SOX2 Promotes Invasion in Human Bladder Cancers through MMP2 Upregulation and FOXO1 Downregulation
Abstract
1. Introduction
2. Results
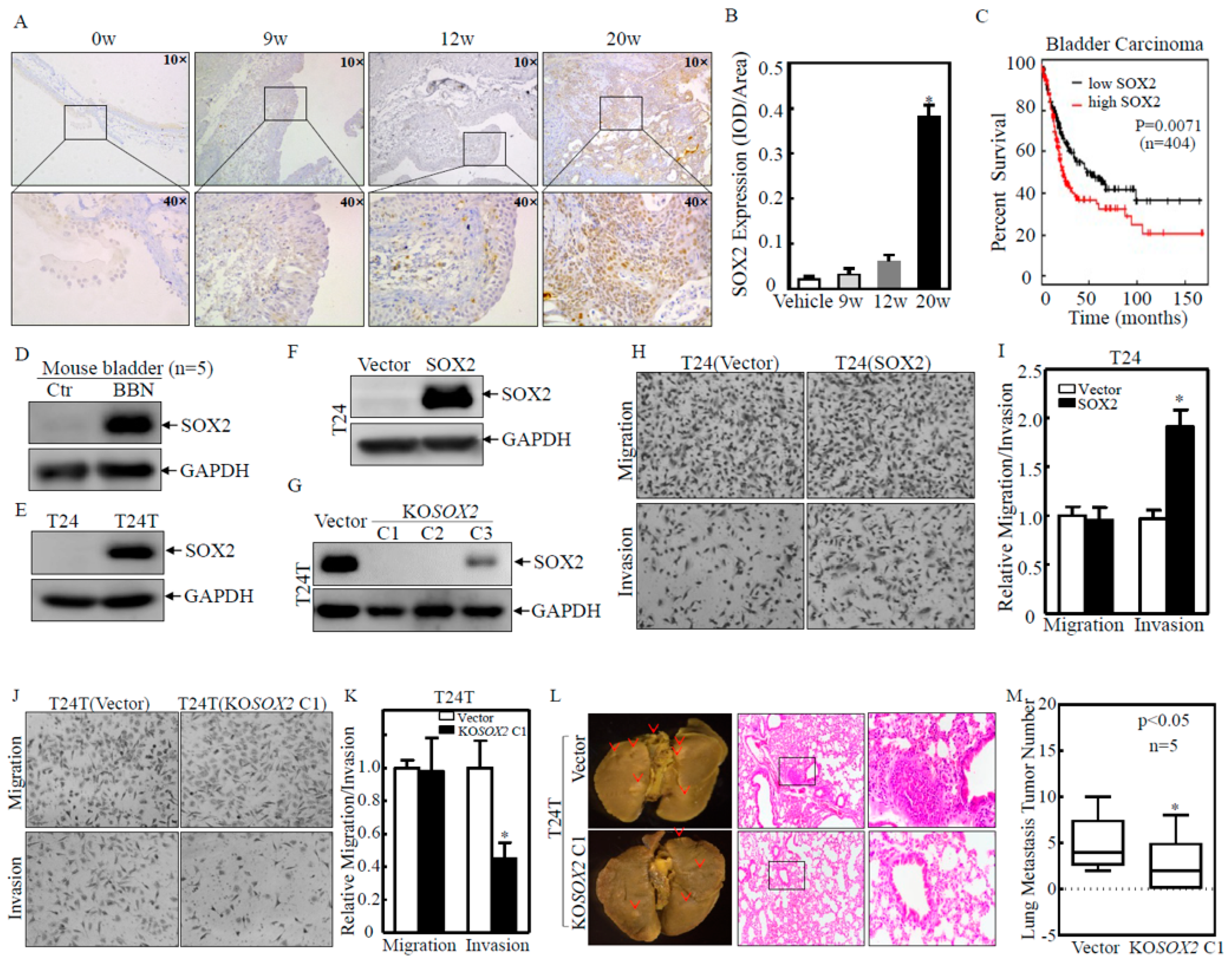
2.1. SOX2 was Overexpressed in Human and Mouse BC Tissues and Contributed to BC Cell Invasion
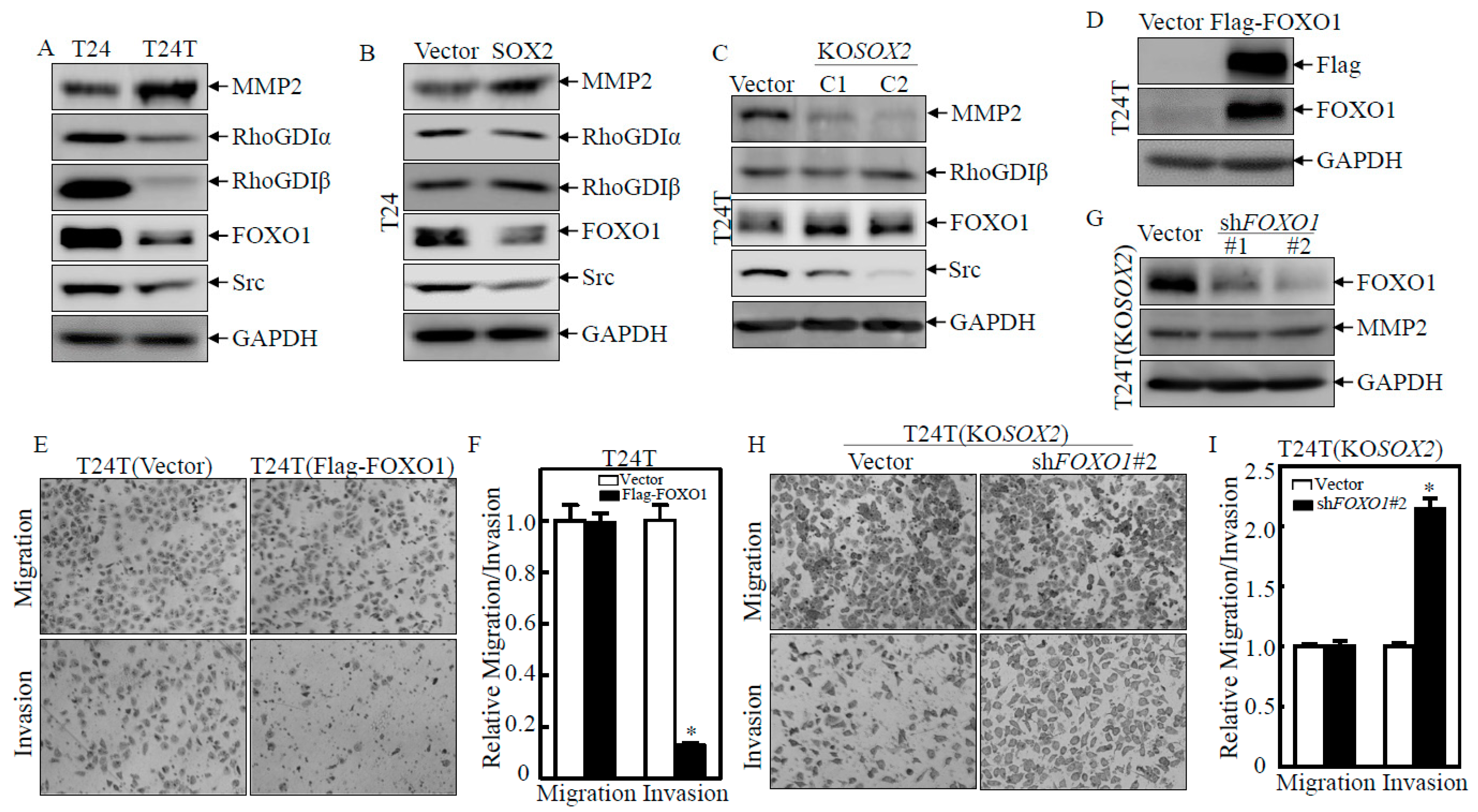
2.2. Upregulation of MMP2 and Downregulation of FOXO1 Play a Crucial Role in SOX2 Promoting the Invasive Ability of BC Cells
2.3. HUR Downregulation Mediated SOX2 Destabilization of FOXO1 mRNA
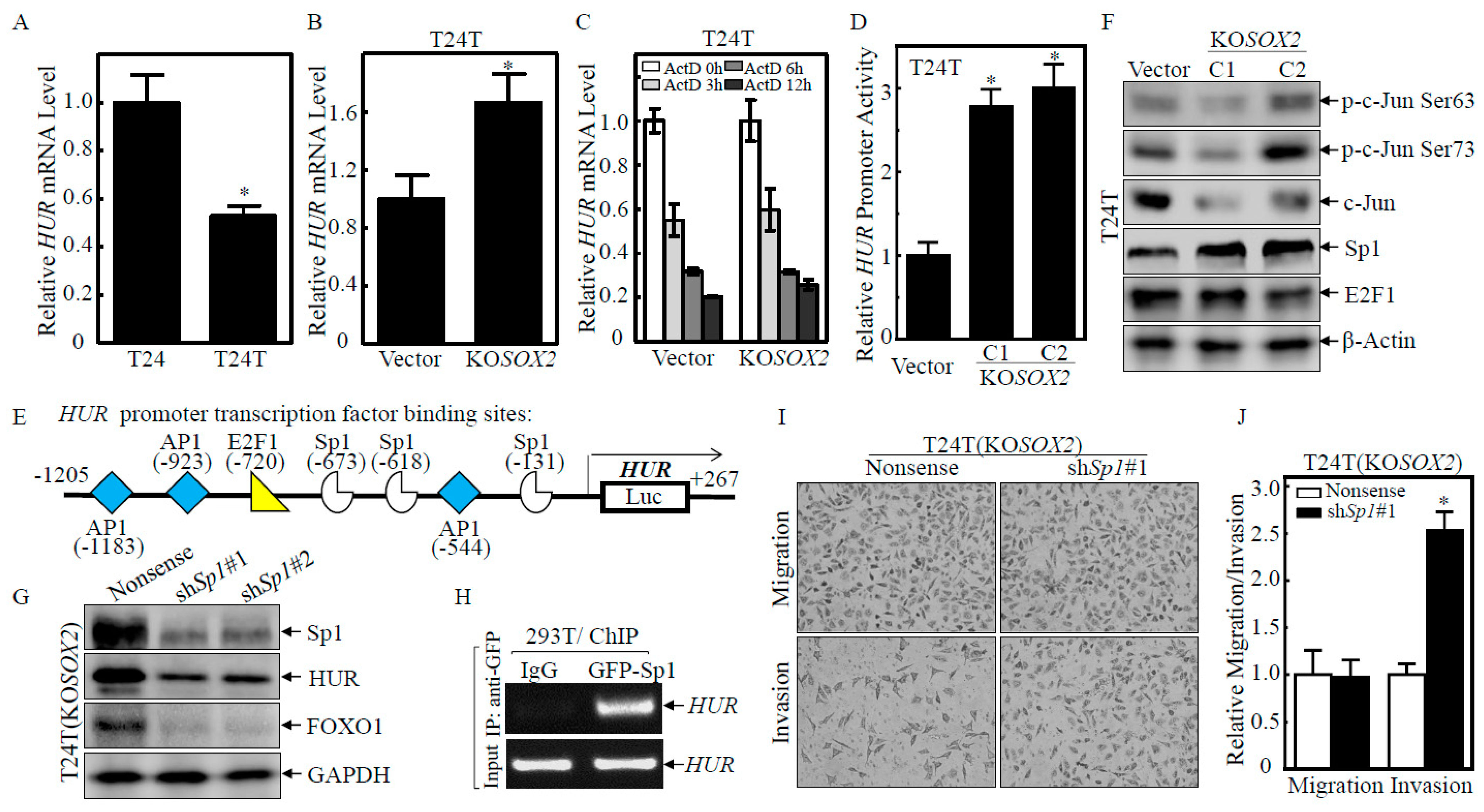
2.4. SOX2 Inhibited HUR Transcription by Downregulating Sp1 Protein Expression
2.5. SOX2-Promoted SKP2 Transcription and in Turn Mediated Sp1 Protein Degradation
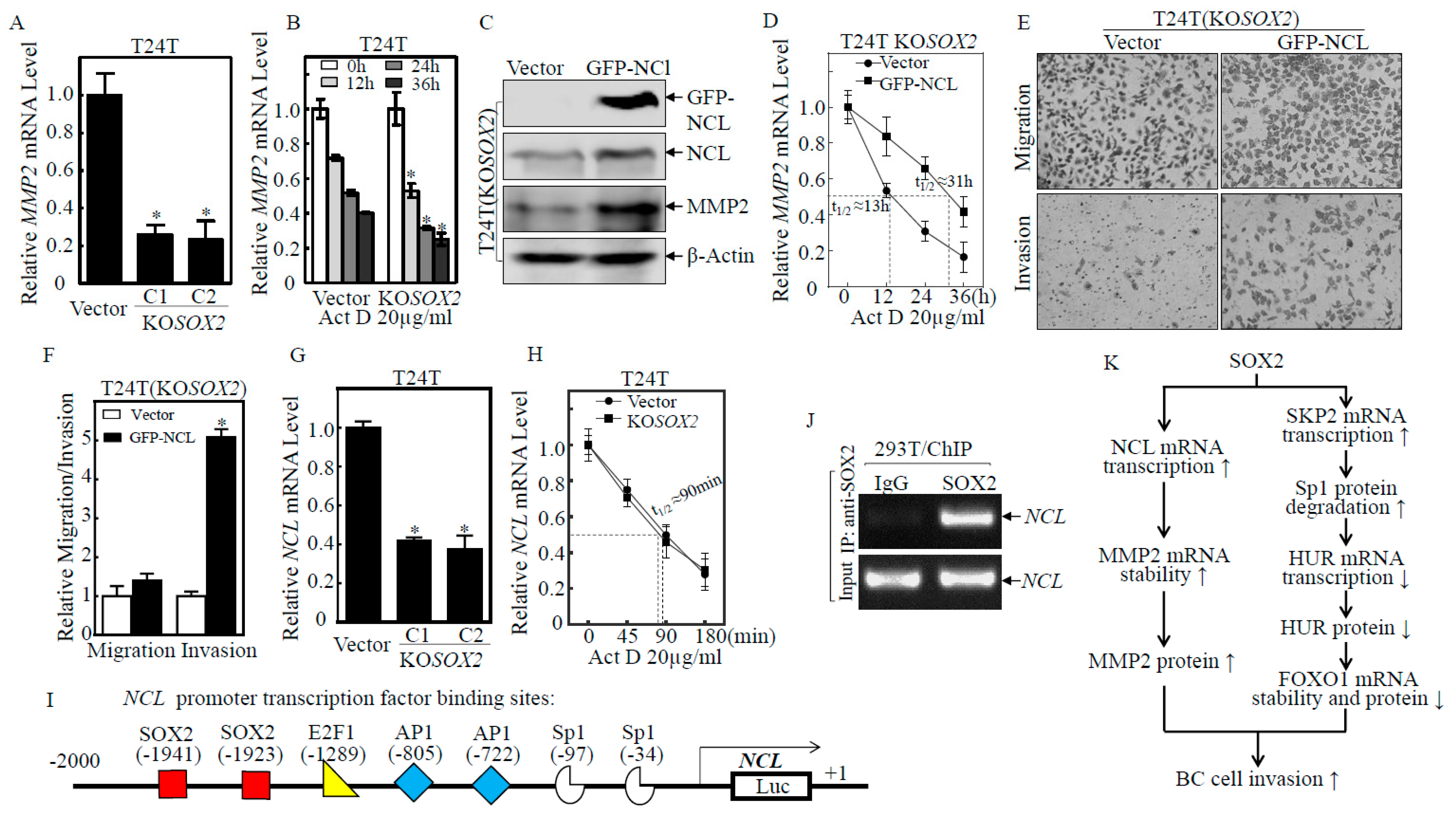
2.6. SOX2 Promoted the Transcription of NCL, which Increased MMP2 mRNA Stability and Protein Expression, Thereby Promoting BC Cell Invasion
3. Discussion
4. Materials and Methods
4.1. Reagents, Antibodies, and Plasmids
4.2. N-butyl-N-(4-hydroxybutyl) Nitrosamine-Induced Mouse Bladder Cancer Model and Immunohistochemistry (IHC)
4.3. Cell Culture and Transfection
4.4. Dual-Luciferase Reporter Assay
4.5. Real-Time Quantitative Reverse Transcription PCR (RT-qPCR)
4.6. Western Blot
4.7. mRNA Degradation Experiment
4.8. Cell Invasion Assay
4.9. RNA-IP Assay
4.10. Chromatin Immunoprecipitation Assay (ChIP)
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sjodahl, G.; Lauss, M.; Lovgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Ferno, M.; Ringner, M.; et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef] [PubMed]
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110–3115. [Google Scholar] [CrossRef]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef]
- Choi, W.; Czerniak, B.; Ochoa, A.; Su, X.; Siefker-Radtke, A.; Dinney, C.; McConkey, D.J. Intrinsic basal and luminal subtypes of muscle-invasive bladder cancer. Nat. Rev. Urol. 2014, 11, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Ferone, G.; Song, J.Y.; Sutherland, K.D.; Bhaskaran, R.; Monkhorst, K.; Lambooij, J.P.; Proost, N.; Gargiulo, G.; Berns, A. