Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia
Abstract
:1. Introduction
2. Results
2.1. Predicted Functional Outputs for BC-Associated SNPs
2.1.1. Non-Synonymous (nsSNP) and Regulatory (regSNP) Impact
2.1.2. Expression (eSNP) and Splicing (sSNP) Impact
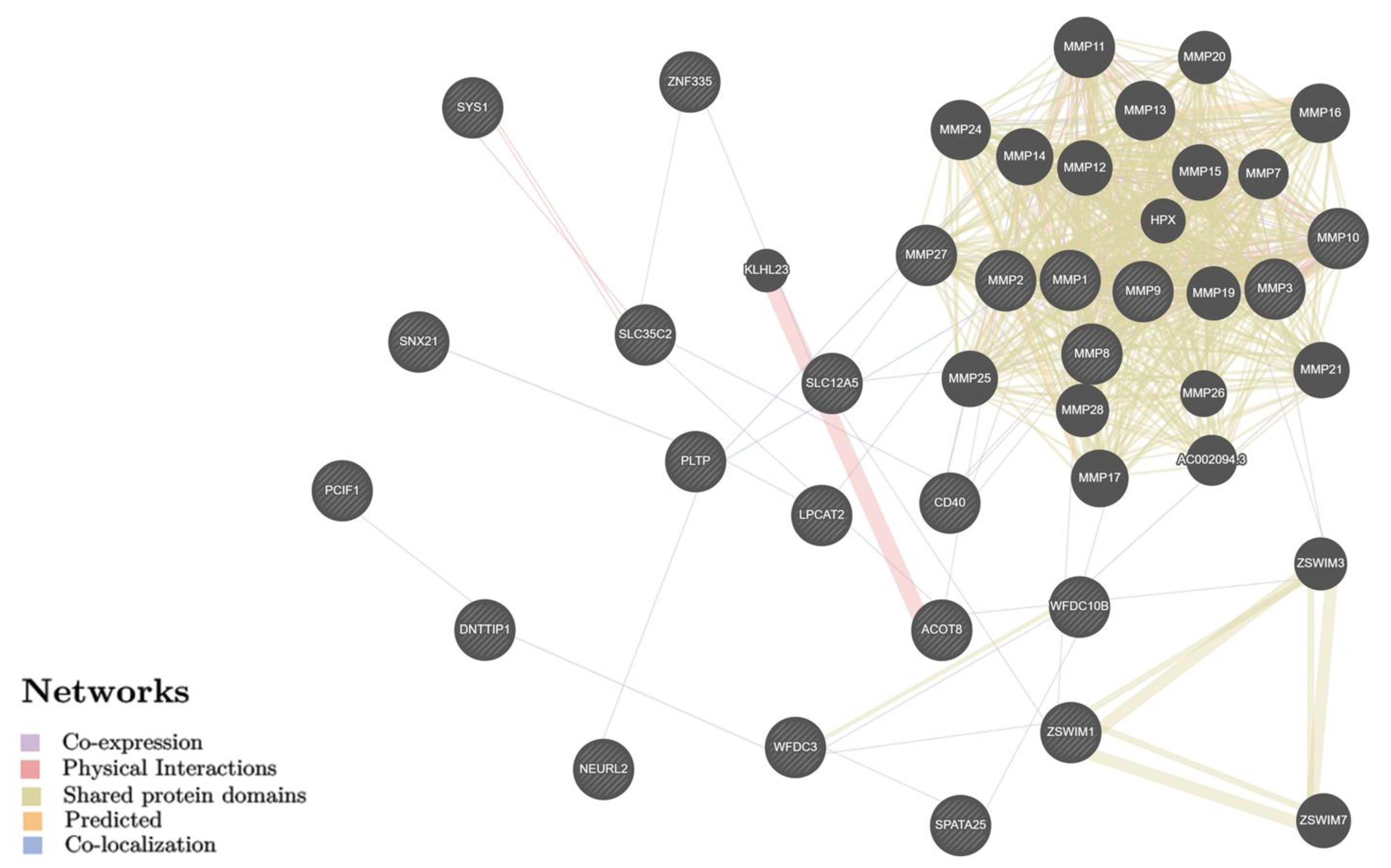
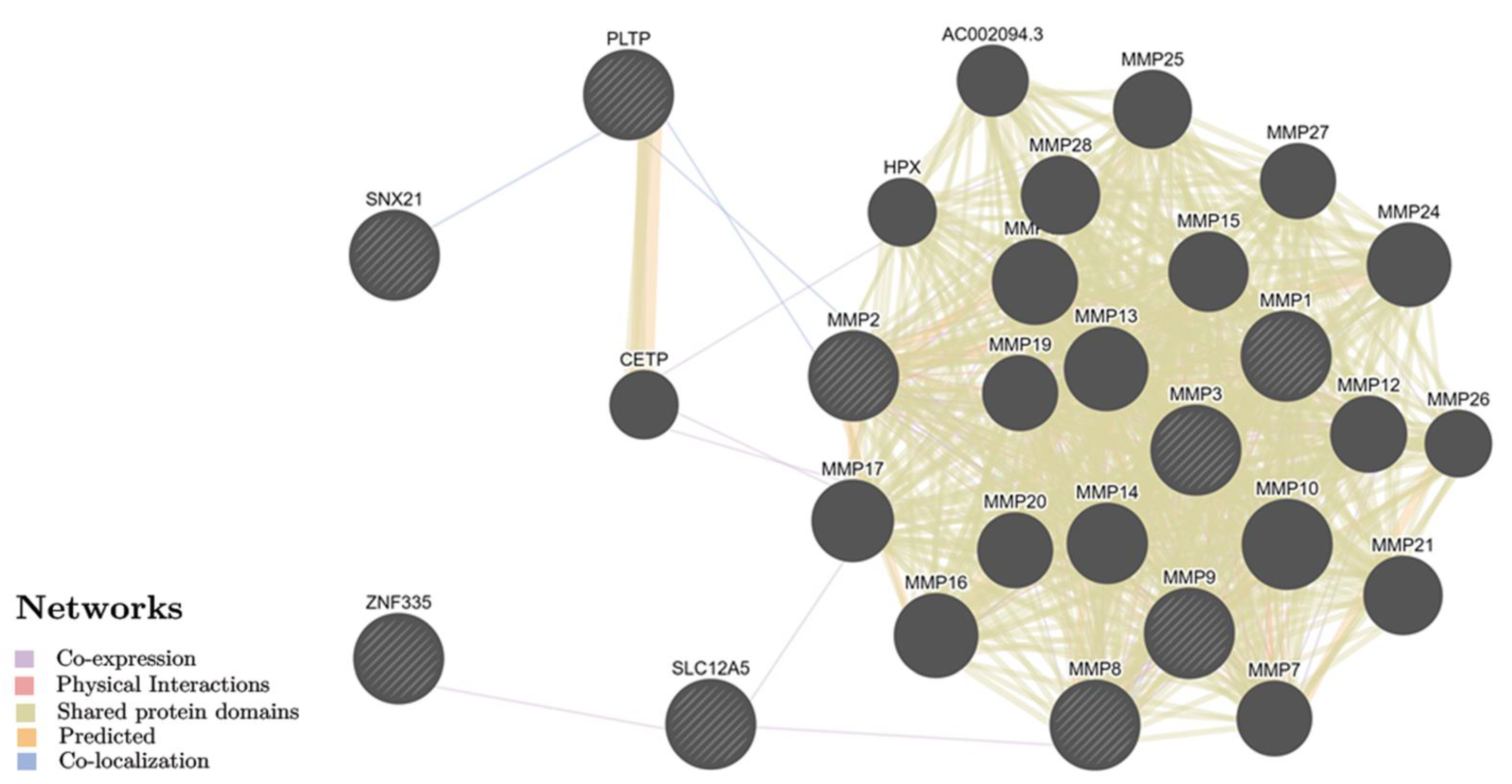
2.2. Identification of Biological Pathways for BC Putative Target Genes
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. SNP Selection and Genotyping
4.3. Statistical Analysis
4.4. SNPs and Genes Predict Functions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gradishar, W.J.; Anderson, B.O.; Blair, S.L.; Burstein, H.J.; Cyr, A.; Elias, A.D.; Farrar, W.B.; Forero, A.; Giordano, S.H.; Goldstein, L.J.; et al. National Comprehensive Cancer Network Breast Cancer Panel. Breast cancer version 3.2014. J. Natl. Compr. Canc. Netw. 2014, 12, 542–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Federal State Statistics Service. Healthcare in Russia 2021: Statistical Compendium; Federal State Statistics Service: Moscow, Russia, 2021; 171p.
- Kaprin, A.D.; Starinskij, V.V.; Petrova, G.V. Malignant Neoplasms in Russia in 2018; MNIOI im. P.A. Gercena: Moscow, Russia, 2019; 250p. (In Russian) [Google Scholar]
- Lilyquist, J.; Ruddy, K.J.; Vachon, C.M.; Couch, F.J. Common Genetic Variation and Breast Cancer Risk-Past, Present, and Future. Cancer Epidemiol. Biomark. Prev. 2018, 27, 380–394. [Google Scholar] [CrossRef] [Green Version]
- Shiovitz, S.; Korde, L.A. Genetics of breast cancer: A topic in evolution. Ann. Oncol. 2015, 26, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.ebi.ac.uk/gwas/search?query=breast%20carcinoma (accessed on 18 June 2022).
- Michailidou, K.; Lindström, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemaçon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef] [Green Version]
- Mucci, L.A.; Hjelmborg, J.B.; Harris, J.R.; Czene, K.; Havelick, D.J.; Scheike, T.; Graff, R.E.; Holst, K.; Möller, S.; Unger, R.H.; et al. Nordic Twin Study of Cancer (NorTwinCan) Collaboration. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries [published correction appears in JAMA. 2016 Feb 23;315(8):822]. JAMA 2016, 315, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Dofara, S.G.; Chang, S.L.; Diorio, C. Gene polymorphisms and circulating levels of MMP-2 and MMP-9: A review of their role in breast cancer risk. Anticancer Res. 2020, 40, 3619–3631. [Google Scholar] [CrossRef]
- Przybylowska, K.; Kluczna, A.; Zadrozny, M.; Krawczyk, T.; Kulig, A.; Rykala, J.; Kolacinska, A.; Morawiec, Z.; Drzewoski, J.; Blasiak, J. Polymorphisms of the promoter regions of matrix metalloproteinases genes MMP-1 and MMP-9 in breast cancer. Breast Cancer Res. Treat. 2006, 95, 65–72. [Google Scholar] [CrossRef]
- Radisky, E.S.; Radisky, D.C. Matrix metalloproteinases as breast cancer drivers and therapeutic targets. Front. Biosci. (Landmark Ed.) 2015, 20, 1144–1163. [Google Scholar] [CrossRef]
- Eiro, N.; Gonzalez, L.O.; Fraile, M.; Cid, S.; Schneider, J.; Vizoso, F.J. Breast Cancer Tumor Stroma: Cellular Components, Phenotypic Heterogeneity, Intercellular Communication, Prognostic Implications and Therapeutic Opportunities. Cancers 2019, 11, 664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, E.A.; Stephenson, T.J.; Reed, M.W.; Brown, N.J. Expression of proteinases and inhibitors in human breast cancer progression and survival. Mol. Pathol. 2002, 55, 300–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McColgan, P.; Sharma, P. Polymorphisms of matrix metalloproteinases 1, 2, 3 and 9 and susceptibility to lung, breast and colorectal cancer in over 30,000 subjects. Int. J. Cancer 2009, 125, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Du, L.F.; Lv, G.Q.; Yu, X.M.; Gu, Y.L.; Li, J.P.; Zhang, C. Current evidence on the relationship between four polymorphisms in the matrix metalloproteinases (MMP) gene and breast cancer risk: A meta-analysis. Breast Cancer Res. Treat. 2011, 127, 813–818. [Google Scholar] [CrossRef]
