Lipid Droplets in Lung Cancers Are Crucial for the Cell Growth and Starvation Survival
Abstract
:1. Introduction
2. Results
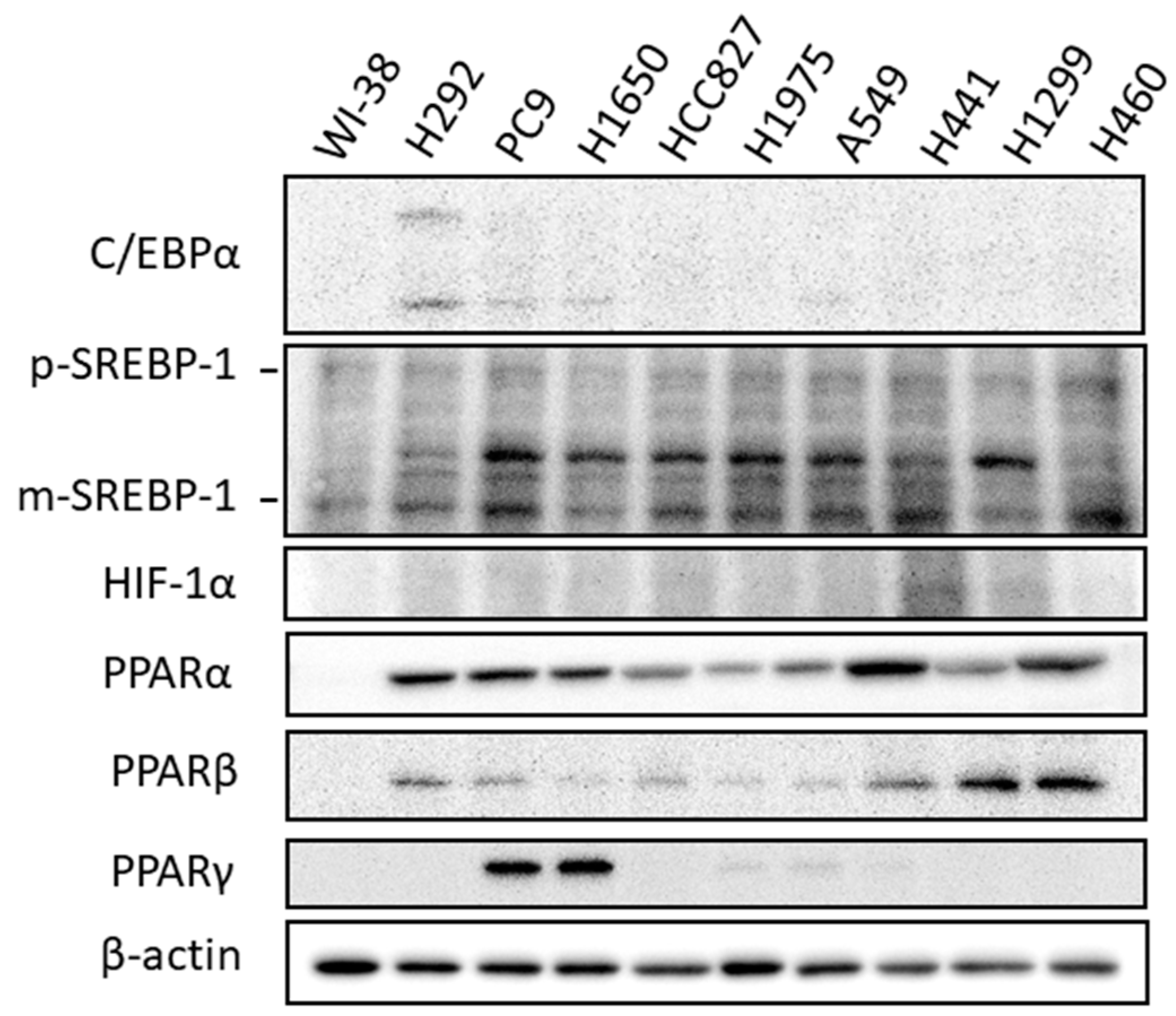
2.1. Expression of Molecules Involved in Lipid Biosynthesis and Uptake in Lung Cancer Cells
2.2. Expression of Transcriptional Regulators Related to Lipid Biosynthesis
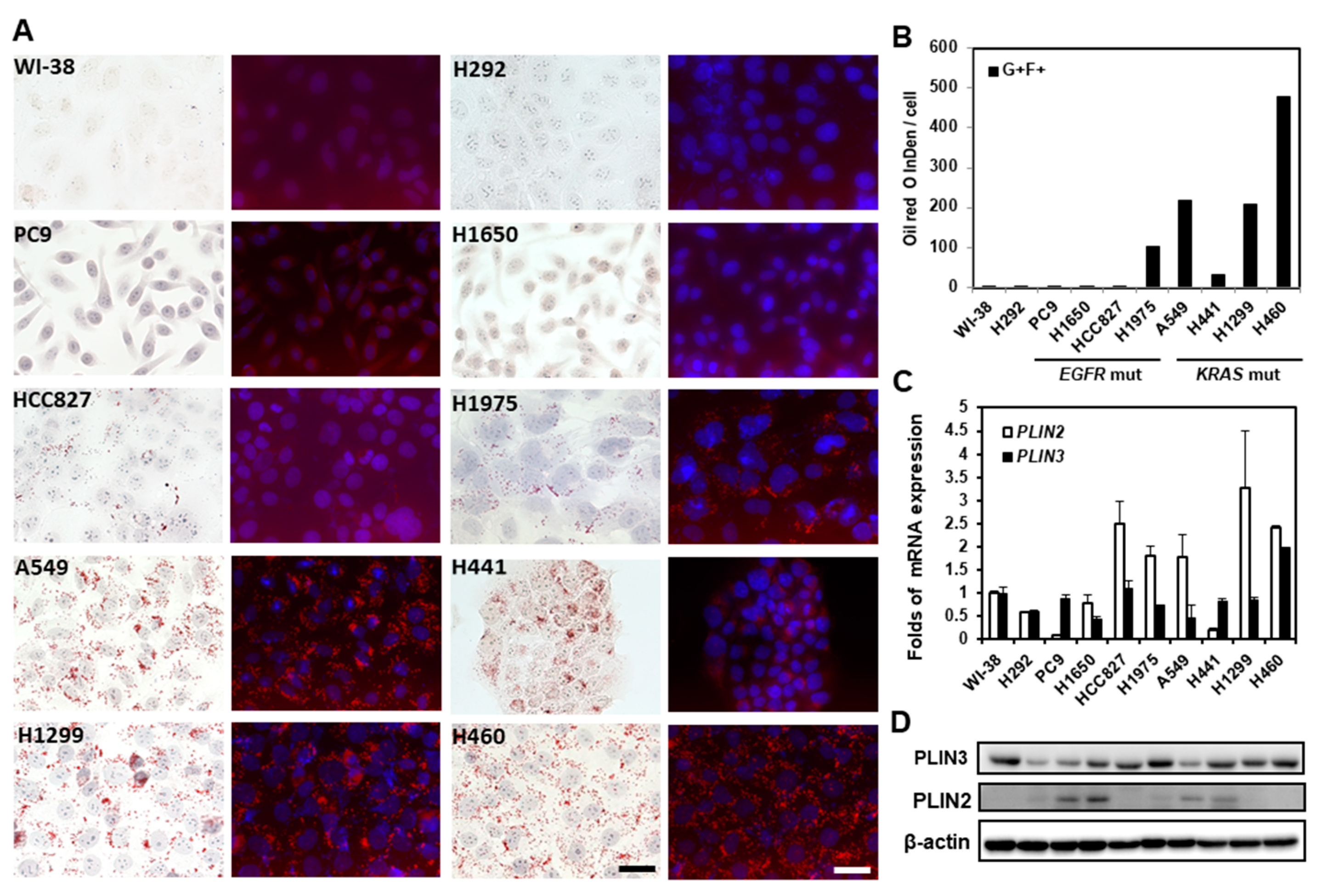
2.3. Lipid Droplets in Lung Cancer Cells
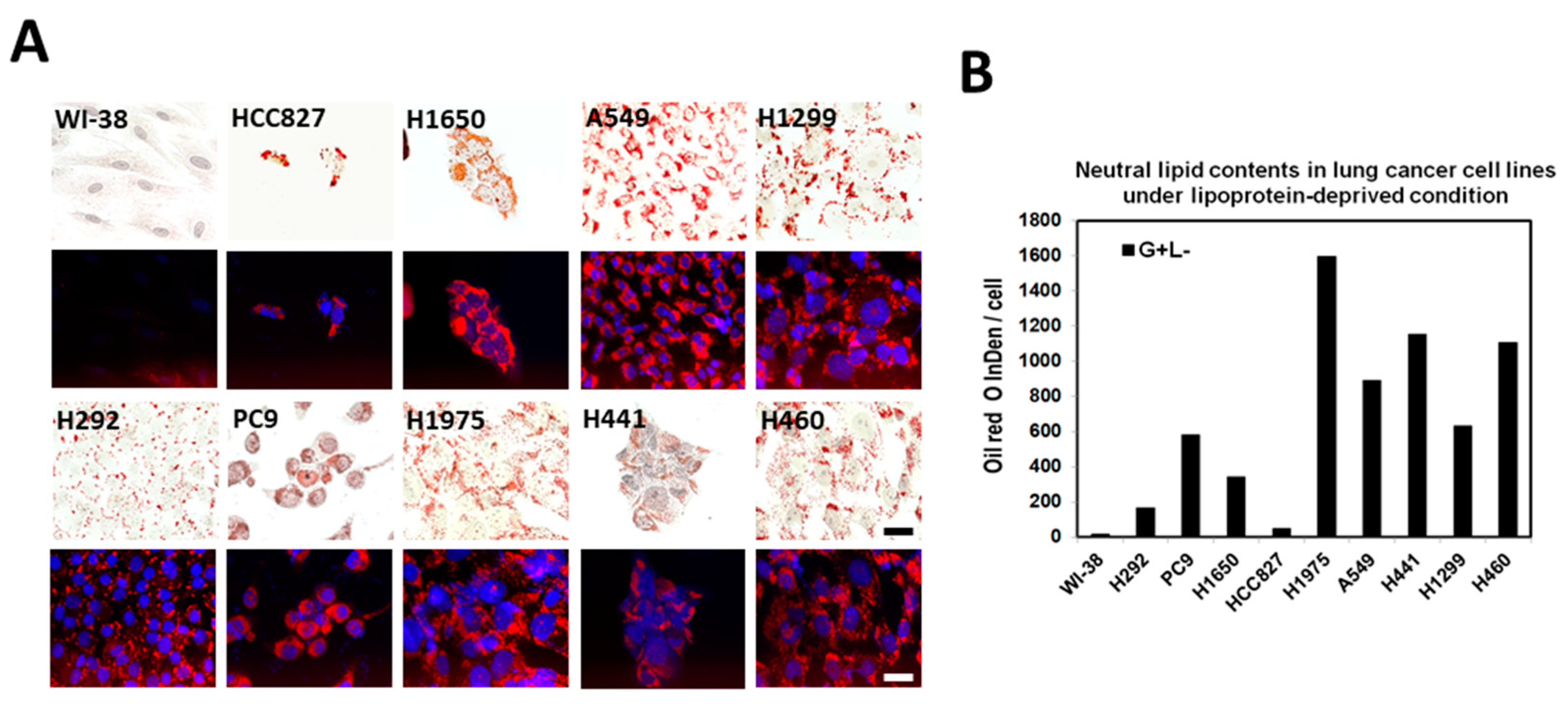
2.4. Lipid Droplet Formation Induced in Lung Cancer Cells under Lipoprotein-Deficient Growth Medium
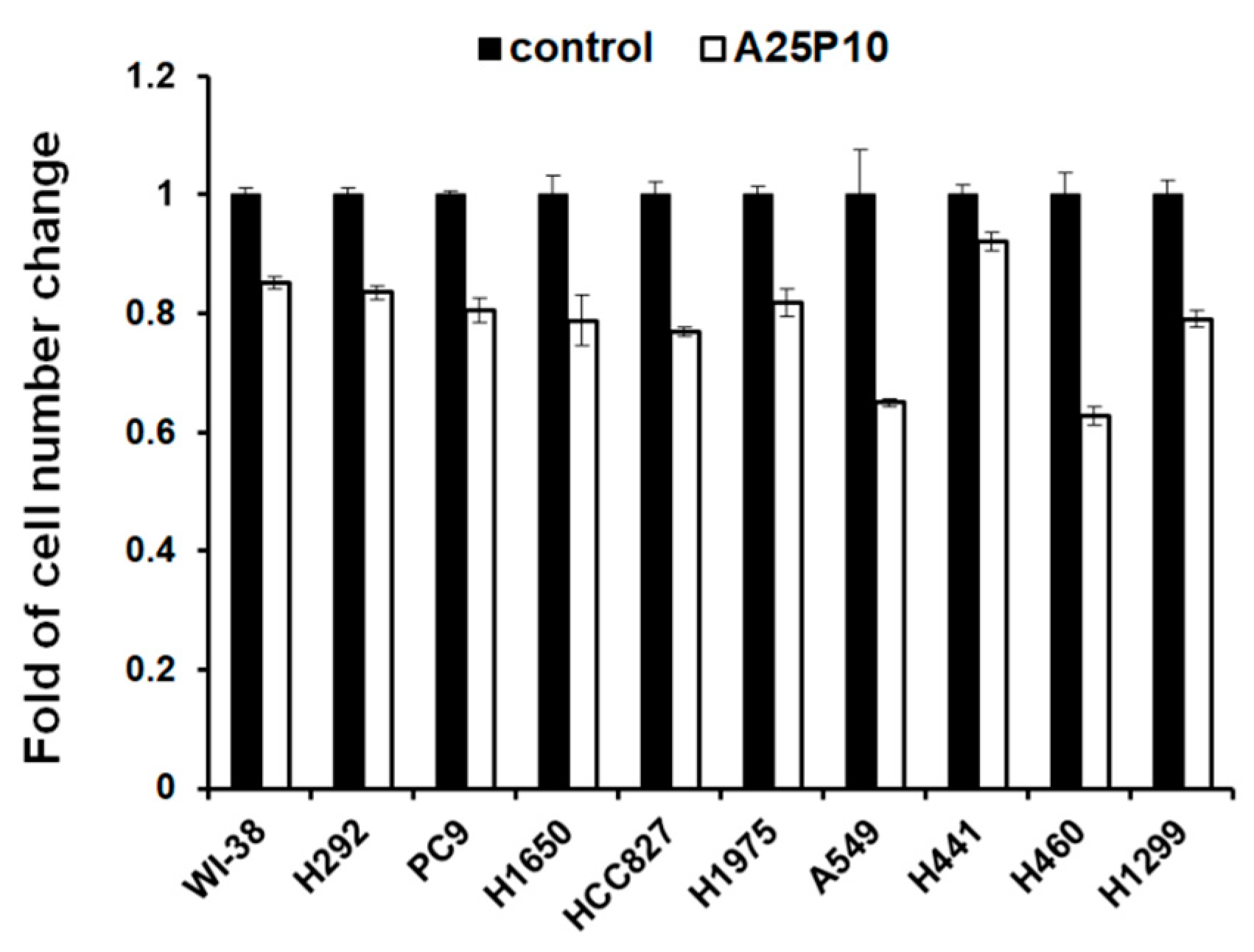
2.5. Blocking of Lipid Drops’ Formation Slows down the Cell Proliferation of the Lung Cancer Cells
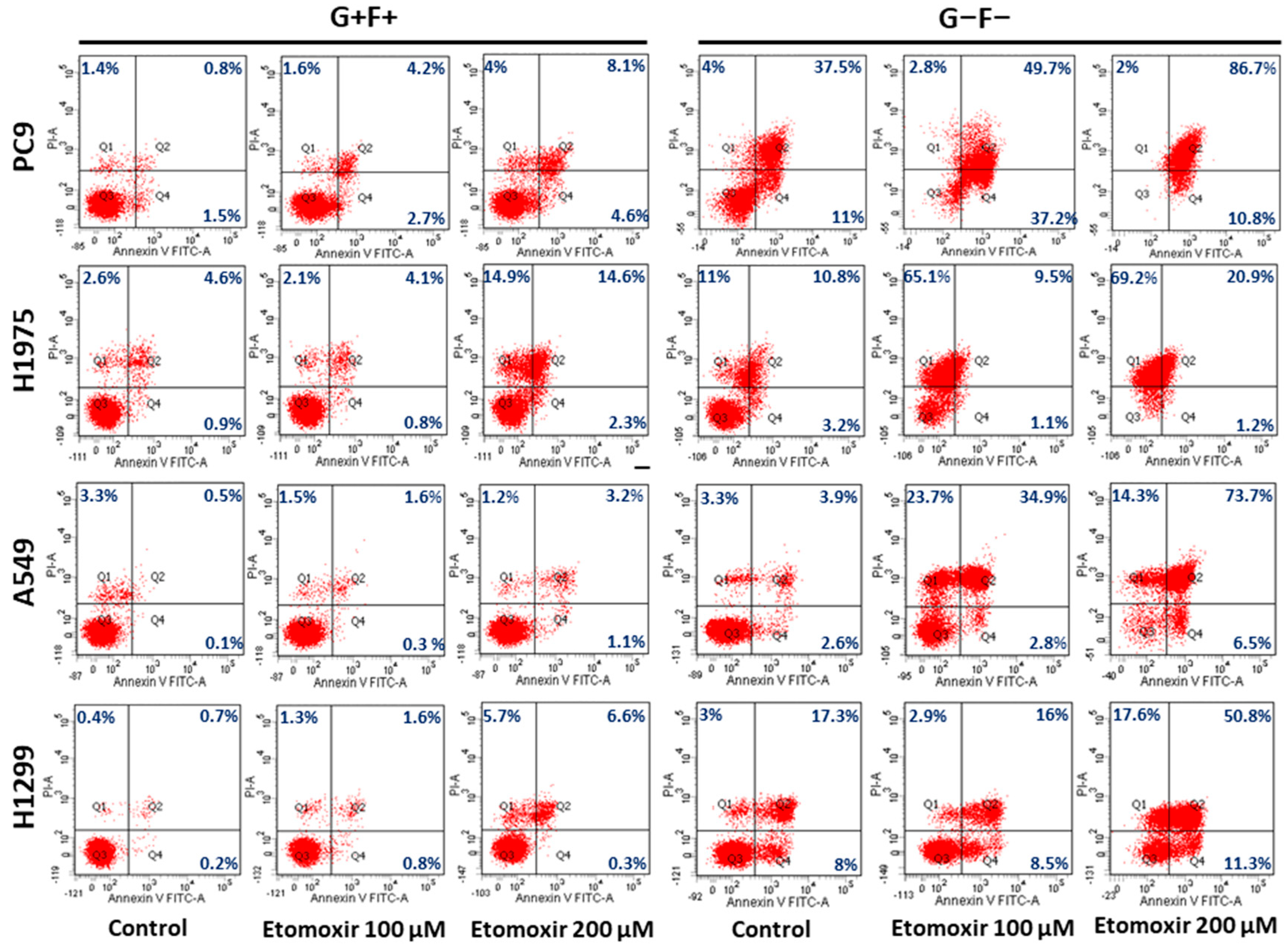
2.6. Lipid Droplets Increase Survival of Lung Cancer Cells under Starvation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Oil Red O Staining and Quantitation
4.3. Annexin V/propidium Iodide (PI) Staining Assay
4.4. Immunoblot Analysis
4.5. Chemicals and Antibodies
4.6. Immunohistochemistry
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structuRes. and tools. Biochim. Biophys. Acta 2011, 1811, 637–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, L.P.; Gomez de Cedron, M.; Ramirez de Molina, A. Alterations of Lipid Metabolism in Cancer: Implications in Prognosis and Treatment. Front. Oncol. 