Reduction of Methyltransferase-like 3-Mediated RNA N6-Methyladenosine Exacerbates the Development of Psoriasis Vulgaris in Imiquimod-Induced Psoriasis-like Mouse Model
Abstract
1. Introduction
2. Results
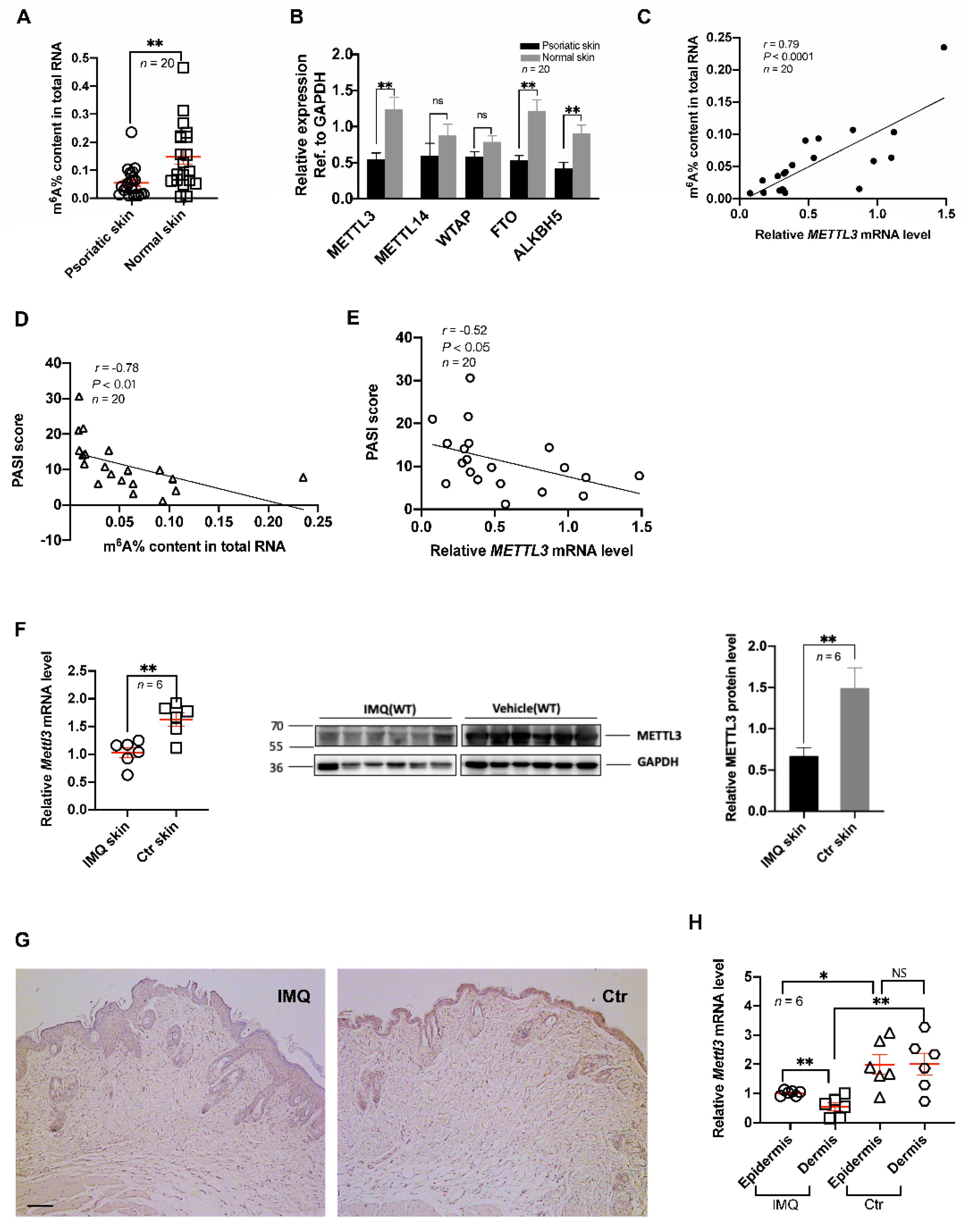
2.1. Downregulation of m6A and METTL3 Level in Psoriatic Skin Lesions
2.2. Psoriasis Area and Severity were Associated with Low Levels of m6A Methylation and METTL3
2.3. Heterozygous knockout of Mettl3 Accelerated the Development of Psoriasis
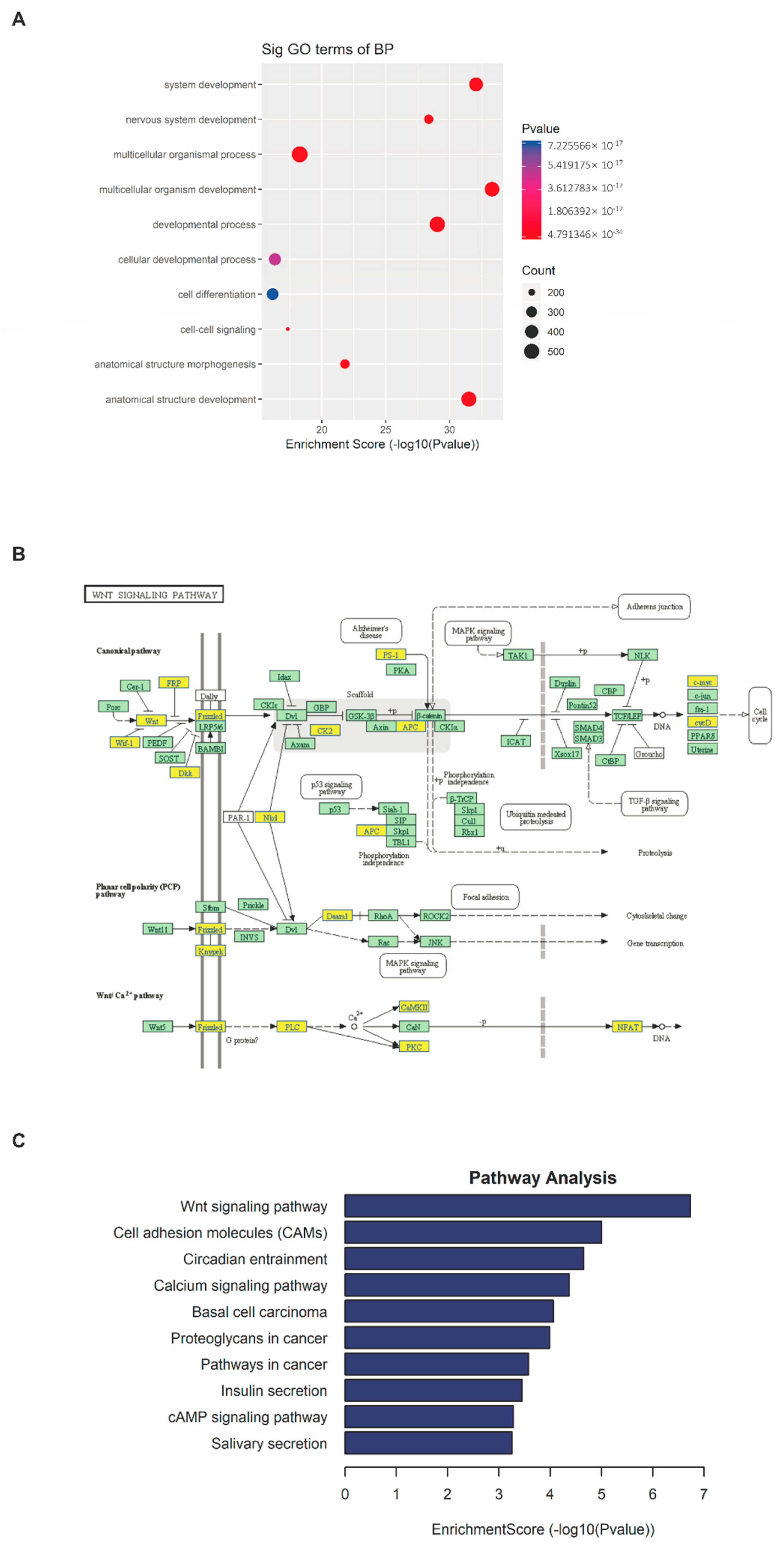
2.4. m6A-seq Identifies Transcripts with Altered Methylation in Psoriasis Vulgaris
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Mice
4.3. IMQ-Induced Psoriasis-like Mouse Model
4.4. Scoring the Severity of Skin Inflammation in Psoriasis-like Mouse Model
4.5. RNA m6A Quantification
4.6. Flow Cytometry
4.7. RT-qPCR
4.8. Western Blotting
4.9. Histological Analysis and Immunohistochemistry
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Greb, J.E.; Goldminz, A.M.; Elder, J.T.; Lebwohl, M.G.; Gladman, D.D.; Wu, J.J.; Mehta, N.N.; Finlay, A.Y.; Gottlieb, A.B. Psoriasis. Nat. Rev. Dis. Prim. 2016, 2, 16082. [Google Scholar] [CrossRef] [PubMed]
- Boehncke, W.H.; Schön, M.P. Psoriasis. Lancet 2015, 386, 983–994. [Google Scholar] [CrossRef]
- Hugh, J.M.; Weinberg, J.M. Update on the pathophysiology of psoriasis. Cutis 2018, 102, 6–12. [Google Scholar] [PubMed]
- Becher, B.; Pantelyushin, S. Hiding under the skin: Interleukin-17-producing γδ T cells go under the skin? Nat. Med. 2012, 18, 1748–1750. [Google Scholar] [CrossRef]
- Gutcher, I.; Becher, B. APC-derived cytokines and T cell polarization in autoimmune inflammation. J. Clin. Invest. 2007, 117, 1119–1127. [Google Scholar] [CrossRef]
- Kagami, S.; Rizzo, H.L.; Lee, J.J.; Koguchi, Y.; Blauvelt, A. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J. Invest. Dermatol. 2010, 130, 1373–1383. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Griffiths, C.E.M.; Armstrong, A.W.; Gudjonsson, J.E.; Barker, J. Psoriasis. Lancet 2021, 397, 1301–1315. [Google Scholar] [CrossRef]