- Liu, D.; Guo, H.; Li, Y.; Xu, X.; Yang, K.; Bai, Y. Association between polymorphisms in the promoter regions of matrix metalloproteinases (MMPs) and risk of cancer metastasis: A meta-analysis. PLoS ONE 2012, 7, e31251. [Google Scholar] [CrossRef] [Green Version]
- Białkowska, K.; Marciniak, W.; Muszyńska, M.; Baszuk, P.; Gupta, S.; Jaworska-Bieniek, K.; Sukiennicki, G.; Durda, K.; Gromowski, T.; Lener, M.; et al. Polymorphisms in MMP-1, MMP-2, MMP-7, MMP-13 and MT2A do not contribute to breast, lung and colon cancer risk in polish population. Hered. Cancer Clin. Pract. 2020, 18, 16. [Google Scholar] [CrossRef]
- Zhang, X.; Jin, G.; Li, J.; Zhang, L. Association between four MMP-9 polymorphisms and breast cancer risk: A meta-analysis. Med. Sci. Monit. 2015, 21, 1115–1123. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Zhang, S.; Qiu, D.; Li, X.; Fan, Y. Association between matrix metalloproteinase 9 polymorphisms and breast cancer risk: An updated meta-analysis and trial sequential analysis. Gene 2020, 759, 144972. [Google Scholar] [CrossRef]
- Yan, C.; Sun, C.; Lu, D.; Zhao, T.; Ding, X.; Zamir, I.; Tang, M.; Shao, C.; Zhang, F. Estimation of associations between MMP9 gene polymorphisms and breast cancer: Evidence from a meta-analysis. Int. J. Biol. Markers 2022, 37, 13–20. [Google Scholar] [CrossRef]
- Lei, H.; Hemminki, K.; Altieri, A.; Johansson, R.; Enquist, K.; Hallmans, G.; Lenner, P.; Försti, A. Promoter polymorphisms in matrix metalloproteinases and their inhibitors: Few associations with breast cancer susceptibility and progression. Breast Cancer Res. Treat. 2007, 103, 61–69. [Google Scholar] [CrossRef]
- Roehe, A.V.; Frazzon, A.P.; Agnes, G.; Damin, A.P.; Hartman, A.A.; Graudenz, M.S. Detection of polymorphisms in the promoters of matrix metalloproteinases 2 and 9 genes in breast cancer in South Brazil: Preliminary results. Breast Cancer Res. Treat. 2007, 102, 123–124. [Google Scholar] [CrossRef] [PubMed]
- Beeghly-Fadiel, A.; Lu, W.; Shu, X.O.; Long, J.; Cai, Q.; Xiang, Y.; Gao, Y.T.; Zheng, W. MMP9 polymorphisms and breast cancer risk: A report from the Shanghai Breast Cancer Genetics Study. Breast Cancer Res. Treat. 2011, 126, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shevchenko, A.V.; Konenkov, V.I.; EIu, G.; Stakheeva, M.N. Associating of polymorphism in the promoter regions of genes of metalloproteinase (MMP2, MMP3, MMP9) with options of the clinical course of breast cancer in Russian women. Vopr. Onkol. 2014, 60, 630–635. [Google Scholar] [PubMed]
- Fu, F.; Wang, C.; Chen, L.M.; Huang, M.; Huang, H.G. The influence of functional polymorphisms in matrix metalloproteinase 9 on survival of breast cancer patients in a Chinese population. DNA Cell Biol. 2013, 32, 274–282. [Google Scholar] [CrossRef]
- Beeghly-Fadiel, A.; Cai, Q.; Lu, W.; Long, J.; Gao, Y.T.; Shu, X.O.; Zheng, W. No association between matrix metalloproteinase-1 or matrix metalloproteinase-3 polymorphisms and breast cancer susceptibility: A report from the Shanghai Breast Cancer Study. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1324–1327. [Google Scholar] [CrossRef] [Green Version]
- Chahil, J.K.; Munretnam, K.; Samsudin, N.; Lye, S.H.; Hashim, N.A.; Ramzi, N.H.; Velapasamy, S.; Wee, L.L.; Alex, L. Genetic polymorphisms associated with breast cancer in malaysian cohort. Indian J. Clin. Biochem. 2015, 30, 134–139. [Google Scholar] [CrossRef] [Green Version]
- Al-Eitan, L.N.; Jamous, R.I.; Khasawneh, R.H. Candidate Gene Analysis of Breast Cancer in the Jordanian Population of Arab Descent: A Case-Control Study. Cancer Investig. 2017, 35, 256–270. [Google Scholar] [CrossRef]
- Resler, A.J.; Malone, K.E.; Johnson, L.G.; Malkki, M.; Petersdorf, E.W.; McKnight, B.; Madeleine, M.M. Genetic variation in TLR or NFkappaB pathways and the risk of breast cancer: A case-control study. BMC Cancer 2013, 13, 219. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, V.A.; Chagas, D.C.; Amorim, J.R.; Pereira, R.O.; Nogueira, T.A.; Borges, V.; Campos-Verde, L.M.; Martins, L.M.; Rodrigues, G.P.; Nery Júnior, E.J.; et al. Association between matrix metalloproteinase-9 gene polymorphism and breast cancer in Brazilian women. Clinics (Sao Paulo) 2020, 75, e1762. [Google Scholar] [CrossRef]