2020, 10, 577420. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, M.; Braun, S.M.; Zurkirchen, L.; von Schoultz, C.; Zamboni, N.; Arauzo-Bravo, M.J.; Kovacs, W.J.; Karalay, O.; Suter, U.; Machado, R.A.; et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 2013, 493, 226–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Models Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Rohrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Amemiya-Kudo, M.; Shimano, H.; Hasty, A.H.; Yahagi, N.; Yoshikawa, T.; Matsuzaka, T.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Ohashi, K.; et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J. Lipid Res. 2002, 43, 1220–1235. [Google Scholar] [CrossRef] [Green Version]
- Engin, A.B. What Is Lipotoxicity? Adv. Exp. Med. Biol. 2017, 960, 197–220. [Google Scholar]
- Zhang, P.; Reue, K. Lipin proteins and glycerolipid metabolism: Roles at the ER membrane and beyond. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1583–1595. [Google Scholar] [CrossRef]
- Itabe, H.; Yamaguchi, T.; Nimura, S.; Sasabe, N. Perilipins: A diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017, 16, 83. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Gao, Q.; Goodman, J.M. The lipid droplet-a well-connected organelle. Front. Cell Dev. Biol. 2015, 3, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koizume, S.; Miyagi, Y. Lipid Droplets: A Key Cellular Organelle Associated with Cancer Cell Survival under Normoxia and Hypoxia. Int. J. Mol. Sci. 2016, 17, 1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.W.; Cravatt, B.F. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 2010, 140, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniels, V.W.; Machiels, J.; et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef] [Green Version]
- Zhang, I.; Cui, Y.; Amiri, A.; Ding, Y.; Campbell, R.E.; Maysinger, D. Pharmacological inhibition of lipid droplet formation enhances the effectiveness of curcumin in glioblastoma. Eur. J. Pharm. Biopharm. 2016, 100, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Kuramoto, K.; Okamura, T.; Yamaguchi, T.; Nakamura, T.Y.; Wakabayashi, S.; Morinaga, H.; Nomura, M.; Yanase, T.; Otsu, K.; Usuda, N.; et al. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 2012, 287, 23852–23863. [Google Scholar] [CrossRef] [Green Version]