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell 2016, 30, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Leis, O.; Eguiara, A.; Lopez-Arribillaga, E.; Alberdi, M.J.; Hernandez-Garcia, S.; Elorriaga, K.; Pandiella, A.; Rezola, R.; Martin, A.G. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, I.V.; Edin, S.; Eklof, V.; Oberg, A.; Palmqvist, R.; Wikberg, M.L. SOX2 expression is associated with a cancer stem cell state and down-regulation of CDX2 in colorectal cancer. BMC Cancer 2016, 16, 471. [Google Scholar] [CrossRef]
- Ferreira-Teixeira, M.; Parada, B.; Rodrigues-Santos, P.; Alves, V.; Ramalho, J.S.; Caramelo, F.; Sousa, V.; Reis, F.; Gomes, C.M. Functional and molecular characterization of cancer stem-like cells in bladder cancer: A potential signature for muscle-invasive tumors. Oncotarget 2015, 6, 36185–36201. [Google Scholar] [CrossRef]
- Chen, X.; Xie, R.; Gu, P.; Huang, M.; Han, J.; Dong, W.; Xie, W.; Wang, B.; He, W.; Zhong, G.; et al. Long Noncoding RNA lbcs Inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin. Cancer Res. 2019, 25, 1389–1403. [Google Scholar] [CrossRef] [PubMed]
- Girouard, S.D.; Laga, A.C.; Mihm, M.C.; Scolyer, R.A.; Thompson, J.F.; Zhan, Q.; Widlund, H.R.; Lee, C.W.; Murphy, G.F. SOX2 contributes to melanoma cell invasion. Lab. Investig. 2012, 92, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Han, X.; Jin, C.; Tian, W.; Yu, W.; Ding, D.; Cheng, L.; Huang, B.; Jiang, H.; Lin, B. SOX2 targets fibronectin 1 to promote cell migration and invasion in ovarian cancer: New molecular leads for therapeutic intervention. OMICS 2013, 17, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Sun, L.; Li, Y.; Kang, X.; Zhang, S.; Liu, Y. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating Slug. Med. Oncol. 2013, 30, 503. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Hui, L.; Wang, Y.; Yang, H.; Jiang, X. SOX2 promotes the migration and invasion of laryngeal cancer cells by induction of MMP-2 via the PI3K/Akt/mTOR pathway. Oncol. Rep. 2014, 31, 2651–2659. [Google Scholar] [CrossRef]