- Van den Steen, P.E.; Dubois, B.; Nelissen, I.; Rudd, P.M.; Dwek, R.A.; Opdenakker, G. Biochemistry and Molecular biology of gelatinase B or matrix metalloproteinase-9 (gelatinase B/MMP-9). Crit. Rev. Biochem. Mol. Biol. 2002, 37, 375–536. [Google Scholar] [CrossRef]
- Farina, A.R.; Mackay, A.R. Gelatinase B/MMP-9 in Tumour Pathogenesis and Progression. Cancers 2014, 6, 240–296. [Google Scholar] [CrossRef] [PubMed]
- Joseph, C.; Alsaleem, M.; Orah, N.; Narasimha, P.L.; Miligy, I.M.; Kurozumi, S.; Ellis, I.O.; Mongan, N.P.; Green, A.R.; Rakha, E.A. Elevated MMP9 expression in breast cancer is a predictor of shorter patient survival. Breast Cancer Res. Treat. 2020, 182, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Su, H.; Zhou, Y.Y.; Guo, L.L. Prognostic value of matrix metalloproteinase 9 expression in breast cancer patients: A meta-analysis. Asian Pac. J. Cancer Prev. 2013, 14, 1615–1621. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Li, H. Prognostic values of tumoral MMP2 and MMP9 overexpression in breast cancer: A systematic review and meta-analysis. BMC Cancer 2021, 21, 149. [Google Scholar] [CrossRef]
- Kalavska, K.; Cierna, Z.; Karaba, M.; Minarik, G.; Benca, J.; Sedlackova, T.; Kolekova, D.; Mrvova, I.; Pindak, D.; Mardiak, J.; et al. Prognostic role of matrix metalloproteinase 9 in early breast cancer. Oncol. Lett. 2021, 21, 78. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, E.A.; Akulova, L.Y.; Dobrodomova, I.S.; Dvornyk, V.Y.; Polonikov, A.V.; Churnosov, M.I. The insertion-deletion polymorphism of the ACE gene is associated with increased blood pressure in women at the end of pregnancy. J. Renin-Angiotensin-Aldosterone Syst. 2015, 16, 623–632. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with endometriosis. Reprod. Biomed. Online 2020, 41, 943–956. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Zarudskaya, O.; Polonikov, A.; Bushueva, O.; Orlova, V.; Krikun, E.; Dvornyk, V.; Churnosov, M. Genetic markers for inherited thrombophilia are associated with fetal growth retardation in the population of Central Russia. J. Obstet. Gynaecol. Res. 2017, 43, 1139–1144. [Google Scholar] [CrossRef]
- Eliseeva, N.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. LOXL1 gene polymorphism candidates for exfoliation glaucoma are also associated with a risk for primary open-angle glaucoma in a Caucasian population from central Russia. Mol. Vis. 2021, 27, 262–269. [Google Scholar]
- Ponomarenko, I.; Reshetnikov, E.; Altuchova, O.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Golovchenko, O.; Churnosov, M. Association of genetic polymorphisms with age at menarche in Russian women. Gene 2019, 686, 228–236. [Google Scholar] [CrossRef] [Green Version]
- Moskalenko, M.I.; Milanova, S.N.; Ponomarenko, I.V.; Polonikov, A.V.; Churnosov, M.I. Study of associations of polymorphism of matrix metalloproteinases genes with the development of arterial hypertension in men. Kardiologiia 2019, 59, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Polonikov, A.; Rymarova, L.; Klyosova, E.; Volkova, A.; Azarova, I.; Bushueva, O.; Bykanova, M.; Bocharova, I.; Zhabin, S.; Churnosov, M.; et al. Matrix metalloproteinases as target genes for gene regulatory networks driving molecular and cellular pathways related to a multistep pathogenesis of cerebrovascular disease. J. Cell Biochem. 2019, 10, 16467–16482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minyaylo, O.N. Allele distribution and haploblock structure of matrix metalloproteinase gene polymorphism in patients with H. pylori-negative gastric ulcer and duodenal ulcer. Res. Results Biomed. 2020, 6, 488–502. (In Russian) [Google Scholar] [CrossRef]
- Svinareva, D.I. The contribution of gene-gene interactions of polymorphic loci of matrix metalloproteinases to susceptibility to primary open-angle glaucoma in men. Res. Results Biomed. 2020, 6, 63–77. [Google Scholar] [CrossRef]
