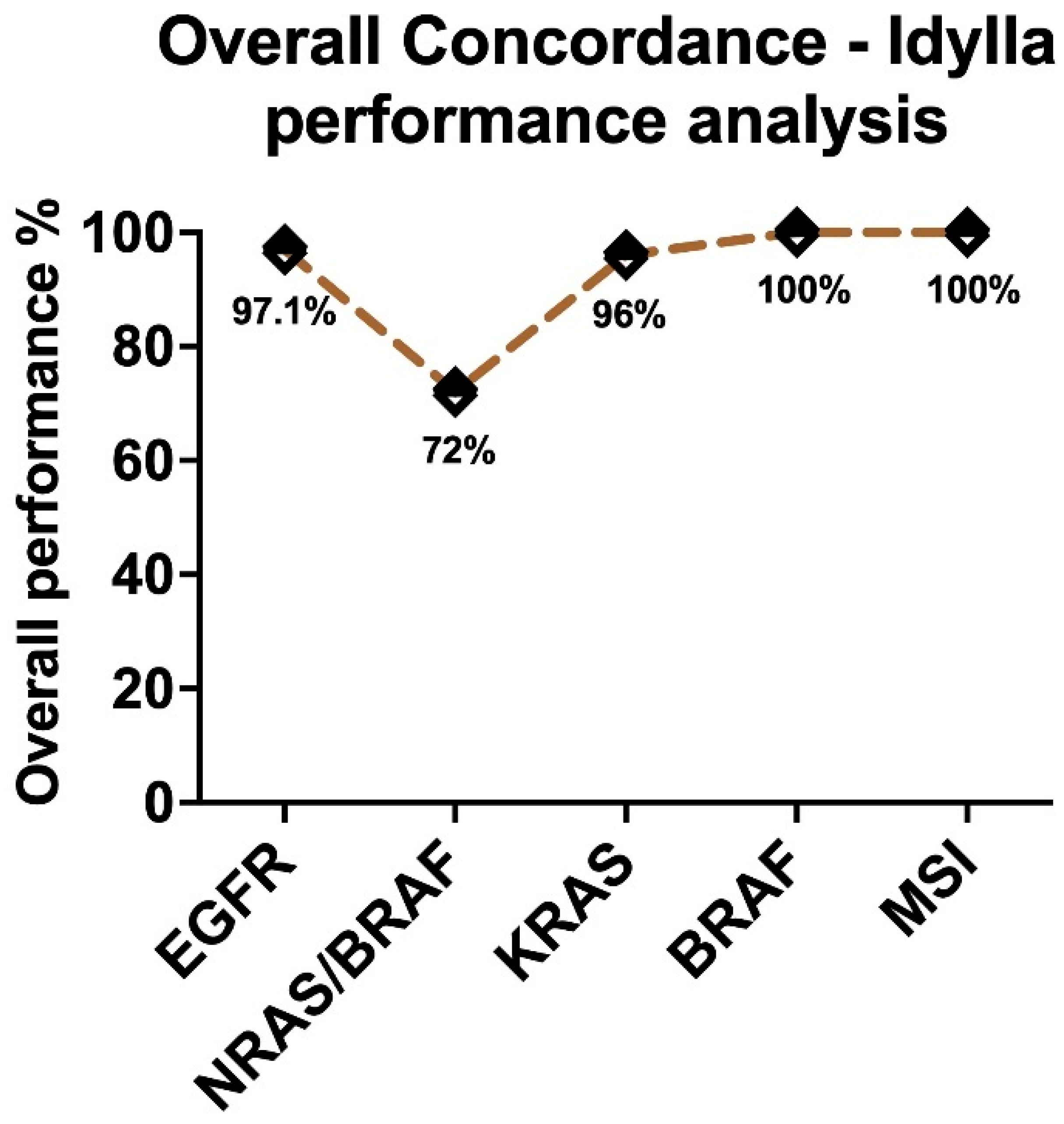
2.1. Assessment of Sensitivity and Specificity of IVD-Labelled Assays
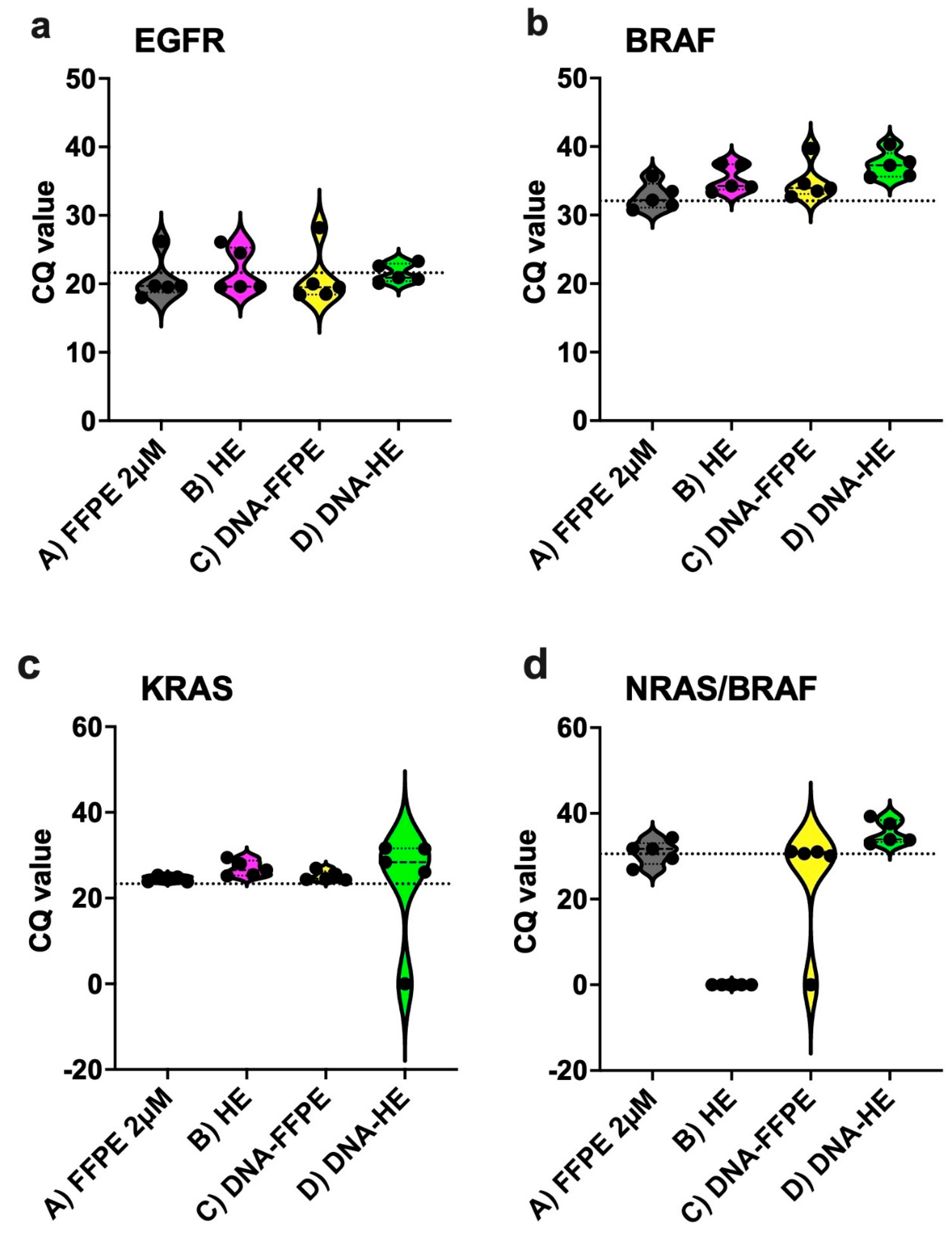
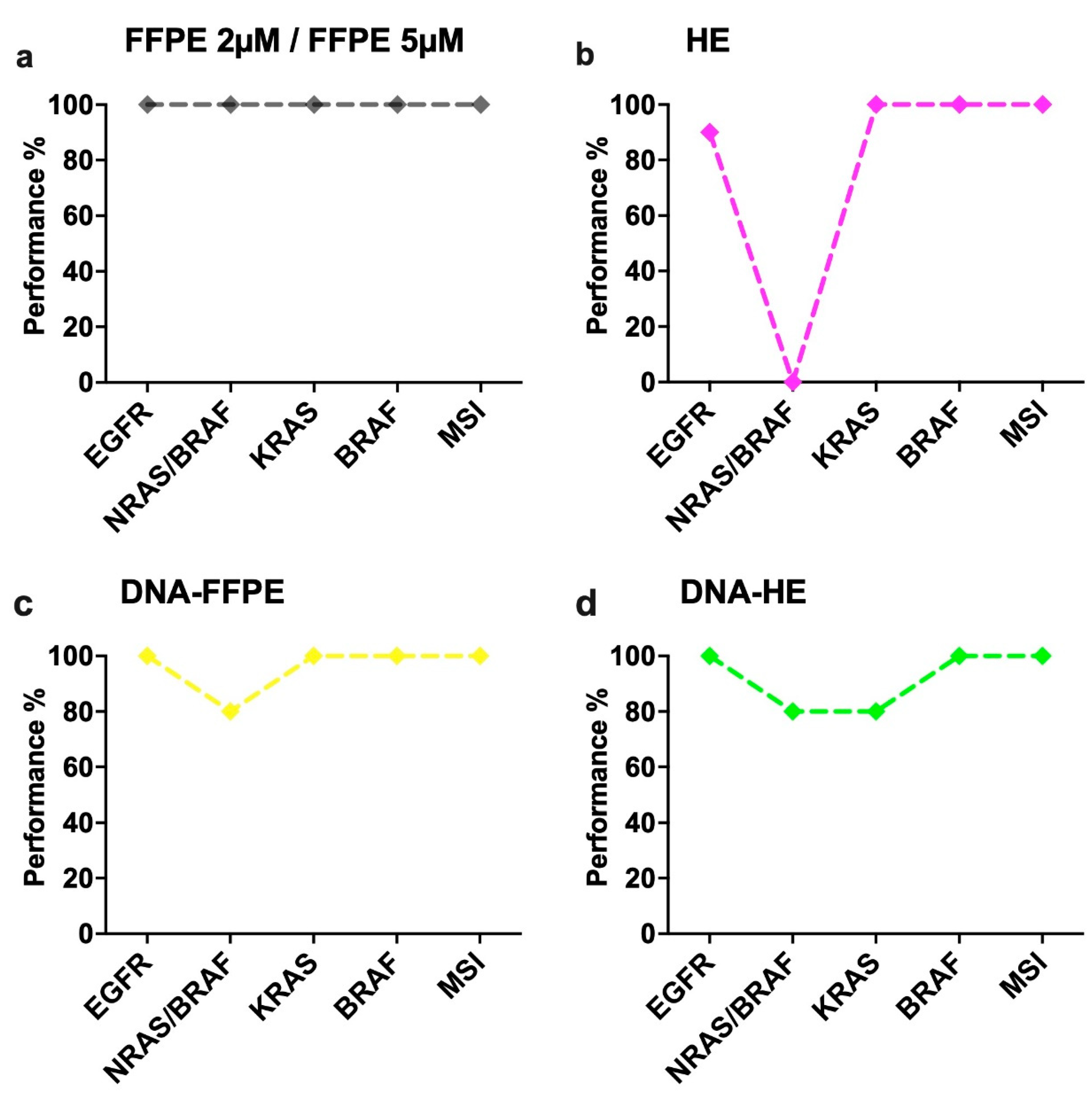
2.2. Performance of Different Sample Types in the IVD-Assays
2.3. Assessment of Sensitivity and Specificity of Gene Fusion Assay
- For HD784, all three specific fusions (ALK, ROS1, RET) were detected by specific PCR, and two-thirds expression imbalance (ALK, ROS1) could also be detected. The RET expression imbalance had a delta Cq (5′–3′) value of −0.58, which is on the borderline and the Cq of HKG is around 30 at the upper cutoff, meaning more input material is required for accurate analysis.
- For the CP-Positive control, ROS1 was detected both by specific detection and expression imbalance with a good input with HKG Cq at 28.6. In both of these controls, NTRK2/3 were invalid, and no borderline invalid curve was present, with a possible explanation that NGS controls mostly use artificial NTRK fragments, which cannot be detected by a full-length PCR design.
- For the Seraseq v4 material, all three specific fusions (ALK, ROS1, RET) along with MET Ex14 Skipping were detected by specific PCR, and the NTRK3 expression imbalance was also detected. The ALK expression imbalance could not be detected despite a high delta Cq (−5.29) value. The delta Cq cut-off is often dynamic and is increasing with high Cq HKG and is at a Cq 5′PCR of 38 at −5.3835. Thus, this sample tested represents a borderline negative case, and with the hypothesis of higher input, the assay would pick up the expression imbalance too. For Seraseq v4 material, the ROS1, RET and NTRK1 expression imbalance could not be detected, since, as mentioned in the instruction manual from Biocartis, the Seraseq version 4 reference material is an In-Vitro Transcript (IVT) RNAs with a sequence around the breakpoint of the fusion and doesn’t cover the 3′ regions, as a consequence, it is not suited for several of the expression imbalance detections by the GeneFusion Assay.