- Jiang, G.; Wu, A.D.; Huang, C.; Gu, J.; Zhang, L.; Huang, H.; Liao, X.; Li, J.; Zhang, D.; Zeng, X.; et al. Isorhapontigenin (ISO) inhibits invasive bladder cancer formation in vivo and human bladder cancer invasion in vitro by targeting STAT1/FOXO1 axis. Cancer Prev. Res. 2016, 9, 567–580. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Seraj, M.J.; Harding, M.A.; Gildea, J.J.; Welch, D.R.; Theodorescu, D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin. Exp. Metastasis 2000, 18, 519–525. [Google Scholar] [CrossRef]
- Yamada, T.; Nagahama, M.; Akimitsu, N. Interplay between transcription and RNA degradation. In Gene Expression and Regulation in Mammalian Cells-Transcription From General Aspects; InTech: London, UK, 2018. [Google Scholar]
- Yu, Y.; Jin, H.; Xu, J.; Gu, J.; Li, X.; Xie, Q.; Huang, H.; Li, J.; Tian, Z.; Jiang, G.; et al. XIAP overexpression promotes bladder cancer invasion in vitro and lung metastasis in vivo via enhancing nucleolin-mediated Rho-GDIbeta mRNA stability. Int. J. Cancer 2018, 142, 2040–2055. [Google Scholar] [CrossRef]
- Pan, H.; Strickland, A.; Madhu, V.; Johnson, Z.I.; Chand, S.N.; Brody, J.R.; Fertala, A.; Zheng, Z.; Shapiro, I.M.; Risbud, M.V. RNA binding protein HuR regulates extracellular matrix gene expression and pH homeostasis independent of controlling HIF-1alpha signaling in nucleus pulposus cells. Matrix Biol. 2019, 77, 23–40. [Google Scholar] [CrossRef]
- Li, X.; Tian, Z.; Jin, H.; Xu, J.; Hua, X.; Yan, H.; Liufu, H.; Wang, J.; Li, J.; Zhu, J. Decreased c-Myc mRNA stability via the microRNA 141–3p/AUF1 axis is crucial for p63α inhibition of cyclin D1 gene transcription and bladder cancer cell tumorigenicity. Mol. Cell. Biol. 2018, 38, e00273-18. [Google Scholar] [CrossRef]
- Burger, A.M.; Seth, A.K. The ubiquitin-mediated protein degradation pathway in cancer: Therapeutic implications. Eur. J. Cancer 2004, 40, 2217–2229. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Vucic, D. Ubiquitin ligases in cancer: Ushers for degradation. Cancer Investig. 2007, 25, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Barral, J.M.; Broadley, S.A.; Schaffar, G.; Hartl, F.U. Roles of Molecular Chaperones in Protein Misfolding Diseases, Seminars in Cell & Developmental Biology, 2004; Elsevier: Amsterdam, The Netherlands, 2004; pp. 17–29. [Google Scholar]