- Minyaylo, O.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of Central Russia. Sci. Rep. 2021, 11, 13515. [Google Scholar] [CrossRef] [PubMed]
- Moskalenko, M.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Polymorphisms of the matrix metalloproteinase genes are associated with essential hypertension in a Caucasian population of Central Russia. Sci. Rep. 2021, 11, 5224. [Google Scholar] [CrossRef] [PubMed]
- Starikova, D.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Novel data about association of the functionally significant polymorphisms of the MMP9 gene with exfoliation glaucoma in the caucasian population of Central Russia. Ophthalmic Res. 2021, 64, 458–464. [Google Scholar] [CrossRef]
- Tikunova, E.; Ovtcharova, V.; Reshetnikov, E.; Dvornyk, V.; Polonikov, A.; Bushueva, O.; Churnosov, M. Genes of tumor necrosis factors and their receptors and the primary open angle glaucoma in the population of Central Russia. Int. J. Ophthalmol. 2017, 10, 1490–1494. [Google Scholar] [CrossRef]
- Golovchenko, O.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Polonikov, A.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of ESR1and PGR and risk of intrauterine growth restriction in population of Central Russia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 52–57. [Google Scholar] [CrossRef]
- Bushueva, O.; Solodilova, M.; Churnosov, M.; Ivanov, V.; Polonikov, A. The Flavin-Containing Monooxygenase 3 Gene and Essential Hypertension: The Joint Effect of Polymorphism E158K and Cigarette Smoking on Disease Susceptibility. Int. J. Hypertens. 2014, 2014, 712169. [Google Scholar] [CrossRef] [Green Version]
- Sirotina, S.; Ponomarenko, I.; Kharchenko, A.; Bykanova, M.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Churnosov, M.; Solodilova, M.; Polonikov, A. A Novel Polymorphism in the Promoter of the CYP4A11 Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis. Markers 2018, 2018, 5812802. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, E.; Ponomarenko, I.; Golovchenko, O.; Sorokina, I.; Batlutskaya, I.; Yakunchenko, T.; Dvornyk, V.; Polonikov, A.; Churnosov, M. The VNTR polymorphism of the endothelial nitric oxide synthase gene and blood pressure in women at the end of pregnancy. Taiwan. J. Obstet. Gynecol. 2019, 58, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Polonikov, A.V.; Bushueva, O.Y.; Bulgakova, I.V.; Freidin, M.B.; Churnosov, M.I.; Solodilova, M.A.; Shvetsov, Y.D.; Ivanov, V.P. A comprehensive contribution of genes for aryl hydrocarbon receptor signaling pathway to hypertension susceptibility. Pharmacogenet. Genom. 2017, 2, 57–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Churnosov, M.; Abramova, M.; Reshetnikov, E.; Lyashenko, I.V.; Efremova, O.; Churnosova, M.; Ponomarenko, I. Polymorphisms of hypertension susceptibility genes as a risk factors of preeclampsia in the Caucasian population of central Russia. Placenta 2022, 129, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Jack, J.R.; Motsinger-Reif, A.A.; Brown, C.C. An adaptive permutation approach for genome-wide association study: Evaluation and recommendations for use. BioData Min. 2014, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with endometrial hyperplasia. Gene 2020, 757, 4933. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Verzilina, I.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with uterine leiomyoma. Front. Genet. 2021, 11, 512940. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 36, 1318–1330. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic. Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 7, 1073–1081. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Ibrahim, F.A.E.R.; Elfeky, S.E.; Haroun, M.; Ahmed, M.A.E.; Elnaggar, M.; Ismail, N.A.E.; El Moneim, N.A.A. Association of matrix metalloproteinases 3 and 9 single nucleotide polymorphisms with breast cancer risk: A case-control study. Mol. Clin. Oncol. 2020, 13, 54–62. [Google Scholar] [CrossRef] [PubMed]