2.4. Determination of Repeatability of the Assays
2.5. Performance of the Idylla Assays on Pan-Cancer Tissues
2.6. Performance of the Idylla Assays on Archived FFPE Tissues
2.7. Performance of the Idylla Assays on Cytological Fluids
3. Discussion
4. Materials and Methods
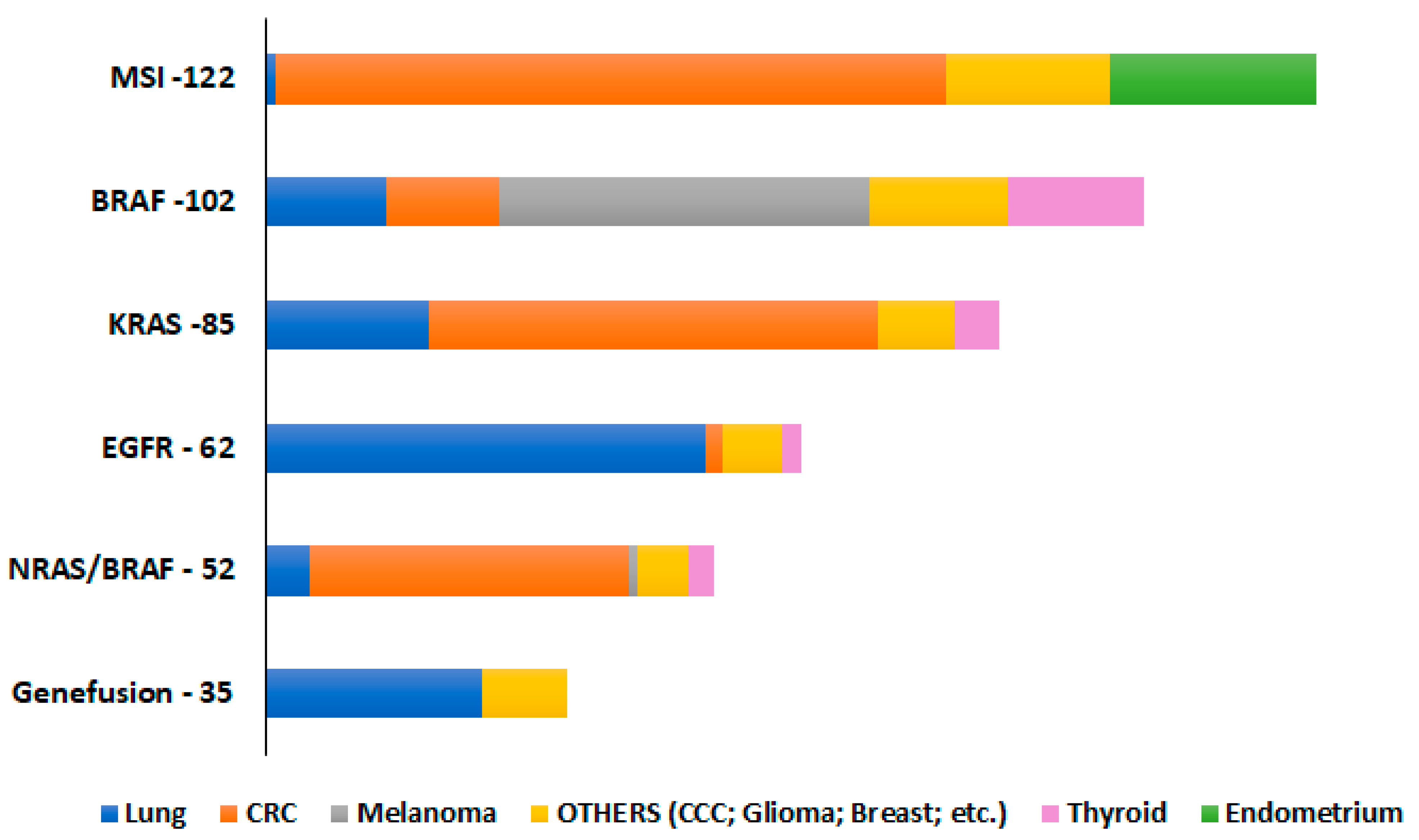
4.1. Sample Selection
4.2. Sample Source Materials
4.3. Molecular Reference Standards
- Two targeted FFPE RNA fusion reference standards—HD784 (Horizon Discovery Inc., Cambridge, UK) and Seraseq FFPE tumor fusion RNA reference material v4 (SeraCare Life Sciences, Milford, MA, USA).
- One CP-positive control (PC; PC-RNA/PC-DNA), which is included in AmoyDx® HANDLE NGS Classic Panel (Amoy Diagnostics Co., Ltd., Xiamen, China).
4.4. Nucleic Acid Isolations and Quantification
4.5. Idylla Platform–Molecular Assays
4.6. Alternative Analytical Methods
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Verma, M. Personalized medicine and cancer. J. Pers. Med. 2012, 2, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luthra, R.; Chen, H.; Roy-Chowdhuri, S.; Singh, R. Next-Generation Sequencing in Clinical Molecular Diagnostics of Cancer: Advantages and Challenges. Cancers 2015, 7, 2023–2036. [Google Scholar] [CrossRef] [PubMed]
- Tan, O.; Shrestha, R.; Cunich, M.; Schofield, D. Application of next-generation sequencing to improve cancer management: A review of the clinical effectiveness and cost-effectiveness. Clin. Genet. 2018, 93, 533–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morash, M.; Mitchell, H.; Beltran, H.; Elemento, O.; Pathak, J. The Role of Next-Generation Sequencing in Precision Medicine: A Review of Outcomes in Oncology. J. Pers. Med. 2018, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, L.; Power, M.; Sloan, P.; Long, A.; Silmon, A.; Chaffey, B.; Lisgo, A.J.; Little, L.; Vercauteren, E.; Steiniche, T.; et al. Clinical performance evaluation of the Idylla NRAS-BRAF mutation test on retrospectively collected for-malin-fixed paraffin-embedded colorectal cancer tissue. J. Clin. Pathol. 2018, 71, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Van Haele, M.; Vander Borght, S.; Ceulemans, A.; Wieërs, M.; Metsu, S.; Sagaert, X.; Weynand, B. Rapid clinical mutational testing of KRAS, BRAF and EGFR: A prospective comparative analysis of the Idylla technique with high-throughput next-generation sequencing. J. Clin. Pathol. 2020, 73, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Offerman, S.; Prinsen, C.F.; Knol, A.; Methorst, N.; Kamphorst, J.; Niemantsverdriet, M. Short report: Performance evaluation of the Idylla™ KRAS and EGFR mutation tests on paraffin-embedded cytological NSCLC samples. Diagn. Pathol. 2021, 16, 70. [Google Scholar] [CrossRef]
- Pécriaux, A.; Favre, L.; Calderaro, J.; Charpy, C.; Derman, J.; Pujals, A. Detection of microsatellite instability in a panel of solid tumours with the Idylla MSI Test using extracted DNA. J. Clin. Pathol. 2021, 74, 36–42. [Google Scholar] [CrossRef]
- Gilson, P.; Franczak, C.; Dubouis, L.; Husson, M.; Rouyer, M.; Demange, J.; Perceau, M.; Leroux, A.; Merlin, J.-L.; Harlé, A. Evaluation of KRAS, NRAS and BRAF hotspot mutations detection for patients with metastatic colorectal cancer using direct DNA pipetting in a fully-automated platform and Next-Generation Sequencing for laboratory workflow optimisation. PLoS ONE 2019, 14, e0219204. [Google Scholar] [CrossRef]
- De Luca, C.; Gragnano, G.; Pisapia, P.; Vigliar, E.; Malapelle, U.; Bellevicine, C.; Troncone, G. EGFR mutation detection on lung cancer cytological specimens by the novel fully automated PCR-based Idylla EGFR Mutation Assay. J. Clin. Pathol. 2017, 70, 295–300. [Google Scholar] [CrossRef]
- Hamadou, M.; Lopez, J.; Benzerdjeb, N.; Cugnet-Anceau, C.; Schnoering, G.; Besançon, J.; Mezrag, S.; Lapras, V.; Denier, M.-L.; Descotes, F.; et al. Feasibility and performance of the Idylla™ NRAS/BRAF cartridge mutation assay on thyroid liquid-based fine-needle aspiration. Diagn. Cytopathol. 2021, 49, 1265–1271. [Google Scholar] [CrossRef]
- D’Ardia, A.; Caputo, A.; Fumo, R.; Ciaparrone, C.; Gaeta, S.; Picariello, C.; Zeppa, P.; D’Antonio, A. Advanced non-small cell lung cancer: Rapid evaluation of EGFR status on fine-needle cytology samples using Idylla. Pathol. Res. Pract. 2021, 224, 153547. [Google Scholar] [CrossRef]
- de Biase, D.; de Luca, C.; Gragnano, G.; Visani, M.; Bellevicine, C.; Malapelle, U.; Tallini, G.; Troncone, G. Fully automated PCR detection of KRAS mutations on pancreatic endoscopic ultrasound fine-needle aspi-rates. J. Clin. Pathol. 2016, 69, 986–991. [Google Scholar] [CrossRef]
- Caputo, A.; D’Ardia, A.; Sabbatino, F.; Picariello, C.; Ciaparrone, C.; Zeppa, P.; D’Antonio, A. Testing EGFR with Idylla on Cytological Specimens of Lung Cancer: A Review. Int. J. Mol. Sci. 2021, 22, 4852. [Google Scholar] [CrossRef]
- Delgado-García, M.; Weynand, B.; Gómez-Izquierdo, L.; Hernández, M.J.; Blanco, Á.M.; Varela, M.; Matias-Guiu, X.; Nadal, E.; Márquez-Lobo, B.; Alarcão, A.; et al. Clinical performance evaluation of the Idylla™ EGFR Mutation Test on formalin-fixed paraf-fin-embedded tissue of non-small cell lung cancer. BMC Cancer 2020, 20, 275. [Google Scholar] [CrossRef] [Green Version]