- Jin, H.; Yu, Y.; Hu, Y.; Lu, C.; Li, J.; Gu, J.; Zhang, L.; Huang, H.; Zhang, D.; Wu, X.R.; et al. Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget 2015, 6, 522–536. [Google Scholar] [CrossRef]
- Sarkar, A.; Hochedlinger, K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 2013, 12, 15–30. [Google Scholar] [CrossRef]
- Stolzenburg, S.; Rots, M.G.; Beltran, A.S.; Rivenbark, A.G.; Yuan, X.; Qian, H.; Strahl, B.D.; Blancafort, P. Targeted Silencing of the Oncogenic Transcription Factor SOX2 in Breast Cancer; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Ji, J.; Zheng, P.S. Expression of Sox2 in human cervical carcinogenesis. Hum. Pathol. 2010, 41, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Fang, X.; Lou, X.; Hua, D.; Ding, W.; Foltz, G.; Hood, L.; Yuan, Y.; Lin, B. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PLoS ONE 2012, 7, e41335. [Google Scholar] [CrossRef]
- Hussenet, T.; Dali, S.; Exinger, J.; Monga, B.; Jost, B.; Dembele, D.; Martinet, N.; Thibault, C.; Huelsken, J.; Brambilla, E.; et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS ONE 2010, 5, e8960. [Google Scholar] [CrossRef]
- Kitamura, H.; Torigoe, T.; Hirohashi, Y.; Asanuma, H.; Inoue, R.; Nishida, S.; Tanaka, T.; Fukuta, F.; Masumori, N.; Sato, N.; et al. Prognostic impact of the expression of ALDH1 and SOX2 in urothelial cancer of the upper urinary tract. Mod. Pathol. 2013, 26, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Migita, T.; Ueda, A.; Ohishi, T.; Hatano, M.; Seimiya, H.; Horiguchi, S.I.; Koga, F.; Shibasaki, F. Epithelial-mesenchymal transition promotes SOX2 and NANOG expression in bladder cancer. Lab. Investig. 2017, 97, 567. [Google Scholar] [CrossRef]