- AbdRaboh, N.R.; Bayoumi, F.A. Gene polymorphism of matrix metalloproteinases 3 and 9 in breast cancer. Gene Rep. 2016, 5, 151–156. [Google Scholar] [CrossRef]
- Balkhi, S.; Mashayekhi, F.; Salehzadeh, A.; Saedi, H.S. Matrix metalloproteinase (MMP)-1 and MMP-3 gene variations affect MMP-1 and -3 serum concentration and associates with breast cancer. Mol. Biol. Rep. 2020, 47, 9637–9644. [Google Scholar] [CrossRef] [PubMed]
- Bargostavan, M.H.; Eslami, G.; Esfandiari, N.; Shahemabadi, A.S. MMP9 Promoter Polymorphism (−1562 C/T) Does not Affect the Serum Levels of Soluble MICB and MICA in Breast Cancer. Iran. J. Immunol. IJI 2016, 13. [Google Scholar]
- Beeghly-Fadiel, A.; Lu, W.; Long, J.R.; Shu, X.O.; Zheng, Y.; Cai, Q.; Gao, Y.; Zheng, W. Matrix metalloproteinase-2 polymorphisms and breast cancer susceptibility. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1770–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biondi, M.L.; Turri, O.; Leviti, S.; Seminati, R.; Cecchini, F.; Bernini, M.; Ghilardi, G.; Guagnellini, E. MMP1 and MMP3 polymorphisms in promoter regions and cancer. Clin. Chem. 2000, 46, 2023–2024. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.C. Identification and analysis of single nucleotide polymorphisms in matrix metallopeptidase 2 and 3 genes in Malaysian breast cancer patients. Ph.D. Thesis, Universiti Putra Malaysia, Serdang, Malaysia, 2013. [Google Scholar]
- Chiranjeevi, P.; Spurthi, K.M.; Rani, N.S.; Kumar, G.R.; Aiyengar, T.M.; Saraswati, M.; Srilatha, G.; Kumar, G.K.; Sinha, S.; Kumari, C.S.; et al. Gelatinase B (−1562C/T) polymorphism in tumor progression and invasion of breast cancer. Tumor Biol. 2013, 35, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Decock, J.; Long, J.-R.; Laxton, R.C.; Shu, X.-O.; Hodgkinson, C.; Hendrickx, W.; Pearce, E.G.; Gao, Y.-T.; Pereira, A.C.; Paridaens, R.; et al. Association of Matrix Metalloproteinase-8 Gene Variation with Breast Cancer Prognosis. Cancer Res. 2007, 67, 10214–10221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Enciso, I.; Cepeda-Lopez, F.R.; Monrroy-Guizar, E.A.; Bautista-Lam, J.R.; Andrade-Soto, M.; Jonguitud-Olguin, G.; Rodriguez-Hernandez, A.; Anaya-Ventura, A.; Baltazar-Rodriguez, L.M.; Orozco-Ruiz, M.; et al. Matrix Metalloproteinase-2 Promoter Polymorphism Is Associated with Breast Cancer in a Mexican Population. Gynecol. Obstet. Investig. 2007, 65, 68–72. [Google Scholar] [CrossRef]
- Felizi, R.T.; Veiga, M.G.; Filho, I.C.; Souto, R.P.D.; Fernandes, C.E.; Oliveira, E. Association between Matrix Metallopeptidase 9 Polymorphism and Breast Cancer Risk. Rev Bras Ginecol Obstet 2018, 40, 620–624. [Google Scholar] [CrossRef] [Green Version]
- Ghilardi, G.; Biondi, M.L.; Caputo, M.; Leviti, S.; DeMonti, M.; Guagnellini, E.; Scorza, R. A single nucleotide polymorphism in the matrix metalloproteinase-3 promoter enhances breast cancer susceptibility. Clin. Cancer Res. 2002, 8, 3820–3823. [Google Scholar]
- Habel, A.F.; Ghali, R.M.; Bouaziz, H.; Daldoul, A.; Hadj-Ahmed, M.; Mokrani, A.; Zaied, S.; Hechiche, M.; Rahal, K.; Yacoubi-Loueslati, B.; et al. Common matrix metalloproteinase-2 gene variants and altered susceptibility to breast cancer and associated features in Tunisian women. Tumor Biol. 2019, 41. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, C.-L.; Liu, L.-C.; Shih, T.-C.; Lai, Y.-L.; Hsu, S.-W.; Wang, H.-C.; Pan, S.-Y.; Shen, T.-C.; Tsai, C.-W.; Chang, W.-S.; et al. The Association of Matrix Metalloproteinase-1 Promoter Polymorphisms with Breast Cancer. Vivo 2018, 32, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Hughes, S.; Agbaje, O.; Bowen, R.L.; Holliday, D.L.; Shaw, J.A.; Duffy, S.; Jones, J.L. Matrix Metalloproteinase Single-Nucleotide Polymorphisms and Haplotypes Predict Breast Cancer Progression. Clin. Cancer Res. 2007, 13, 6673–6680. [Google Scholar] [CrossRef] [Green Version]
- Ledwon, J.; Hennig, E.E.; Maryan, N.; Goryca, K.; Nowakowska, D.; Niwińska, A.; Ostrowski, J. Common low-penetrance risk variants associated with breast cancer in Polish women. BMC Cancer 2013, 13, 510. [Google Scholar] [CrossRef] [Green Version]