- Schwartz, S. Cracking the epitranscriptome. RNA 2016, 22, 169–174. [Google Scholar] [CrossRef]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m(6)A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bögler, O.; et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e6. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, C.; Zhang, G. m6A RNA Methylation Controls Proliferation of Human Glioma Cells by Influencing Cell Apoptosis. Cytogenet. Genome Res. 2019, 159, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Maity, A.; Das, B. N6-methyladenosine modification in mRNA: Machinery, function and implications for health and diseases. FEBS J. 2016, 283, 1607–1630. [Google Scholar] [CrossRef]
- Wang, Y.N.; Yu, C.Y.; Jin, H.Z. RNA N(6)-Methyladenosine Modifications and the Immune Response. J. Immunol. Res. 2020, 2020, 6327614. [Google Scholar] [CrossRef]
- Feng, Z.; Li, Q.; Meng, R.; Yi, B.; Xu, Q. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell Mol. Med. 2018, 22, 2558–2568. [Google Scholar] [CrossRef]
- Lv, X.; Liu, X.; Zhao, M.; Wu, H.; Zhang, W.; Lu, Q.; Chen, X. RNA Methylation in Systemic Lupus Erythematosus. Front. Cell Dev. Biol. 2021, 9, 696559. [Google Scholar] [CrossRef]
- Wang, Y.N.; Jin, H.Z. Transcriptome-Wide m(6)A Methylation in Skin Lesions From Patients With Psoriasis Vulgaris. Front. Cell Dev. Biol. 2020, 8, 591629. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- van der Fits, L.; Mourits, S.; Voerman, J.S.; Kant, M.; Boon, L.; Laman, J.D.; Cornelissen, F.; Mus, A.M.; Florencia, E.; Prens, E.P.; et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 2009, 182, 5836–5845. [Google Scholar] [CrossRef] [PubMed]
- Mattei, P.L.; Corey, K.C.; Kimball, A.B. Psoriasis Area Severity Index (PASI) and the Dermatology Life Quality Index (DLQI): The correlation between disease severity and psychological burden in patients treated with biological therapies. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem. Cell 2014, 15, 707–719. [Google Scholar] [CrossRef]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef]
- Liu, Z.X.; Li, L.M.; Sun, H.L.; Liu, S.M. Link Between m6A Modification and Cancers. Front. Bioeng. Biotechnol. 2018, 6, 89. [Google Scholar] [CrossRef]