- Solassol, J.; Vendrell, J.; Märkl, B.; Haas, C.; Bellosillo, B.; Montagut, C.; Smith, M.; O’Sullivan, B.; D’Haene, N.; Le Mercier, M.; et al. Multi-Center Evaluation of the Fully Automated PCR-Based Idylla™ KRAS Mutation Assay for Rapid KRAS Mutation Status Determination on Formalin-Fixed Paraffin-Embedded Tissue of Human Colorectal Cancer. PLoS ONE 2016, 11, e0163444. [Google Scholar] [CrossRef]
- Murase, T.; Inagaki, H.; Eimoto, T. Influence of Histochemical and Immunohistochemical Stains on Polymerase Chain Reaction. Mod. Pathol. 2000, 13, 147–151. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, T.; Shima, K.; Kuchiba, A.; Yamauchi, M.; Tanaka, N.; Imamura, Y.; Liao, X.; Qian, Z.R.; Brahmandam, M.; Longtine, J.A.; et al. No evidence for interference of H&E staining in DNA testing: Usefulness of DNA extraction from H&E-stained archival tissue sections. Am. J. Clin. Pathol. 2012, 138, 122–129. [Google Scholar]
- Franczak, C.; Dubouis, L.; Gilson, P.; Husson, M.; Rouyer, M.; Demange, J.; Leroux, A.; Merlin, J.-L.; Harlé, A. Integrated routine workflow using next-generation sequencing and a fully-automated platform for the detection of KRAS, NRAS and BRAF mutations in formalin-fixed paraffin embedded samples with poor DNA quality in patients with colorectal carcinoma. PLoS ONE 2019, 14, e0212801. [Google Scholar] [CrossRef]
- Aisner, D.L.; Sams, S.B. The role of cytology specimens in molecular testing of solid tumors: Techniques, limitations, and opportunities. Diagn. Cytopathol. 2012, 40, 511–524. [Google Scholar] [CrossRef]
- Chen, J.T.; Lane, M.A.; Clark, D.P. Inhibitors of the polymerase chain reaction in Papanicolaou stain. Removal with a simple destaining procedure. Acta Cytol. 1996, 40, 873–877. [Google Scholar] [CrossRef]
- Ercolani, C.; Di Benedetto, A.; Bonomo, C.; Visca, P.; Palange, A.; Assisi, D.; Forcella, D.; Terrenato, I.; Pescarmona, E.; Ciliberto, G.; et al. Not enough can be enough: Feasibility of the Idylla EGFR mutation test when reuse of stained tissue slides is the only option available. J. Clin. Pathol. 2021, 207726. [Google Scholar] [CrossRef]
- Malapelle, U.; Parente, P.; Pepe, F.; De Luca, C.; Pisapia, P.; Sgariglia, R.; Nacchio, M.; Gragnano, G.; Russo, G.; Conticelli, F.; et al. Evaluation of Micro Satellite Instability and Mismatch Repair Status in Different Solid Tumors: A Multi-center Analysis in a Real World Setting. Cells 2021, 10, 1878. [Google Scholar] [CrossRef]
- Gilson, P.; Levy, J.; Rouyer, M.; Demange, J.; Husson, M.; Bonnet, C.; Salleron, J.; Leroux, A.; Merlin, J.-L.; Harlé, A. Evaluation of 3 molecular-based assays for microsatellite instability detection in formalin-fixed tissues of patients with endometrial and colorectal cancers. Sci. Rep. 2020, 10, 16386. [Google Scholar] [CrossRef]
- Favre, L.; Chen, R.; Bellahsen-Harrar, Y.; Ortonne, N.; Pujals, A. Idylla MSI test as a new tool for microsatellite instability detection in sebaceous tumours and keratoacanthomas. J. Clin. Pathol. 2021, 75, 675–680. [Google Scholar] [CrossRef]
- Chu, Y.-H.; Barbee, J.; Yang, S.-R.; Chang, J.C.; Liang, P.; Mullaney, K.; Chan, R.; Salazar, P.; Benayed, R.; Offin, M.; et al. Clinical Utility and Performance of an Ultrarapid Multiplex RNA-Based Assay for Detection of ALK, ROS1, RET, and NTRK1/2/3 Rearrangements and MET Exon 14 Skipping Alterations. J. Mol. Diagn. 2022, 24, 642–654. [Google Scholar] [CrossRef]
- Bocciarelli, C.; Cohen, J.; Pelletier, R.; Van Nhieu, J.T.; Derman, J.; Favre, L.; Bourgogne, A.; Monnet, I.; Chouaid, C.; Pujals, A. Evaluation of the Idylla system to detect the EGFR(T790M) mutation using extracted DNA. Pathol. Res. Pract. 2020, 216, 152773. [Google Scholar] [CrossRef] [PubMed]
- Dietmaier, W.; Wallinger, S.; Bocker, T.; Kullmann, F.; Fishel, R.; Rüschoff, J. Diagnostic microsatellite instability: Definition and correlation with mismatch repair protein expression. Cancer Res. 1997, 57, 4749–4756. [Google Scholar] [PubMed]
| Case No. # | Year of Tissue Prepared | Tumor Cells (%) | Cancer Type | Previous Result | VAF % | Alternate Method (s) | Idylla Cartridge Used | Input Material (s) in Cartridge | gDNA Input-FFPE Scratched Slide | gDNA Input-HE Scratched Slide | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA Conc. (ng/μL) | DNA Quality Score * | DNA Input in the Cartridge-µL | DNA Conc. (ng/μL) | DNA Quality Score * | DNA Input in the Cartridge-µL | |||||||||
| 1 | 2019 | 90% | NSCLC | EGFR-Ex19:p.L747_S7522del | 38.9% | NGS; Pyro | EGFR | FFPE; gDNA | 26.4 | 1 | 10 | 34.7 | 2 | 15 |
| EGFR-Ex19:p.A755G | 39.4% | |||||||||||||
| 2 | 2020 | 30–40% | NSCLC | EGFR-Ex20:p.T790M | 8.4% | NGS; Pyro | EGFR | FFPE; gDNA | 24.1 | 1 | 10 | 23.2 | 3 | 25 |
| EGFR-Ex21:p.L858R | 29.7% | |||||||||||||
| 3 | 2020 | 70% | NSCLC | EGFR-Ex 20:p.S768I | 31.6% | NGS; Pyro | EGFR | FFPE; gDNA | 54.2 | 1 | 15 | 70.5 | 3 | 20 |
| EGFR-Ex21:p.L858R | 14.8% | |||||||||||||
| 4 | 2012 | 50% | NSCLC | EGFR-Ex 19: p.E746_T753>V | 51.5% | NGS; Pyro | EGFR | FFPE; gDNA | 31.6 | 2 | 20 | 29.1 | 3 | 20 |
| 5 | 2021 | 75% | NSCLC | EGFR-Ex19: p.L747_T751del | 34.4% | NGS | EGFR | FFPE; gDNA | 143.5 | 1 | 10 | 52 | 2 | 15 |