- Manshadi, Z.D.; Hamid, M.; Kosari, F.; Tayebinia, H.; Khodadadi, I. The Relationship between Matrix Metalloproteinase Gene Polymorphisms and Tumor Type, Tumor Size, and Metastasis in Women with Breast Cancer in Central Iran. Middle East J. Cancer 2018, 9, 123–131. [Google Scholar] [CrossRef]
- Mavaddat, N.; Dunning, A.M.; Ponder, B.A.; Easton, D.F.; Pharoah, P.D. Common Genetic Variation in Candidate Genes and Susceptibility to Subtypes of Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2009, 18, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Ben Néjima, D.; Ben Zarkouna, Y.; Gammoudi, A.; Manai, M.; Boussen, H. Prognostic impact of polymorphism of matrix metalloproteinase-2 and metalloproteinase tissue inhibitor-2 promoters in breast cancer in Tunisia: Case-control study. Tumor Biol. 2015, 36, 3815–3822. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.-X.; Bi, R. Meta-analysis on the relationship between the SNP of MMP-2-1306 C>T and susceptibility to breast cancer. Eur. Rev. Med. Pharmacl. Sci. 2020, 24, 1264–1270. [Google Scholar]
- Padala, C.; Tupurani, M.A.; Puranam, K.; Gantala, S.; Shyamala, N.; Kondapalli, M.S.; Gundapaneni, K.K.; Mudigonda, S.; Galimudi, R.K.; Kupsal, K.; et al. Synergistic effect of collagenase-1 (MMP1), stromelysin-1 (MMP3) and gelatinase-B (MMP9) gene polymorphisms in breast cancer. PLoS ONE 2017, 12, e0184448. [Google Scholar] [CrossRef] [PubMed]
- Pharoah, P.D.P.; Tyrer, J.; Dunning, A.M.; Easton, D.F.; Ponder, B.A.J. SEARCH Investigators Association between Common Variation in 120 Candidate Genes and Breast Cancer Risk. PLoS Genet. 2007, 3, e42. [Google Scholar] [CrossRef] [PubMed]
- Przybylowska, K.; Zielinska, J.; Zadrozny, M.; Krawczyk, T.; Kulig, A.; Wozniak, P.; Rykala, J.; Kolacinska, A.; Morawiec, Z.; Drzewoski, J.; et al. An association between the matrix metalloproteinase 1 promoter gene polymorphism and lymphnode metastasis in breast cancer. J. Exp. Clin. Cancer Res. 2004, 23, 121–125. [Google Scholar]
- Rahimi, Z.; Yari, K.; Rahimi, Z. Matrix Metalloproteinase-9 -1562T Allele and its Combination with MMP-2 -735 C Allele are Risk Factors for Breast Cancer. Asian Pac. J. Cancer Prev. 2015, 16, 1175–1179. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, M.; Motovali Bashi, M.; Hojati, Z. MMP-9 promoter polymorphism associated with tumor progression of breast cancer in Iranian population. Int. J. Integr. Biology. 2009, 6, 33–37. [Google Scholar]
- Saeed, H.M.; Alanazi, M.S.; Alshahrani, O.; Parine, N.R.; Alabdulkarim, H.A.; Shalaby, M. Matrix metalloproteinase-2 C(-1306)T promoter polymorphism and breast cancer risk in the Saudi population. Acta Biochim. Pol. 2013, 60, 405–409. [Google Scholar] [CrossRef] [Green Version]
- Slattery, M.L.; John, E.; Torres-Mejia, G.; Stern, M.; Lundgreen, A.; Hines, L.; Giuliano, A.; Baumgartner, K.; Herrick, J.; Wolff, R.K. Matrix Metalloproteinase Genes Are Associated with Breast Cancer Risk and Survival: The Breast Cancer Health Disparities Study. PLoS ONE 2013, 8, e63165. [Google Scholar] [CrossRef] [Green Version]
- Su, C.-H.; Lane, H.-Y.; Hsiao, C.-L.; Liu, L.-C.; Ji, H.-X.; Li, H.-T.; Yen, S.-T.; Su, C.-H.; Hsia, T.-C.; Chang, W.-S.; et al. Matrix Metalloproteinase-1 Genetic Polymorphism in Breast Cancer in Taiwanese. Anticancer Res. 2016, 36, 3341–3345. [Google Scholar] [PubMed]
- Sui, J.; Huang, J.; Zhang, Y. The MMP-1 Gene rs1799750 Polymorphism Is Associated with Breast Cancer Risk. Genet. Test. Mol. Biomark. 2021, 25, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Toroghi, F.; Mashayekhi, F.; Montazeri, V.; Saedi, H.S.; Salehi, Z. Association of MMP-9 promoter polymorphism and breast cancer among Iranian patients. Eur. J. Oncol. 2017, 1, 38–42. [Google Scholar]