- Orita, H.; Coulter, J.; Lemmon, C.; Tully, E.; Vadlamudi, A.; Medghalchi, S.M.; Kuhajda, F.P.; Gabrielson, E. Selective inhibition of fatty acid synthase for lung cancer treatment. Clin. Cancer Res. 2007, 13, 7139–7145. [Google Scholar] [CrossRef] [Green Version]
- Espirito Santo, S.M.; Rensen, P.C.; Goudriaan, J.R.; Bensadoun, A.; Bovenschen, N.; Voshol, P.J.; Havekes, L.M.; van Vlijmen, B.J. Triglyceride-rich lipoprotein metabolism in unique VLDL receptor, LDL receptor, and LRP triple-deficient mice. J. Lipid Res. 2005, 46, 1097–1102. [Google Scholar] [CrossRef] [Green Version]
- Roy-Chowdhury, N.; Roy-Chowdhury, J. Chapter 72—Liver Physiology and Energy Metabolism. In Sleisenger and Fordtran’s Gastrointestinal and Liver Disease (Ninth Edition); Feldman, M., Friedman, L.S., Brandt, L.J., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2010; pp. 1207–1225.e3. [Google Scholar]
- Bergen, W.G.; Burnett, D.D. Topics in transcriptional control of lipid metabolism: From transcription factors to gene-promoter polymorphisms. J. Genom. 2013, 1, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Franco, C.; Britto, K.; Wong, E.; Hou, G.; Zhu, S.N.; Chen, M.; Cybulsky, M.I.; Bendeck, M.P. Discoidin domain receptor 1 on bone marrow-derived cells promotes macrophage accumulation during atherogenesis. Circ. Res. 2009, 105, 1141–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; DeBose-Boyd, R.A. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb. Perspect. Biol. 2011, 3, a004754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, R.J.; Krueger, E.W.; Weller, S.G.; Johnson, K.M.; Casey, C.A.; Schott, M.B.; McNiven, M.A. Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc. Natl. Acad. Sci. USA 2020, 117, 32443–32452. [Google Scholar] [CrossRef]
- Yen, C.L.; Stone, S.J.; Koliwad, S.; Harris, C.; Farese, R.V., Jr. Thematic review series: Glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008, 49, 2283–2301. [Google Scholar] [CrossRef] [Green Version]
- Buckley, D.; Duke, G.; Heuer, T.S.; O’Farrell, M.; Wagman, A.S.; McCulloch, W.; Kemble, G. Fatty acid synthase—Modern tumor cell biology insights into a classical oncology target. Pharmacol. Ther. 2017, 177, 23–31. [Google Scholar] [CrossRef]
- Fhu, C.W.; Ali, A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules 2020, 25, 3935. [Google Scholar] [CrossRef]
- Marks, J.L.; Broderick, S.; Zhou, Q.; Chitale, D.; Li, A.R.; Zakowski, M.F.; Kris, M.G.; Rusch, V.W.; Azzoli, C.G.; Seshan, V.E.; et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J. Thorac. Oncol. 2008, 3, 111–116. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.M.; Zhu, Q.G.; Ding, X.X.; Lin, S.; Zhao, J.; Guan, L.; Li, T.; He, B.; Zhang, H.Q. Prognostic value of EGFR and KRAS in resected non-small cell lung cancer: A systematic review and meta-analysis. Cancer Manag. Res. 2018, 10, 3393–3404. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, K.; Fukasaka, Y.; Yoshimoto, R.; Nambu, H.; Yukioka, H. Diacylglycerol acyltransferase 1/2 inhibition induces dysregulation of fatty acid metabolism and leads to intestinal barrier failure and diarrhea in mice. Physiol. Rep. 2020, 8, e14542. [Google Scholar] [CrossRef]
- Kichenadasse, G.; Miners, J.O.; Mangoni, A.A.; Rowland, A.; Hopkins, A.M.; Sorich, M.J. Association Between Body Mass Index and Overall Survival With Immune Checkpoint Inhibitor Therapy for Advanced Non-Small Cell Lung Cancer. JAMA Oncol. 2020, 6, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Laird, B.J.A.; Skipworth, R.J.E. The Obesity Paradox in Cancer: Is Bigger Better? J. Cachexia Sarcopenia Muscle 2022, 13, 1440–1441. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Tran, T.M.; Ware, T.B.; Luse, M.A.; Prevost, C.T.; Ferguson, A.N.; Kashatus, J.A.; Hsu, K.L.; Kashatus, D.F. RalA and PLD1 promote lipid droplet growth in response to nutrient withdrawal. Cell Rep. 2021, 36, 109451. [Google Scholar] [CrossRef] [PubMed]