| 6 | 2020 | 80% | CRC | NRAS-Ex2-c:49delA | 28.40% | NGS | NRAS/BRAF | FFPE; gDNA | 91.3 | 1 | 10 | 38.2 | 2 | 20 |
| BRAF-Ex15:p.V600E | 41.70% | NGS | NRAS/BRAF | FFPE; gDNA | 10 | 20 | ||||||||
| MSI High (5/10 System Unstable) | NGS; FA | MSI | FFPE; gDNA | 10 | 20 | |||||||||
| 7 | 2019 | 80% | CRC | NRAS-Ex 3:p.Q61L BRAF-WT | 22.10% | NGS; Pyro | NRAS/BRAF | FFPE; gDNA | 139 | 1 | 12 | 21.3 | 3 | 20 |
| 8 | 2019 | 100% | PTC | NRAS-Ex 3:p.Q61R BRAF-WT | 34.20% | NGS; Pyro | NRAS/BRAF | FFPE; gDNA | 36 | 2 | 20 | 88.8 | 2 | 15 |
| 9 | 2019 | 80% | CRC | NRAS-Ex 3:p.Q61R BRAF-WT | 20.20% | NGS; Pyro | NRAS/BRAF | FFPE; gDNA | 12.4 | 4 | 25 | 24.5 | 4 | 25 |
| 10 | 2020 | 95% | Melanoma | BRAF-Ex15:p.V600E, NRAS-WT | 14.40% | Pyro | NRAS/BRAF | FFPE; gDNA | 275 | 1 | 5 | 35.3 | 4 | 25 |
| 11 | 2004 | 70% | CRC | KRAS-Ex2:p.G12D | 14.1% | Pyro | KRAS | FFPE; gDNA | 196.1 | 1 | 12 | 50.9 | 1 | 15 |
| KRAS-Ex2:p.G12V | 2.1% | |||||||||||||
| 12 | 2019 | 30–40% | NSCLC | KRAS-Ex 2:p.G12A | 26.1% | NGS; Pyro | KRAS | FFPE; gDNA | 101.1 | 1 | 10 | 33.5 | 1 | 15 |
| 13 | 2019 | 80% | Oesophageal Carcinoma | KRAS-Ex 2:p.G13D | 55.94% | NGS; Pyro | KRAS | FFPE; gDNA | 86.5 | 1 | 10 | 64.7 | 3 | 25 |
| 14 | 2018 | 65% | NSCLC | KRAS-Ex4:p.Ala146Thr | 63.86% | NGS; Pyro | KRAS | FFPE; gDNA | 295.1 | 1 | 5 | 9.7 | 2 | 25 |
| 15 | 2022 | 60% | Pancreatic cancer | KRAS- Ex 2:p.G12R | 9.50% | Pyro | KRAS | FFPE; gDNA | 59.6 | 1 | 12 | 167.2 | 1 | 10 |
| 16 | 2021 | 90% | Thyroid cancer | BRAF-Ex15:p.V600E | 19.70% | Pyro | BRAF | FFPE; gDNA | 10 | 1 | 15 | 13.2 | 1 | 10 |
| 17 | 2018 | 75% | Melanoma | BRAF-Ex15:p.V600K | 32.90% | NGS; Pyro | BRAF | FFPE; gDNA | 259 | 1 | 8 | 96.2 | 1 | 10 |
| 18 | 2021 | 50% | Thyroid cancer | BRAF-Ex15:p.V600E | 36.70% | Pyro | BRAF | FFPE; gDNA | 113.9 | 1 | 10 | 38.5 | 3 | 25 |
| 19 | 2021 | 90% | Thyroid cancer | BRAF-Ex15:p.V600E | 24.10% | Pyro | BRAF | FFPE; gDNA | 95 | 1 | 10 | 118.1 | 2 | 15 |
| 20 | 2022 | 50% | Melanoma | BRAF-Ex15:p.V600E | 44.70% | Pyro | BRAF | FFPE; gDNA | 45.3 | 2 | 15 | 43.8 | 2 | 20 |
| 21 | 2020 | 80% | CRC | MSI High (3/10 System unstable) | FA | MSI | FFPE; gDNA | 190 | 1 | 8 | 8.9 | 1 | 15 | |
| 22 | 2010 | 80% | CRC | MSS | FA | MSI | FFPE; gDNA | 141 | 1 | 10 | 5.35 | 1 | 15 | |
| 23 | 2021 | 70% | CRC | MSI High (7/10 System unstable) | FA | MSI | FFPE; gDNA | 132 | 1 | 10 | 125.3 | 2 | 10 | |
| 24 | 2020 | 90% | CRC | MSI High (8/10 System unstable) | NGS; FA | MSI | FFPE; gDNA | 322 | 1 | 8 | 140.5 | 1 | 10 | |
| 25 | 2017 | 30% | Melanoma | BRAF-WT | Pyro | BRAF | FFPE-HE | N/A | N/A | N/A | N/A | N/A | N/A | |
| 26 | 2021 | 60% | CRC | NRAS-WT; BRAF-WT | Pyro | NRAS-BRAF | FFPE; gDNA-HE | N/A | N/A | N/A | 11.5 | 1 | 10 | |
| 27 | 2021 | 70% | NSCLC | EGFR-Ex18: p.G721S | 5.80% | NGS | EGFR | FFPE-HE | N/A | N/A | N/A | N/A | N/A | N/A |
| 28 | 2020 | 90% | NSCLC | EGFR-Ex19: p.E746_S752delins | 57.20% | NGS | EGFR | FFPE-HE | N/A | N/A | N/A | N/A | N/A | N/A |
| 29 | 2021 | 80% | NSCLC | EGFR-Ex21:p.L858R | 14.80% | NGS | EGFR | gDNA-FFPE | 5.5 | 1 | 15 | N/A | N/A | N/A |
| 30 | 2022 | 80% | CRC | KRAS-Ex2:p.G12D KRAS-Ex2:p.G12C | 25.8% 1.43% | NGS | KRAS | FFPE-HE + FFPE | N/A | N/A | N/A | N/A | N/A | N/A |
| 31 | 2021 | 20% | Thyroid cancer | BRAF-Ex15:p.V600E | 6.90% | Pyro | BRAF | gDNA-HE | N/A | N/A | N/A | 6.7 | 4 | 10–25 µL |
| 32 | 2021 | 60% | cholangiocellular carcinoma | EGFR-Ex19:PV742I | 46.70% | NGS | EGFR | Cytological fluid; gDNA | 20 µL Cytofluid material input in the cartridge | |||||
| KRAS | 20–500 µL Cytofluid material input in the cartridge and gDNA prepared from FFPE-cytoblock with 30 µL input and gDNA prepared from Cytofluid material with 40 µL input | |||||||||||||
| KRAS:Ex2:G12D | 3.10% | |||||||||||||
| 33 | 2021 | 30% | NSCLC | EGFR-WT | NGS | EGFR | Cytofluid pleural effusion | 20 µL Cytofluid material input in the cartridge | ||||||
| 34 | 2022 | 10% | IPMN | KRAS-Ex2: G12D | 26.50% | Pyro | KRAS | pleural effusion fluid; gDNA | gDNA prepared from Cytopleural effusion-with 40 µL input | |||||
| 35 | 2015 | 70% | Melanoma | BRAF-WT | Pyro | BRAF | FFPE | N/A | ||||||
| 36 | 2011 | 20% | CRC | MSS | FA | MSI | FFPE | N/A | ||||||
| 37 | 2018 | 50% | CRC | NRAS-WT; BRAF-WT | Pyro | NRAS-BRAF | FFPE | N/A | ||||||
| 38 | 2022 | Oesophageal Carcinoma | MSS | FA | MSI | FFPE-HE + PAS | N/A | |||||||
| 39 | 2021 | 90% | NSCLC | KRAS-WT | NGS | KRAS | FFPE | N/A | ||||||
| EGFR-WT | EGFR | FFPE | N/A | |||||||||||
| NRAS-WT; BRAF-WT | NRAS-BRAF | FFPE | N/A | |||||||||||
| 40 | N/A | CP-Positive Control (Reference Standard) | EGFR-Ex20: p.T790M EGFR-Ex19:p.E746_A750del EGFR-Ex21:p.L858R | 7.46% 7.12% 8.42% | NGS | EGFR; BRAF; KRAS; MSI; NRAS-BRAF | gDNA | 6.25 ng/µL–10 µL direct input in each cartridge | ||||||
| KRAS-Ex 2:p.G13D | 20.89% | |||||||||||||
| NRAS-WT BRAF-WT MSI-High | ||||||||||||||