- Wang, K.; Zhou, Y.; Li, G.; Wen, X.; Kou, Y.; Yu, J.; He, H.; Zhao, Q.; Xue, F.; Wang, J.; et al. MMP8 and MMP9 gene polymorphisms were associated with breast cancer risk in a Chinese Han population. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Li, N.; Wang, S.; Kong, Y.; Tang, H.; Xie, X.; Xie, X. Lack of association between the matrix metalloproteinase-2 -1306C>T polymorphism and breast cancer susceptibility: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 4823–4827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zagouri, F.; Sergentanis, T.N.; Gazouli, M.; Dimitrakakis, C.; Tsigginou, A.; Papaspyrou, I.; Chrysikos, D.; Lymperi, M.; Zografos, G.C.; Antsaklis, A.; et al. MMP-2 −1306C > T polymorphism in breast cancer: A case-control study in a South European population. Mol. Biol. Rep. 2013, 40, 5035–5040. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yu, C.; Miao, X.; Tan, W.; Liang, G.; Xiong, P.; Sun, T.; Lin, N. Substantial reduction in risk of breast cancer associated with genetic polymorphisms in the promoters of the matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 genes. Carcinogenesis 2003, 25, 399–404. [Google Scholar] [CrossRef]
- Zhou, Z.; Ma, X.; Wang, F.; Sun, L.; Zhang, G. A Matrix Metalloproteinase-1 Polymorphism, MMP1–1607(1G>2G), Is Associated with Increased Cancer Risk: A Meta-Analysis Including 21,327 Patients. Dis. Mrk. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
| Parameters | BC Patients, % (n) | Controls,% (n) | p |
|---|---|---|---|
| N | 358 | 746 | - |
| Age, years (min–max) | 55.74 ± 12.79 (28–84) | 55.29 ± 12.27 (30–82) | 0.54 |
| <50 years | 33.80 (121) | 35.12 (262) | 0.72 |
| ≥50 years | 66.20 (237) | 64.88 (484) | |
| BMI, kg/m2 | 30.27 ± 6.13 | 28.19 ± 5.73 | 0.003 |
| Obesity (BMI ≥ 30) (yes) | 33.24 (119) | 25.47 (190) | 0.01 |
| Age at menarche, years | 12.42 ± 1.12 | 12.64 ± 1.14 | 0.58 |
| Age at menopause, years | 48.27 ± 5.02 | 47.97 ± 4.91 | 0.48 |
| Mensuration status | |||
| Premenopause | 31.84 (114) | 34.05 (254) | 0.51 |
| Postmenopause | 68.16 (244) | 65.95 (492) | |
| Smoker (yes) | 22.07 (79) | 18.77 (140) | 0.22 |
| Clinicopathological parameters of BC patients | |||
| Stage of the cancer | T0–T2—74%, T3–T4—26% | ||
| Lymph node involvement (N) | negative—47%, positive—53% | ||
| Estrogen receptor (ER) | negative—34%, positive—66% | ||
| Progesterone receptor (PR) | negative—41%, positive—59% | ||
| Human epidermal growth factor receptor 2 (HER2) | negative—64%, positive—36% | ||
| Triple negative | 22% | ||
| Tumor histological type | ductal—94%, lobular—6% | ||
| Tumor histological grade (G) | G1/G2—68%, G3—32% | ||
| Progression | absent—66%, present—34% | ||
| Metastasis | absent—78%, present—22% | ||
| Death | absent—81%, present—19% | ||
| SNP | Gene | Minor Allele | n | Allelic Model | Additive Model | Dominant Model | Recessive Model | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | ||||||||
| L95 | U95 | L95 | U95 | L95 | U95 | L95 | U95 | ||||||||||||
| rs1799750 | MMP-1 | 2G | 1077 | 1.02 | 0.85 | 1.22 | 0.831 | 1.05 | 0.87 | 1.25 | 0.626 | 1.16 | 0.87 | 1.54 | 0.325 | 0.96 | 0.70 | 1.32 | 0.811 |
| rs243865 | MMP-2 | T | 1082 | 0.93 | 0.75 | 1.15 | 0.491 | 0.95 | 0.77 | 1.18 | 0.651 | 1.01 | 0.78 | 1.32 | 0.922 | 0.68 | 0.38 | 1.20 | 0.185 |
| rs679620 | MMP-3 | T | 1095 | 0.90 | 0.76 | 1.08 | 0.267 | 0.87 | 0.72 | 1.05 | 0.135 | 0.88 | 0.65 | 1.18 | 0.381 | 0.78 | 0.57 | 1.06 | 0.117 |
| rs1940475 | MMP-8 | T | 1096 | 0.97 | 0.81 | 1.16 | 0.742 | 1.00 | 0.84 | 1.20 | 0.989 | 1.22 | 0.91 | 1.64 | 0.189 | 0.81 | 0.60 | 1.10 | 0.185 |
| rs3918242 | MMP-9 | T | 1089 | 1.02 | 0.80 | 1.30 | 0.869 | 1.04 | 0.82 | 1.33 | 0.744 | 0.96 | 0.73 | 1.28 | 0.802 | 1.82 | 0.90 | 3.70 | 0.097 |
| rs3918249 | MMP-9 | C | 1083 | 0.88 | 0.73 | 1.06 | 0.180 | 0.90 | 0.75 | 1.09 | 0.294 | 0.83 | 0.63 | 1.08 | 0.161 | 0.97 | 0.68 | 1.40 | 0.890 |
| rs17576 | MMP-9 | G | 1095 | 0.82 | 0.68 | 0.99 | 0.035 | 0.84 | 0.70 | 1.01 | 0.068 | 0.78 | 0.60 | 1.01 | 0.063 | 0.82 | 0.57 | 1.19 | 0.302 |
| rs3787268 | MMP-9 | A | 1089 | 1.11 | 0.89 | 1.37 | 0.352 | 1.11 | 0.89 | 1.38 | 0.350 | 1.05 | 0.80 | 1.36 | 0.746 | 1.68 | 0.95 | 2.96 | 0.074 |
| rs2250889 | MMP-9 | G | 1090 | 0.71 | 0.52 | 0.97 | 0.033 | 0.69 | 0.51 | 0.95 | 0.024 | 0.67 | 0.47 | 0.95 | 0.026 | 0.54 | 0.18 | 1.67 | 0.286 |