- Zhu, F.; Qian, W.; Zhang, H.; Liang, Y.; Wu, M.; Zhang, Y.; Zhang, X.; Gao, Q.; Li, Y. SOX2 Is a marker for stem-like tumor cells in bladder cancer. Stem Cell Rep. 2017, 9, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Eijkelenboom, A.; Burgering, B.M. FOXOs: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013, 14, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Link, W.; Fernandez-Marcos, P.J. FOXO transcription factors at the interface of metabolism and cancer. Int. J. Cancer 2017, 141, 2379–2391. [Google Scholar] [CrossRef]
- Stone, R.C.; Pastar, I.; Ojeh, N.; Chen, V.; Liu, S.; Garzon, K.I.; Tomic-Canic, M. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 2016, 365, 495–506. [Google Scholar] [CrossRef]
- Jablonska, B.; Scafidi, J.; Aguirre, A.; Vaccarino, F.; Nguyen, V.; Borok, E.; Horvath, T.L.; Rowitch, D.H.; Gallo, V. Oligodendrocyte regeneration after neonatal hypoxia requires FoxO1-mediated p27Kip1 expression. J. Neurosci. 2012, 32, 14775–14793. [Google Scholar] [CrossRef]
- Melnik, B.C. FoxO1–the key for the pathogenesis and therapy of acne? J. Dtsch. Dermatol. Ges. 2010, 8, 105–114. [Google Scholar] [CrossRef]
- Cheng, C.; Jiao, J.T.; Qian, Y.; Guo, X.Y.; Huang, J.; Dai, M.C.; Zhang, L.; Ding, X.P.; Zong, D.; Shao, J.F. Curcumin induces G2/M arrest and triggers apoptosis via FoxO1 signaling in U87 human glioma cells. Mol. Med. Rep. 2016, 13, 3763–3770. [Google Scholar] [CrossRef]
- Matsuzaki, H.; Lee, S.; Maeda, M.; Kumagai-Takei, N.; Nishimura, Y.; Otsuki, T. FoxO1 regulates apoptosis induced by asbestos in the MT-2 human T-cell line. J. Immunotoxicol. 2016, 13, 620–627. [Google Scholar] [CrossRef]
- Huang, H.; Tindall, D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007, 120, 2479–2487. [Google Scholar] [CrossRef]
- Yang, Y.; Blee, A.M.; Wang, D.; An, J.; Pan, Y.; Yan, Y.; Ma, T.; He, Y.; Dugdale, J.; Hou, X.; et al. Loss of FOXO1 cooperates with TMPRSS2-ERG overexpression to promote prostate tumorigenesis and cell invasion. Cancer Res. 2017, 77, 6524–6537. [Google Scholar] [CrossRef]
- Jiang, G.; Huang, C.; Li, J.; Huang, H.; Jin, H.; Zhu, J.; Wu, X.R.; Huang, C. Role of STAT3 and FOXO1 in the divergent therapeutic responses of non-metastatic and metastatic bladder cancer cells to miR-145. Mol. Cancer Ther. 2017, 16, 924–935. [Google Scholar] [CrossRef]