- Bansal, H.; Yihua, Q.; Iyer, S.P.; Ganapathy, S.; Proia, D.A.; Penalva, L.O.; Uren, P.J.; Suresh, U.; Carew, J.S.; Karnad, A.B.; et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 2014, 28, 1171–1174. [Google Scholar] [CrossRef]
- Jin, D.I.; Lee, S.W.; Han, M.E.; Kim, H.J.; Seo, S.A.; Hur, G.Y.; Jung, S.; Kim, B.S.; Oh, S.O. Expression and roles of Wilms’ tumor 1-associating protein in glioblastoma. Cancer Sci. 2012, 103, 2102–2109. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018, 37, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tang, J.; Huang, W.; Wang, F.; Li, P.; Qin, C.; Qin, Z.; Zou, Q.; Wei, J.; Hua, L.; et al. The M6A methyltransferase METTL3: Acting as a tumor suppressor in renal cell carcinoma. Oncotarget 2017, 8, 96103–96116. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Guo, J.; Lv, Y.; Zheng, Y.; Feng, T.; Gao, Q.; Zeng, W. Meclofenamic acid represses spermatogonial proliferation through modulating m(6)A RNA modification. J. Anim. Sci. Biotechnol. 2019, 10, 63. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Kong, B.; Song, C.; Cong, J.; Hou, J.; Wang, S. Reduced m(6)A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget 2017, 8, 98918–98930. [Google Scholar] [CrossRef]
- Yang, J.; Liu, J.; Zhao, S.; Tian, F. N(6)-Methyladenosine METTL3 Modulates the Proliferation and Apoptosis of Lens Epithelial Cells in Diabetic Cataract. Mol. Ther. Nucleic Acids 2020, 20, 111–116. [Google Scholar] [CrossRef]
- Gu, S.; Sun, D.; Dai, H.; Zhang, Z. N(6)-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol. Lett. 2018, 292, 1–11. [Google Scholar] [CrossRef]
- Li, H.B.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef]
- Yang, J.; Wang, H.; Zhang, W. Regulation of Virus Replication and T Cell Homeostasis by N(6)-Methyladenosine. Virol. Sin. 2019, 34, 22–29. [Google Scholar] [CrossRef]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, M.; Jiang, L.; Jiang, R.; Fu, B. METTL3 promotes experimental osteoarthritis development by regulating inflammatory response and apoptosis in chondrocyte. Biochem. Biophys. Res. Commun. 