| rs17577 | MMP-9 | A | 1079 | 0.97 | 0.76 | 1.23 | 0.798 | 0.98 | 0.77 | 1.25 | 0.850 | 0.92 | 0.69 | 1.22 | 0.556 | 1.41 | 0.69 | 2.85 | 0.343 |
| SNPs | Haplotype | Frequency | OR | praw value | pperm | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| risk effect | ||||||
| rs17576-rs3787268 | AA | 0.0377 | 0.0186 | 2.46 | 0.004 | 0.020 |
| rs17576-rs3787268-rs2250889 | AAC | 0.0358 | 0.0179 | 2.53 | 0.001 | 0.012 |
| rs3918249-rs17576-rs3787268 | TAA | 0.0254 | 0.0110 | 2.89 | 0.004 | 0.034 |
| rs17576-rs3787268-rs2250889-rs17577 | AACG | 0.0375 | 0.0171 | 2.68 | 0.003 | 0.020 |
| rs3918249-rs17576-rs3787268-rs2250889 | TAAC | 0.0246 | 0.0102 | 3.21 | 0.004 | 0.031 |
| rs3918242-rs3918249-rs17576-rs3787268 | CTAA | 0.0252 | 0.0101 | 3.07 | 0.005 | 0.032 |
| rs3918249-rs17576-rs3787268-rs2250889-rs17577 | TAACG | 0.0263 | 0.0095 | 3.63 | 0.003 | 0.016 |
| rs3918242-rs3918249-rs17576-rs3787268-rs2250889 | CTAAC | 0.0247 | 0.0092 | 3.55 | 0.002 | 0.016 |
| rs3918242-rs3918249-rs17576-rs3787268-rs2250889-rs17577 | CTAACG | 0.0247 | 0.0090 | 3.26 | 0.003 | 0.046 |
| protective effect | ||||||
| rs3787268-rs2250889 | GG | 0.0693 | 0.1011 | 0.63 | 0.013 | 0.032 |
| rs3787268-rs2250889-rs17577 | GGG | 0.0710 | 0.0994 | 0.66 | 0.017 | 0.050 |
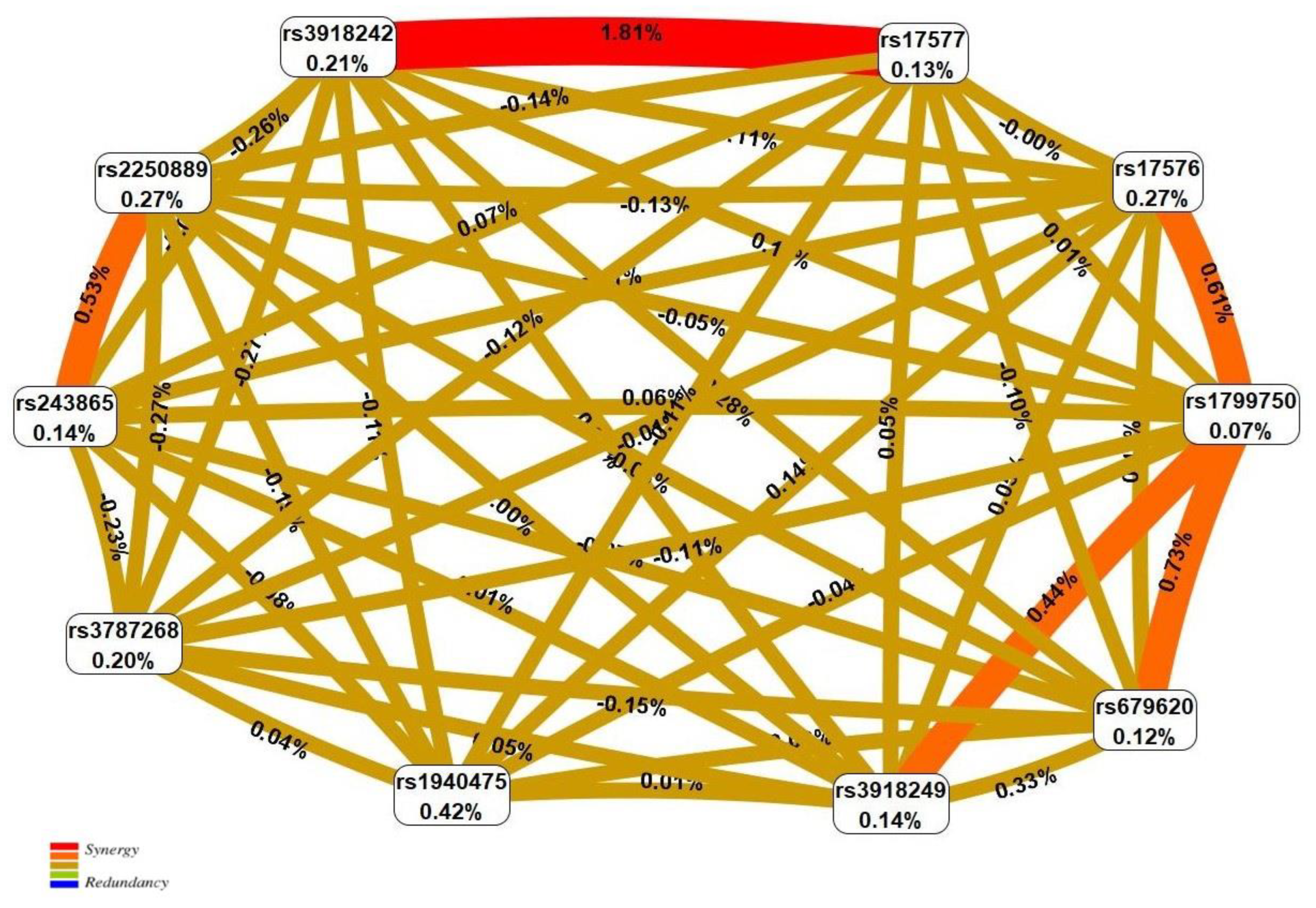
| N | SNP × SNP Interaction Models | NH | beta H | WH | NL | beta L | WL | pperm |
|---|---|---|---|---|---|---|---|---|
| Two-order interaction models (p < 1.02 × 10−3) | ||||||||
| 1 | rs17577 MMP9 × rs3918242 MMP9 | 2 | 1.589 | 29.73 | 1 | −0.434 | 7.15 | <0.001 |
| 2 | rs1799750 MMP1 × rs17576 MMP9 | 2 | 0.502 | 13.58 | 1 | −0.608 | 8.24 | 0.004 |
| 3 | rs17577 MMP9 × rs17576 MMP9 | 2 | 1.256 | 12.47 | 0 | - | - | 0.005 |
| 4 | rs1799750 MMP1 × rs679620 MMP3 | 2 | 0.413 | 7.38 | 1 | −0.788 | 12.65 | 0.011 |
| 5 | rs2250889 MMP9 × rs243865 MMP2 | 0 | - | - | 2 | −0.674 | 10.79 | 0.011 |
| Three-order interaction models (p < 2.15 × 10−7) | ||||||||
| 1 | rs17577 MMP9 × rs3918242 MMP9 × rs1940475 MMP8 | 6 | 1.620 | 32.65 | 1 | −0.536 | 3.11 | <0.001 |
| 2 | rs17577 MMP9 × rs3918242 MMP9 × rs243865 MMP2 | 4 | 1.654 | 31.11 | 1 | −0.702 | 7.13 | <0.001 |
| 3 | rs17577 MMP9 × rs3787268 MMP9 × rs3918242 MMP9 | 4 | 1.545 | 27.67 | 2 | −0.452 | 7.67 | <0.001 |
| 4 | rs17577 MMP9 × rs1799750 MMP1 × rs3918242 MMP9 | 5 | 1.640 | 27.52 | 2 | −0.918 | 14.91 | <0.001 |
| 5 | rs17577 MMP9 × rs3918242 MMP9 × rs679620 MMP3 | 5 | 1.538 | 27.38 | 1 | −1.189 | 10.40 | <0.001 |
| 6 | rs17577 MMP9 × rs2250889 MMP9 × rs3918242 MMP9 | 4 | 1.620 | 26.90 | 1 | −0.352 | 4.22 | <0.001 |
| Four-order interaction models (p < 9.93 × 10−8) | ||||||||
| 1 | rs17577 MMP9 × rs3787268 MMP9 × rs3918242 MMP9 × rs243865 MMP2 | 7 | 1.341 | 28.39 | 2 | −0.702 | 7.13 | <0.001 |
| 2 | rs1799750 MMP1 × rs3918249 MMP9 × rs194047 MMP8 × rs243865 MMP2 | 6 | 0.993 | 30.90 | 1 | −1.086 | 3.94 | <0.001 |
| Five-order interaction models (p = 2.96 × 10−12) | ||||||||
| 1 | rs2250889 MMP9 × rs1799750 MMP1 × rs3918249 MMP9 × rs1940475 MMP8 × rs243865 MMP2 | 9 | 1.308 | 48.71 | 0 | - | - | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. Int. J. Mol. Sci. 2022, 23, 12638. https://doi.org/10.3390/ijms232012638
Pavlova N, Demin S, Churnosov M, Reshetnikov E, Aristova I, Churnosova M, Ponomarenko I. Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. International Journal of Molecular Sciences. 2022; 23(20):12638. https://doi.org/10.3390/ijms232012638
Chicago/Turabian StylePavlova, Nadezhda, Sergey Demin, Mikhail Churnosov, Evgeny Reshetnikov, Inna Aristova, Maria Churnosova, and Irina Ponomarenko. 2022. "Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia" International Journal of Molecular Sciences 23, no. 20: 12638. https://doi.org/10.3390/ijms232012638
APA StylePavlova, N., Demin, S., Churnosov, M., Reshetnikov, E., Aristova, I., Churnosova, M., & Ponomarenko, I. (2022). Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. International Journal of Molecular Sciences, 23(20), 12638. https://doi.org/10.3390/ijms232012638