| Case No. # | Year of Tissue Prepared | Tumor Cells (%) | Cancer Type | Previous Result | Alternate Method (s) | Input Material (s) | RNA Input-FFPE Scratched Slide | RNA Input-HE Scratched Slide | RNA Input-Other Materials | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA Conc. (ng/μL) | RNA Quality Score-DV 200 | RNA Input in the Cartridge-µL | RNA Conc. (ng/μL) | RNA Quality Score-DV 200 | RNA Input in the Cartridge-µL | RNA Conc. (ng/μL) | RNA Quality Score-DV 200 | RNA Input in the Cartridge-µL | |||||||
| 41 | 2020 | 30% | Neck level I-V -Oropharynx Carcinoma | ALK fusion ALK::EML4 | NGS; FISH | FFPE; RNA | 48.8 | 50–70% | 12 | 14.7 | 30–50% | 20 | N/A | N/A | N/A |
| 42 | 2020 | 70% | PTC | RET fusion RET::NCOA4 | NGS; FISH | FFPE; RNA | 350 | >70% | 6 | 46.4 | 50–70% | 25 | N/A | N/A | N/A |
| 43 | 2020 | 60% | NSCLC | ALK fusion ALK::EML4 | NGS; FISH | FFPE; RNA | 20.4 | 30–50% | 15 | 5.63 | 30–50% | 25 | N/A | N/A | N/A |
| 44 | 2019 | 90% | NSCLC | MET Ex14 Skipping | NGS; FISH | FFPE; RNA | 91.1 | 50–70% | 12 | 48.5 | 30–50% | 20 | N/A | N/A | N/A |
| 45 | 2019 | 80% | NSCLC | ROS1 fusion ROS1::SDC4 | NGS; FISH | FFPE; RNA | 12.5 | 30–50% | 25 | 10 | <30% | 25 | N/A | N/A | N/A |
| 46 | N/A | Horizon HD 784 | ALK, RET, ROS1 fusions EML4::ALK, CCDC6::RET and SLC34A2::ROS1 | NGS | RNA | N/A | N/A | N/A | N/A | N/A | N/A | 18.2 | >70% | 5.8 | |
| 47 | N/A | SeraSeq V4 RNA reference material | ALK, RET, ROS1, NTRK3, NTRK1 fusions; MET Ex14 Skipping | NGS | RNA | N/A | N/A | N/A | N/A | N/A | N/A | 10.2 | >70% | 9 | |
| 48 | N/A | CP-Positive Control RNA | ROS1 fusion SLC34A2::ROS1 | NGS | RNA | N/A | N/A | N/A | N/A | N/A | N/A | 4 | >70% | 13 | |
| 49 | 2014 | 90% | NSCLC | ALK fusion ALK::EML4 | NGS; FISH | FFPE | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 50 | 2010 | 50% | Leiomyosarkom | Wild-type | NGS; FISH | FFPE | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Case No. # | Alternative Method Result | Conclusion of Idylla Test-Mutation Detected YES/NO | Conclusion of Idylla Test-Cq Value−EGFR Control/NRAS Control/KRAS Target Cq Value/BRAF Target Cq Value | Concordance | Idylla Performance Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | A | B | C | D | E | ||||
| EGFR | |||||||||||||
| 1 | EGFR-Ex 19: p.L747_S7522del | YES | YES | YES | YES | YES | 18 | 19.6 | 19.5 | 20.7 | 18.5 | YES | 100% |
| EGFR-Ex 19:p.A755G | N/A | N/A | N/A | N/A | N/A | N/A | |||||||
| 2 | EGFR-Ex 20: p.T790M | YES | YES | YES | YES | YES | 19.7 | 19.5 | 20 | 22.6 | 21.9 | YES | 90% |
| EGFR-Ex 21:p.L858R | YES | NO | YES | YES | YES | Partial | |||||||
| 3 | EGFR-Ex 20:p.S768I | YES | YES | YES | YES | YES | 19.7 | 19.6 | 18.5 | 20.1 | 19.1 | YES | 100% |
| EGFR-Ex 21:p.L858R | YES | YES | YES | YES | YES | YES | |||||||
| 4 | EGFR-Ex 19: p.E746_T753>V | YES | YES | YES | YES | YES | 26.2 | 26.1 | 28.2 | 23.3 | 29.8 | YES | 100% |
| 5 | EGFR-Ex 19: p.L747_T751del | YES | YES | YES | YES | YES | 19.5 | 24.5 | 18.4 | 20.9 | 18.7 | YES | 100% |
| NRAS/BRAF | |||||||||||||
| 6 | BRAF-Ex 15:p.V600E | YES | NO | YES | YES | YES | 29.5 | 0 | 31 | 32.9 | 28.4 | Partial | 90% |
| NRAS-Ex 2-c:49delA | N/A | N/A | N/A | N/A | N/A | N/A | |||||||
| 7 | NRAS-Ex 3-p.Q61L | YES | NO | YES | YES | YES | 31.7 | 0 | 30.1 | 33.9 | 29.3 | Partial | 80% |
| BRAF-WT | YES | NO | YES | YES | YES | Partial | |||||||
| 8 | NRAS-Ex 3-p.Q61R | YES | NO | YES | YES | YES | 31.8 | 0 | 31.1 | 33.8 | 30.5 | Partial | 80% |
| BRAF-WT | YES | NO | YES | YES | YES | Partial | |||||||
| 9 | NRAS-Ex 3-p.Q61R | YES | NO | NO | NO | YES | 34.3 | 0 | 0 | 39.3 | 37.3 | Partial | 40% |
| BRAF-WT | YES | NO | NO | NO | YES | Partial | |||||||
| 10 | BRAF-Ex 15:p.V600E | YES | NO | YES | YES | YES | 26.9 | 0 | 30.6 | 37.6 | 28.4 | Partial | 80% |
| NRAS-WT | YES | NO | YES | YES | YES | Partial | |||||||
| KRAS | |||||||||||||
| 11 | KRAS-Ex 2:p.G12D/V | YES | YES | YES | YES | YES | 23.84 | 25.19 | 24.27 | 28.39 | 23 | YES | 100% |
| 12 | KRAS- Ex 2:p.G12A | YES | YES | YES | YES | YES | 25.33 | 29.45 | 25.57 | 31.68 | 23.98 | YES | 100% |
| 13 | KRAS- Ex 2:p.G13D | YES | YES | YES | YES | YES | 23.79 | 25.43 | 24.54 | 26.06 | 23.32 | YES | 100% |
| 14 | KRAS -Ex 4:p.A146T | YES | YES | YES | NO | YES | 24.8 | 26.54 | 24.38 | 0 | 23.95 | Partial | 90% |
| 15 | KRAS- Ex 2:p.G12R | YES | YES | YES | YES | YES | 24.93 | 28.01 | 26.98 | 31.5 | 24.28 | YES | 100% |
| BRAF | |||||||||||||
| 16 | BRAF-Ex 15:p.V600E | YES | YES | YES | YES | YES | 35.69 | 37.30 | 39.72 | 40.31 | 32.5 | YES | 100% |
| 17 | BRAF-Ex 15:p.V600K | YES | YES | YES | YES | YES | 33.47 | 37.57 | 34.57 | 37.77 | 33.56 | YES | 100% |
| 18 | BRAF-Ex 15:p.V600E | YES | YES | YES | YES | YES | 31.43 | 34.25 | 32.7 | 37.26 | 32.4 | YES | 100% |
| 19 | BRAF-Ex 15:p.V600E | YES | YES | YES | YES | YES | 30.75 | 34.12 | 33.5 | 35.47 | 32.3 | YES | 100% |
| 20 | BRAF-Ex 15:p.V600E | YES | YES | YES | YES | YES | 32.19 | 33.35 | 33.96 | 35.76 | 31.2 | YES | 100% |
| Case No. # | Alternative Method Result | Input Material (s) in Idylla Cartridge | Idylla-Sample MSI Status | Idylla Quality Status | ACVR2A | BTBD7 | DIDO1 | MRE11 | RYR3 | SEC31A | SULF2 | Concordance | Idylla Performance Analysis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | MSI High (5/10 System unstable) | A | MSI High (6/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 0.7 | 0.38 | 0.98 | 1 | 0.98 | YES | 100% |