2019, 516, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, S.; Lu, H.; Wang, S.; Xu, D. METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-κB Signaling Pathway. Mediat. Inflamm. 2019, 2019, 3120391. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Li, Q.; Feng, Z.; Cai, L.; Xu, Q. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int. J. Mol. Sci. 2019, 20, 1323. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Su, Q.; Li, B.; Lan, L.; Wang, C.; Li, W.; Wang, G.; Chen, W.; He, Y.; Zhang, C. High expression of WTAP leads to poor prognosis of gastric cancer by influencing tumour-associated T lymphocyte infiltration. J. Cell Mol. Med. 2020, 24, 4452–4465. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.E.; Hess, S.; Meyer, K.D.; Verhagen, L.A.; Koch, L.; Brönneke, H.S.; Dietrich, M.O.; Jordan, S.D.; Saletore, Y.; Elemento, O.; et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013, 16, 1042–1048. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Zhang, S.W.; Fan, X.N.; Meng, J.; Chen, Y.; Gao, S.J.; Huang, Y. Global analysis of N6-methyladenosine functions and its disease association using deep learning and network-based methods. PLoS Comput. Biol. 2019, 15, e1006663. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Sun, G.; Wu, Q.; Ma, J.; Zhang, X.; Huang, N.; Bian, Z.; Gu, S.; Xu, M.; et al. m(6)A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol. Cancer 2019, 18, 188. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Ge, S.; Huang, W.; Lin, X.; Gao, J.; Gong, J.; Shen, L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019, 8, 4766–4781. [Google Scholar] [CrossRef]
- Fukumoto, T.; Zhu, H.; Nacarelli, T.; Karakashev, S.; Fatkhutdinov, N.; Wu, S.; Liu, P.; Kossenkov, A.V.; Showe, L.C.; Jean, S.; et al. N(6)-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance. Cancer Res. 2019, 79, 2812–2820. [Google Scholar] [CrossRef]
- Duchartre, Y.; Kim, Y.M.; Kahn, M. The Wnt signaling pathway in cancer. Crit. Rev. Oncol. Hematol. 2016, 99, 141–149. [Google Scholar] [CrossRef]