- Larigauderie, G.; Cuaz-Perolin, C.; Younes, A.B.; Furman, C.; Lasselin, C.; Copin, C.; Jaye, M.; Fruchart, J.C.; Rouis, M. Adipophilin increases triglyceride storage in human macrophages by stimulation of biosynthesis and inhibition of beta-oxidation. FEBS J. 2006, 273, 3498–3510. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lung, J.; Hung, M.-S.; Wang, T.-Y.; Chen, K.-L.; Luo, C.-W.; Jiang, Y.-Y.; Wu, S.-Y.; Lee, L.-W.; Lin, P.-Y.; Chen, F.-F.; et al. Lipid Droplets in Lung Cancers Are Crucial for the Cell Growth and Starvation Survival. Int. J. Mol. Sci. 2022, 23, 12533. https://doi.org/10.3390/ijms232012533
Lung J, Hung M-S, Wang T-Y, Chen K-L, Luo C-W, Jiang Y-Y, Wu S-Y, Lee L-W, Lin P-Y, Chen F-F, et al. Lipid Droplets in Lung Cancers Are Crucial for the Cell Growth and Starvation Survival. International Journal of Molecular Sciences. 2022; 23(20):12533. https://doi.org/10.3390/ijms232012533
Chicago/Turabian StyleLung, Jrhau, Ming-Szu Hung, Ting-Yao Wang, Kuan-Liang Chen, Chi-Wen Luo, Yuan-Yuan Jiang, Shin-Yi Wu, Li-Wen Lee, Paul-Yann Lin, Fen-Fen Chen, and et al. 2022. "Lipid Droplets in Lung Cancers Are Crucial for the Cell Growth and Starvation Survival" International Journal of Molecular Sciences 23, no. 20: 12533. https://doi.org/10.3390/ijms232012533