| B | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 0.64 | 0.52 | 0.96 | 1 | 0.91 | YES | 100% | ||
| C | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 0.88 | 0.52 | 1 | 1 | 1 | YES | 100% | ||
| D | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 0.73 | 0.75 | 0.99 | 1 | 0.98 | YES | 100% | ||
| E | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 0.8 | 0.62 | 1 | 1 | 1 | YES | 100% | ||
| 21 | MSI High (3/10 System unstable) | A | MSI High (4/7 System unstable) | 7/7 biomarkers amplified | 1 | 0.92 | 0.79 | 0.4 | 0.15 | 0.15 | 0.62 | YES | 100% |
| B | MSI High (2/7 System unstable) | 6/7 biomarkers amplified | 1 | - | 0.8 | 0.25 | 0.03 | 0.19 | 0.5 | YES | 100% | ||
| C | MSI High (3/7 System unstable) | 7/7 biomarkers amplified | 1 | 0.89 | 0.74 | 0.32 | 0.12 | 0.13 | 0.33 | YES | 100% | ||
| D | MSI High (3/7 System unstable) | 7/7 biomarkers amplified | 0.98 | 0.79 | 0.75 | 0.32 | 0.09 | 0.1 | 0.22 | YES | 100% | ||
| E | MSI High (4/7 System unstable) | 7/7 biomarkers amplified | 1 | 0.92 | 0.79 | 0.4 | 0.15 | 0.15 | 0.62 | YES | 100% | ||
| 22 | MSS | A | MSS-No mutation detected (7/7 stable) | 7/7 biomarkers amplified | 0 | 0 | 0 | 0 | 0 | 0 | 0 | YES | 100% |
| B | MSS-No mutation detected (7/7 stable) | 7/7 biomarkers amplified | 0 | 0 | 0 | 0 | 0 | 0 | 0 | YES | 100% | ||
| C | MSS-No mutation detected (7/7 stable) | 7/7 biomarkers amplified | 0 | 0 | 0 | 0 | 0 | 0 | 0 | YES | 100% | ||
| D | MSS-No mutation detected (7/7 stable) | 7/7 biomarkers amplified | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | YES | 100% | ||
| E | MSS-No mutation detected (7/7 stable) | 7/7 biomarkers amplified | 0 | 0 | 0 | 0 | 0 | 0 | 0 | YES | 100% | ||
| 23 | MSI High (7/10 System unstable) | A | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 1 | 1 | 1 | 1 | 1 | YES | 100% |
| B | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 0.99 | 1 | 1 | 1 | 1 | YES | 100% | ||
| C | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 1 | 1 | 1 | 1 | 1 | YES | 100% | ||
| D | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 1 | 1 | 1 | 1 | 1 | YES | 100% | ||
| E | MSI High (7/7 System unstable) | 7/7 biomarkers amplified | 1 | 1 | 1 | 1 | 1 | 1 | 1 | YES | 100% | ||
| 24 | MSI High (8/10 System unstable) | A | MSI High (5/7 System unstable) | 7/7 biomarkers amplified | 0.98 | 0.37 | 0.02 | 0.95 | 0.98 | 1 | 0.99 | YES | 100% |
| B | MSI High (5/7 System unstable) | 6/7 biomarkers amplified | 1 | - | 0 | 0.61 | 0.81 | 1 | 1 | YES | 100% | ||
| C | MSI High (4/7 System unstable) | 7/7 biomarkers amplified | 1 | 0.26 | 0 | 1 | 0.21 | 1 | 1 | YES | 100% | ||
| D | MSI High (4/7 System unstable) | 7/7 biomarkers amplified | 1 | 0.24 | 0.02 | 0.99 | 0.15 | 1 | 0.99 | YES | 100% | ||
| E | MSI High (5/7 System unstable) | 7/7 biomarkers amplified | 0.99 | 0.22 | 0.01 | 0.98 | 0.67 | 1 | 1 | YES | 100% |
| Conclusion of Idylla Test | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case No. # | Alternative Method Result | Input Material (s) in Idylla Cartridge | Specific Fusion Detected YES/NO | Expression Imbalance Detected YES/NO | Cq Specific Fusion | ∆Cq Expression Imbalance-(3′–5′) or (3′-RNA Controls) | Cq of the RNA Controls | Cq of the DNA Controls | Invalid Result /Comments | Overall Concordance | Idylla Performance Analysis |
| 41 | ALK fusion ALK::EML4 | A | YES | YES | 27.7 | −6.4 | 26.5 | 27.9 | YES | 100% | |
| B | YES | YES | 32.4 | 1.7 | 32.5 | 30.4 | YES | 100% | |||
| F | YES | YES | 29.4 | −6.4 | 29.5 | 31.9 | YES | 100% | |||
| G | YES | NO | 32.7 | −5.9 | 32.6 | 33.3 | NTRK2-Invalid | Partial | 50% | ||
| 42 | RET fusion RET::NCOA4 | A | N/A | YES | N/A | −0.9 | 24.7 | 25.1 | YES | 100% | |
| B | N/A | YES | N/A | −1.2 | 26.8 | 27.1 | YES | 100% | |||
| F | N/A | YES | N/A | −1.1 | 25.9 | 27.8 | YES | 100% | |||
| G | N/A | YES | N/A | −1.1 | 29.3 | 30.5 | NTRK1-Invalid | YES | 100% | ||
| 43 | ALK fusion ALK::EML4 | A | YES | YES | 31.1 | 1.9 | 30.7 | 29.7 | YES | 100% | |
| B | YES | YES | 33.6 | 2 | 33 | 31.4 | YES | 100% | |||
| F | NO | NO | NO | NO | NO | 35.5 | INVALID Test results | NO | 0% | ||
| G | YES | YES | 32.5 | 0.4 | 32.1 | 33.2 | NTRK1-Invalid | YES | 100% | ||
| 44 | MET Ex14 Skipping | A | YES | N/A | 26.7 | N/A | 27.7 | 27.1 | YES | 100% | |
| B | YES | N/A | 25.9 | N/A | 27.3 | 27.4 | YES | 100% | |||
| F | YES | N/A | 27.1 | N/A | 28.6 | 30.6 | YES | 100% | |||
| G | YES | N/A | 27.3 | N/A | 29.3 | 31.6 | YES | 100% | |||
| 45 | ROS1 fusion ROS1::SDC4 | A | YES | YES | 27.6 | −1.3 | 28.8 | 27.7 | ALK exp. Imbl-Invalid | YES | 100% |
| B | YES | YES | 27.9 | −1.1 | 29 | 29.6 | YES | 100% | |||
| F | YES | YES | 30.2 | −1.1 | 31 | 32.4 | YES | 100% | |||
| G | YES | NO | 31 | −1.1 | 31.5 | 32.4 | ALK exp. Imbl-Indeterminate | Partial | 50% | ||
| 46 | ALK fusion ALK::EML4 | F | YES | YES | 30.3 | 1.2 | 30 | 32 | NTRK2 and NTRK3-Invalid | YES | 100% |
| RET fusion RET::CCDC6 | YES | NO | 28.6 | −0.6 | Partial | 50% | |||||
| ROS1 fusion SLC34A2::ROS1 | YES | YES | 31.3 | −4.7 | YES | 100% | |||||