- Tian, R.; Li, Y.; Yao, X. PGRN Suppresses Inflammation and Promotes Autophagy in Keratinocytes Through the Wnt/β-Catenin Signaling Pathway. Inflammation 2016, 39, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tu, C.; Zhang, D.; Zheng, Y.; Peng, Z.; Feng, Y.; Xiao, S.; Li, Z. Wnt/β-Catenin and Wnt5a/Ca Pathways Regulate Proliferation and Apoptosis of Keratinocytes in Psoriasis Lesions. Cell Physiol. Biochem. 2015, 36, 1890–1902. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yu, X.; Wu, C.; Jin, H. IL-36γ inhibits differentiation and induces inflammation of keratinocyte via Wnt signaling pathway in psoriasis. Int. J. Med. Sci. 2017, 14, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Xu, Z.; Lou, F.; Zhang, L.; Ke, F.; Bai, J.; Liu, Z.; Liu, J.; Wang, H.; Zhu, H.; et al. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 2015, 6, 7652. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Zeng, J.; Yuan, J.; Deng, X.; Huang, Y.; Chen, L.; Zhang, P.; Feng, H.; Liu, Z.; Wang, Z.; et al. MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J. Clin. Invest. 2018, 128, 2551–2568. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Huang, J.; Jin, H. Reduction of Methyltransferase-like 3-Mediated RNA N6-Methyladenosine Exacerbates the Development of Psoriasis Vulgaris in Imiquimod-Induced Psoriasis-like Mouse Model. Int. J. Mol. Sci. 2022, 23, 12672. https://doi.org/10.3390/ijms232012672
Wang Y, Huang J, Jin H. Reduction of Methyltransferase-like 3-Mediated RNA N6-Methyladenosine Exacerbates the Development of Psoriasis Vulgaris in Imiquimod-Induced Psoriasis-like Mouse Model. International Journal of Molecular Sciences. 2022; 23(20):12672. https://doi.org/10.3390/ijms232012672
Chicago/Turabian StyleWang, Yanan, Jiuzuo Huang, and Hongzhong Jin. 2022. "Reduction of Methyltransferase-like 3-Mediated RNA N6-Methyladenosine Exacerbates the Development of Psoriasis Vulgaris in Imiquimod-Induced Psoriasis-like Mouse Model" International Journal of Molecular Sciences 23, no. 20: 12672. https://doi.org/10.3390/ijms232012672
APA StyleWang, Y., Huang, J., & Jin, H. (2022). Reduction of Methyltransferase-like 3-Mediated RNA N6-Methyladenosine Exacerbates the Development of Psoriasis Vulgaris in Imiquimod-Induced Psoriasis-like Mouse Model. International Journal of Molecular Sciences, 23(20), 12672. https://doi.org/10.3390/ijms232012672