- Hao, Z.L.; Huang, S. E3 ubiquitin ligase Skp2 as an attractive target in cancer therapy. Front. Biosci. Landmark 2015, 20, 474–490. [Google Scholar] [CrossRef]
- Ruan, D.; He, J.; Li, C.F.; Lee, H.J.; Liu, J.; Lin, H.K.; Chan, C.H. Skp2 deficiency restricts the progression and stem cell features of castration-resistant prostate cancer by destabilizing Twist. Oncogene 2017, 36, 4299–4310. [Google Scholar] [CrossRef]
- Kitagawa, K.; Kotake, Y.; Kitagawa, M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci. 2009, 100, 1374–1381. [Google Scholar] [CrossRef]
- Kossatz, U.; Dietrich, N.; Zender, L.; Buer, J.; Manns, M.P.; Malek, N.P. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Gene Dev. 2004, 18, 2602–2607. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Nagahama, H.; Minamishima, Y.A.; Miyake, S.; Ishida, N.; Hatakeyama, S.; Kitagawa, M.; Iemura, S.; Natsume, T.; Nakayama, K.I. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev. Cell 2004, 6, 661–672. [Google Scholar] [CrossRef]
- Liang, Y.; Hou, X.; Cui, Q.; Kang, T.B.; Fu, J.H.; Zhang, L.J.; Luo, R.Z.; He, J.H.; Zeng, Y.X.; Yang, H.X. Skp2 expression unfavorably impacts survival in resectable esophageal squamous cell carcinoma. J. Transl. Med. 2012, 10, 73. [Google Scholar] [CrossRef]
- Wang, X.; Ji, X.; Chen, J.; Yan, D.; Zhang, Z.; Wang, Q.; Xi, X.; Feng, Y. SOX2 enhances the migration and invasion of ovarian cancer cells via Src kinase. PLoS ONE 2014, 9, e99594. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.M.; Sun, W.; Wang, Z.H.; Liang, X.; Hua, F.; Li, K.; Lv, X.X.; Zhang, X.W.; Liu, Y.Y.; Yu, J.J.; et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat. Commun. 2019, 10, 5720. [Google Scholar] [CrossRef]
- Ormsbee Golden, B.D.; Wuebben, E.L.; Rizzino, A. Sox2 expression is regulated by a negative feedback loop in embryonic stem cells that involves AKT signaling and FoxO1. PLoS ONE 2013, 8, e76345. [Google Scholar]