| 47 | ALK fusion ALK::EML4 | F | YES | NO | 31.5 | −5.3 | 32.4 | 32 | ROS1 exp. Imbl/RET exp. Imbl/NTRK1 exp. Imbl not detected (invalid result) | Partial | 50% |
| RET fusion CCDC6::RET, KIF5B::RET, NCOA4::RET | YES | N/A | 31.9 | invalid | YES | 100% | |||||
| NTRK1 fusion LMNA::NTRK1, TFG::NTRK1, TPM3::NTRK1 | N/A | N/A | N/A | invalid | N/A | N/A | |||||
| NTRK3 fusion ETV6::NTRK3 | N/A | YES | N/A | −0.2 | YES | 100% | |||||
| ROS1 fusion SLC34A2::ROS1 CD74::ROS1 | YES | N/A | 31.2 | invalid | YES | 100% | |||||
| MET Ex14 Skipping | YES | N/A | 31.8 | N/A | YES | 100% | |||||
| 48 | ROS1 fusion SLC34A2::ROS1 | F | YES | YES | 30.8 | −3.2 | 28.6 | 31.1 | NTRK2 and NTRK3-Invalid | YES | 100% |
| 49 | ALK fusion ALK::EML4 | A | YES | YES | 25.8 | −3.5 | 25.4 | 25.9 | YES | 100% | |
| 50 | Wild-type Gene fusion not detected | A | YES | YES | N/A | N/A | 29.8 | 30.1 | ROS1 exp. Imbl-Invalid | YES | 100% |
| Case No. # | Alternative Method Result | Idylla Cartridge Used | Input Material (s) in Idylla Cartridge | Amount of Input Material | Conclusion of Idylla Test-Mutation Detected YES/NO | Conclusion of Idylla Test − CQ Value-Control/TARGET CQ Value | Overall Concordance | Idylla Performance Analysis |
|---|---|---|---|---|---|---|---|---|
| 25 | BRAF-WT | BRAF | FFPE-HE | 2× slides | NO | Invalid | NO | 0% |
| BRAF | FFPE-2 µM | 4× slides | YES | N/A | YES | 100% | ||
| 26 | NRAS-WT; BRAF-WT | NRAS-BRAF | FFPE-2 µM | 2× slides | YES | 34.6 | YES | 100% |
| gDNA-HE | 10 µL | YES | 38.5 | YES | 100% | |||
| 27 | EGFR-Ex18:p.G721S | EGFR | FFPE-HE | 2× slides | N/A | 29.3 | N/A | N/A |
| 28 | EGFR-Ex19: E746_S752delins | EGFR | FFPE-HE | 3× slides | YES | 23.7 | YES | 100% |
| 29 | EGFR-Ex21:p.L858R | EGFR | gDNA-FFPE | 15 µL | YES | 23 | YES | 100% |
| 30 | KRAS-Ex2:p.G12D | KRAS | FFPE-HE + FFPE-2 µM-broth combined | 1× HE slide + 2× slides FFPE | YES | 20.52 | YES | 100% |
| KRAS-Ex2:p.G12C | Present when raw data was examined. High CQ value −29.3 | NO | 0% | |||||
| 31 | BRAF-Ex15:p.V600E | BRAF | gDNA -HE | 10 µL | NO | Invalid | NO | 0% |
| BRAF | gDNA -HE | 25 µL | NO | Invalid | NO | 0% | ||
| 32 | EGFR-Ex19:PV742I | EGFR | Cytological fluid | 20 µL | N/A | 26.7 | N/A | N/A |
| KRAS-Ex2:G12D | KRAS | Cytological fluid | 20 µL and 500 µL | Present when raw data was examined... High CQ value −34.78 | NO | 0% | ||
| KRAS-Ex2:G12D | KRAS | gDNA from Cytological fluid | 40 µL | YES | 28.67 | YES | 100% | |
| KRAS-Ex2:G12D | KRAS | FFPE-gDNA. Cytoblock | 30 µL | YES | 25.3 | YES | 100% | |
| 33 | EGFR-WT | EGFR | Cytofluid pleural effusion | 20 µL | YES | 15.9 | YES | 100% |
| 34 | KRAS-Ex2:G12D | KRAS | Cytofluid pleural effusion | 500 µL | YES | 28.99 | YES | 100% |
| KRAS-Ex2:G12D | KRAS | gDNA from Cytological fluid | 40 µL | YES | 30.81 | YES | 100% | |
| 35 | BRAF-WT | BRAF | FFPE-2 µM | 2× slides | YES | N/A | YES | 100% |
| 36 | MSS | MSI | FFPE-2 µM | 2× slides | YES | 7/7 System Stabil | YES | 100% |
| 37 | NRAS-WT; BRAF-WT | NRAS-BRAF | FFPE-2 µM | 2× slides | YES | 32.7 | YES | 100% |
| 38 | MSS | MSI | HE + PAS-unstained and both combined | 2× slides | YES | 7/7 System Stabil | YES | 100% |
| 39 | KRAS-WT | KRAS | FFPE-2 µM | 2× slides | YES | N/A | YES | 100% |
| EGFR -WT | EGFR | FFPE-2 µM | 2× slides | YES | 18.9 | YES | 100% | |
| NRAS-WT; BRAF-WT | NRAS-BRAF | FFPE-2 µM | 2× slides | YES | 29.3 | YES | 100% | |
| 40 | EGFR-Ex20: p.T790M | EGFR | gDNA | 10 µL | YES | 21.4 | YES | 100% |
| EGFR-Ex19:p.E746_A750del | YES | YES | 100% | |||||
| EGFR-Ex21:p.L858R | YES | YES | 100% | |||||
| KRAS- Ex 2:p.G13D | KRAS | 10 µL | YES | 24.94 | YES | 100% | ||
| NRAS-WT BRAF-WT | NRAS-BRAF | 10 µL | YES | 33 | YES | 100% | ||
| BRAF-WT | BRAF | 10 µL | YES | N/A | YES | 100% | ||
| MSI-High | MSI | 10 µL | YES | 4/7 System unstable | YES | 100% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boppudi, S.M.; Scheil-Bertram, S.; Faust, E.; Annamneedi, A.; Fisseler-Eckhoff, A. Assessing and Evaluating the Scope and Constraints of Idylla Molecular Assays by Using Different Source Materials in Routine Diagnostic Settings. Int. J. Mol. Sci. 2022, 23, 12515. https://doi.org/10.3390/ijms232012515
Boppudi SM, Scheil-Bertram S, Faust E, Annamneedi A, Fisseler-Eckhoff A. Assessing and Evaluating the Scope and Constraints of Idylla Molecular Assays by Using Different Source Materials in Routine Diagnostic Settings. International Journal of Molecular Sciences. 2022; 23(20):12515. https://doi.org/10.3390/ijms232012515
Chicago/Turabian StyleBoppudi, Sanga Mitra, Stefanie Scheil-Bertram, Elisabeth Faust, Anil Annamneedi, and Annette Fisseler-Eckhoff. 2022. "Assessing and Evaluating the Scope and Constraints of Idylla Molecular Assays by Using Different Source Materials in Routine Diagnostic Settings" International Journal of Molecular Sciences 23, no. 20: 12515. https://doi.org/10.3390/ijms232012515
APA StyleBoppudi, S. M., Scheil-Bertram, S., Faust, E., Annamneedi, A., & Fisseler-Eckhoff, A. (2022). Assessing and Evaluating the Scope and Constraints of Idylla Molecular Assays by Using Different Source Materials in Routine Diagnostic Settings. International Journal of Molecular Sciences, 23(20), 12515. https://doi.org/10.3390/ijms232012515