- Chan, C.H.; Lee, S.W.; Li, C.F.; Wang, J.; Yang, W.L.; Wu, C.Y.; Wu, J.; Nakayama, K.I.; Kang, H.Y.; Huang, H.Y.; et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat. Cell Biol. 2010, 12, 457–467. [Google Scholar] [CrossRef]
- Huang, H.J.; Regan, K.M.; Lou, Z.K.; Chen, J.J.; Tindall, D.J. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 2006, 314, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Absalon, M.J.; McLure, K.G.; Kastan, M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005, 123, 49–63. [Google Scholar] [CrossRef]
- Essaghir, A.; Dif, N.; Marbehant, C.Y.; Coffer, P.J.; Demoulin, J.B. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J. Biol. Chem. 2009, 284, 10334–10342. [Google Scholar] [CrossRef]
- Yang, R.; Xu, J.; Hua, X.; Tian, Z.; Xie, Q.; Li, J.; Jiang, G.; Cohen, M.; Sun, H.; Huang, C. Overexpressed miR-200a promotes bladder cancer invasion through direct regulating Dicer/miR-16/JNK2/MMP-2 axis. Oncogene 2020, 39, 1983–1996. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Xie, Q.; Guo, X.; Xu, J.; Wang, A.; Li, J.; Zhu, J.; Wu, X.-R.; Huang, H.; Huang, C. p63α protein up-regulates heat shock protein 70 expression via E2F1 transcription factor 1, promoting Wasf3/Wave3/MMP9 signaling and bladder cancer invasion. J. Biol. Chem. 2017, 292, 15952–15963. [Google Scholar] [CrossRef]
- Zhang, D.; Li, J.; Zhang, M.; Gao, G.; Zuo, Z.; Yu, Y.; Zhu, L.; Gao, J.; Huang, C. The requirement of c-Jun N-terminal kinase 2 in regulation of hypoxia-inducing factor-1α mRNA stability. J. Biol. Chem. 2012, 287, 34361–34371. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, D.; Huang, H.; Li, J.; Zhang, M.; Wan, Y.; Gao, J.; Huang, C. NF-κB1 p50 promotes p53 protein translation through miR-190 downregulation of PHLPP1. Oncogene 2014, 33, 996–1005. [Google Scholar] [CrossRef]
- Xie, Q.; Guo, X.; Gu, J.; Zhang, L.; Jin, H.; Huang, H.; Li, J.; Huang, C. p85α promotes nucleolin transcription and subsequently enhances EGFR mRNA stability and EGF-induced malignant cellular transformation. Oncotarget 2016, 7, 16636–16649. [Google Scholar] [CrossRef]
- Sun, W.; Li, S.; Yu, Y.; Jin, H.; Xie, Q.; Hua, X.; Wang, S.; Tian, Z.; Zhang, H.; Jiang, G.; et al. MicroRNA-3648 Is upregulated to suppress TCF21, resulting in promotion of invasion and metastasis of human bladder cancer. Mol. Ther. Nucleic Acids 2019, 16, 519–530. [Google Scholar] [CrossRef]
- Zhu, J.; Tian, Z.; Li, Y.; Hua, X.; Zhang, D.; Li, J.; Jin, H.; Xu, J.; Chen, W.; Niu, B.; et al. ATG7 Promotes bladder cancer invasion via autophagy-mediated increased ARHGDIB mRNA stability. Adv. Sci. 2019, 6, 1801927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liang, Y.; Xie, Q.; Gao, G.; Wei, J.; Huang, H.; Li, J.; Gao, J.; Huang, C. A novel post-translational modification of nucleolin, SUMOylation at Lys-294, mediates arsenite-induced cell death by regulating gadd45α mRNA stability. J. Biol. Chem. 2015, 290, 4784–4800. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Q.; Hua, X.; Huang, C.; Liao, X.; Tian, Z.; Xu, J.; Zhao, Y.; Jiang, G.; Huang, H.; Huang, C. SOX2 Promotes Invasion in Human Bladder Cancers through MMP2 Upregulation and FOXO1 Downregulation. Int. J. Mol. Sci. 2022, 23, 12532. https://doi.org/10.3390/ijms232012532
Xie Q, Hua X, Huang C, Liao X, Tian Z, Xu J, Zhao Y, Jiang G, Huang H, Huang C. SOX2 Promotes Invasion in Human Bladder Cancers through MMP2 Upregulation and FOXO1 Downregulation. International Journal of Molecular Sciences. 2022; 23(20):12532. https://doi.org/10.3390/ijms232012532
Chicago/Turabian StyleXie, Qipeng, Xiaohui Hua, Chao Huang, Xin Liao, Zhongxian Tian, Jiheng Xu, Yunping Zhao, Guosong Jiang, Haishan Huang, and Chuanshu Huang. 2022. "SOX2 Promotes Invasion in Human Bladder Cancers through MMP2 Upregulation and FOXO1 Downregulation" International Journal of Molecular Sciences 23, no. 20: 12532. https://doi.org/10.3390/ijms232012532
APA StyleXie, Q., Hua, X., Huang, C., Liao, X., Tian, Z., Xu, J., Zhao, Y., Jiang, G., Huang, H., & Huang, C. (2022). SOX2 Promotes Invasion in Human Bladder Cancers through MMP2 Upregulation and FOXO1 Downregulation. International Journal of Molecular Sciences, 23(20), 12532. https://doi.org/10.3390/ijms232012532






